Abstract
The proto-oncogene c-myc encodes a transcription factor that is implicated in the regulation of cellular proliferation, differentiation, and apoptosis and that has also been found to be deregulated in several forms of human and experimental tumors. We have shown that forced expression of c-myc in epithelial tissues of transgenic mice (K5-Myc) resulted in keratinocyte hyperproliferation and the development of spontaneous tumors in the skin and oral cavity. Although a number of genes involved in cancer development are regulated by c-myc, the actual mechanisms leading to Myc-induced neoplasia are not known. Among the genes regulated by Myc is the cyclin-dependent kinase 4 (CDK4) gene. Interestingly, previous studies from our laboratory showed that the overexpression of CDK4 led to keratinocyte hyperproliferation, although no spontaneous tumor development was observed. Thus, we tested the hypothesis that CDK4 may be one of the critical downstream genes involved in Myc carcinogenesis. Our results showed that CDK4 inhibition in K5-Myc transgenic mice resulted in the complete inhibition of tumor development, suggesting that CDK4 is a critical mediator of tumor formation induced by deregulated Myc. Furthermore, a lack of CDK4 expression resulted in marked decreases in epidermal thickness and keratinocyte proliferation compared to the results obtained for K5-Myc littermates. Biochemical analysis of the K5-Myc epidermis showed that CDK4 mediates the proliferative activities of Myc by sequestering p21Cip1 and p27Kip1 and thereby indirectly activating CDK2 kinase activity. These results show that CDK4 mediates the proliferative and oncogenic activities of Myc in vivo through a mechanism that involves the sequestration of specific CDK inhibitors.
The proto-oncogene c-myc encodes a transcription factor of the basic helix-loop-helix leucine zipper family of proteins and has been implicated in the regulation of cellular proliferation, differentiation, and apoptosis (5, 24, 25, 48, 72). The Myc protein must dimerize with another basic helix-loop-helix leucine zipper protein, Max, to bind the DNA sequence CACGTG (the E box) and activate transcription from adjacent promoters (3, 35). In contrast to Myc, Max can also form homodimers or heterodimers with members of the Mad family of proteins (7). In summary, the Myc, Max, and Mad proteins form a network that regulates gene expression, proliferation, apoptosis, and differentiation. Several target genes of this network have been identified; they include genes for alpha-prothymosin (12, 22, 29), ornithine decarboxylase (9), Cdc25A (27), cyclin D2, cyclin-dependent kinase (CDK) 4 (CDK4), and others (31, 35).
Myc genes are differentially expressed during embryonic development (21) and, with few exceptions, proliferating postnatal tissues express Myc (41). Ectopic expression of the Myc oncoprotein prevents cell cycle arrest in response to growth-inhibitory signals, differentiation stimuli, or mitogen withdrawal. Moreover, Myc activation in quiescent cells is sufficient to induce cell cycle entry in the absence of growth factors. Thus, Myc transduces a potent mitogenic stimulus but, at the same time, induces apoptosis in the absence of survival factors (35).
Deregulated c-myc expression can play a causal role in the genesis of several types of murine and human malignancies (5, 24, 25, 72). The proto-oncogene c-myc has been implicated in the genesis of diverse human tumors, including squamous cell carcinoma (2, 13, 26), lung carcinoma (40), breast carcinoma (42), and rare cases of colon carcinoma (6). The tumorigenic effects of Myc have been generally attributed to sustained effects on cellular proliferation and differentiation (41). In fact, Myc plays a key role in cellular proliferation as a positive regulator of G1-specific CDKs, in particular, cyclin E/CDK2 complexes (44). In addition to activation of the c-myc gene through deregulated gene expression, point mutations in the coding sequence have been found in translocated alleles of c-myc in Burkitt's lymphomas (10).
In the last few years, the role of Myc in carcinogenesis has been widely studied; however, the connection among Myc, Ras, and cell cycle progression is not well understood. The participation of the Ras pathway in the stabilization of the Myc protein was recently established (62, 63). Also, a connection between Myc and cyclin D2 expression and, more recently, with CDK4 expression was reported (17, 18, 36). It has also been reported that in colorectal carcinomas, there is a strong correlation between the overexpression of Myc and the aberrant expression of CDK4 (6, 23, 36). It has been proposed that the proliferative and oncogenic roles of Myc are linked to its ability to induce the transcription of CDK4, cyclin D1, and cyclin D2 (36), which inactivates the product of the Rb tumor suppressor gene; this hypothesis provides a link between Myc and the CDK4/cyclin D1/pRb pathway in some malignant tumors (32).
Several in vivo models of Myc overexpression have demonstrated that the transgenic expression of Myc in the basal and suprabasal cell layers of stratified epithelia and in stem cells leads to hyperplasia, increased epidermal thickness, and proliferation (52, 61, 69, 70). In particular, Myc transgenic mice developed in our laboratory with the K5 promoter (K5-Myc) showed epithelial neoplasia in the skin as well as oral mucosa (60, 61). Also, the overexpression of Myc in B lymphocytes of Eμ-Myc transgenic mice resulted in the development of Burkitt-type lymphoma with a latency of 6 months (1).
A previous report by Miliani de Marval et al. also demonstrated that the forced expression of CDK4 in the epidermis of transgenic mice resulted in a similar phenotype: hyperproliferation of the basal cell layer of the epidermis, hypertrophy, and increased epidermal thickness (46). In addition, these mice showed a high rate of malignant progression of chemically induced tumors (47). Also, the expression of a mutant form (CDK4-R24C) which lacks the capacity to bind p16Ink4a resulted in a wide spectrum of tumors, with the most common being lymphomas, endocrine tumors, and hemangiosarcomas (55, 65). The involvement of CDK4 in the neoplastic process was also suggested by the fact that CDK4 amplification and/or overexpression were detected in human glioblastomas (34). In addition, CDK4 mutations were identified in patients with familial melanoma (73, 76) and, more recently, the amplification and overexpression of CDK4 were detected in sporadic breast carcinomas (4) and sarcomas (39).
Hence, the objective of this project was to examine the role of CDK4 in c-myc-induced tumorigenesis. Here we have demonstrated that the lack of CDK4 expression in Myc transgenic mice results in the complete inhibition of tumor development and suppression of the epidermal phenotype observed in K5-Myc mice. We have also determined that c-myc induces aberrant expression of CDK4, which sequesters the CDK inhibitors (CKIs) p21Cip1 and p27Kip1, leading to an indirect increase in CDK2 kinase activity. These results demonstrate that CDK4 can be a target for therapeutic intervention in Myc-mediated tumorigenesis.
MATERIALS AND METHODS
Mouse experiments.
K5-Myc transgenic mice were developed in an FVB background and backcrossed into an SSIN (Sencar) genetic background (61). CDK4- and cyclin D2-null mice were developed by targeted disruption in a C57BL/6 background and further backcrossed into an SSIN genetic background for two generations (58, 64, 67). K5-Myc transgenic mice (line MM5) were crossed with mice heterozygous for CDK4 (CDK4+/−) to generate K5-Myc transgenic mice heterozygous for CDK4 (K5-Myc/CDK4+/−). These mice were bred with CDK4+/− mice to generate K5-Myc transgenic and nontransgenic mice that were either homozygous, heterozygous, or nullizygous for CDK4. The same strategy was used to generate K5-Myc mice nullizygous for cyclin D2 (64).
BrdU incorporation.
Epithelial cell proliferation was measured by intraperitoneal injection of bromodeoxyuridine (BrdU) (60 mg/g of body weight) 30 min before the mice were sacrificed. BrdU incorporation was detected by immunohistochemical staining of paraffin-embedded sections with mouse anti-BrdU monoclonal antibody (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). The reaction was visualized with a biotin-conjugated anti-mouse antibody (Vector Laboratories, Inc., Burlingame, Calif.) and an avidin-biotin-peroxidase kit (Vectastain Elite; Vector Laboratories, Inc.) with diaminobenzidine as the chromogen. Interfollicular basal cells were examined microscopically to determine the numbers of unstained and stained cells. At least 1,000 cells were counted per section.
Western blot analysis.
The dorsal side of the mice was shaved, treated with a depilatory agent for 1 min, and then washed off. The epidermal tissue was scraped off with a razor blade, placed in homogenization buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 2.5 mM EGTA, 1 mM EDTA, 0.1% Tween 20; 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride, 0.2 U of aprotinin/ml, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate, 1 mM NaF [pH 7.8]), and homogenized by using a manual homogenizer. The epidermal homogenate was centrifuged at 10,000 × g in order to collect the supernatant, which was used directly for Western blotting analysis or stored at −80°C. The protein concentration was measured with a protein assay system from Bio-Rad Laboratories, Richmond, Calif. Sodium dodecyl sulfate sample buffer was added to each sample and boiled for 5 min. Protein lysates (25 μg) were electrophoresed through acrylamide gels and electrophoretically transferred to nitrocellulose membranes (Bio-Rad). After being blocked with 5% nonfat powdered milk in Dulbecco's phosphate-buffered saline (Sigma Chemical Co.), the membranes were incubated with specific antibodies. The following antibodies were used: rabbit polyclonal antibodies against CDK4 (C22), CDK2 (M2), and CDK6 (C21) (all from Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.); rabbit polyclonal antibody against cyclin D1 (Ab-3) (Lab Vision Corp./Neo Markers, Fremont, Calif.); and horseradish peroxidase-conjugated secondary antibody (Amersham Corp., Arlington Heights, Ill.). Enhanced chemiluminescence (Pierce Biotech, Inc., Rockford, Ill.) was used for immunoblotting detection. Bioimage analysis was used to quantify the levels of expression of the proteins.
Immunoprecipitation and kinase assays.
Fresh protein preparations (250 μg per sample) from epidermal tissue were immunoprecipitated for 2 h at 4°C with protein A-agarose beads and specific antibodies and washed three times with homogenization buffer described above. Immunoprecipitates were electrophoresed through acrylamide gels and electrophoretically transferred to nitrocellulose membranes. After being blocked with 5% nonfat powdered milk in Dulbecco's phosphate-buffered saline, the membranes were incubated with specific antibodies. Antibodies to the following were used: CDK4 (C22), CDK2 (M2), p21 (M19), and p27 (M197) (all from Santa Cruz Biotechnology); cyclin D1 (bcl Ab-3) (Lab Vision Corp./Neo Markers); protein A-Sepharose beads (Life Technologies Inc., Grand Island, N.Y.); and horseradish peroxidase-conjugated secondary antibody (Amersham). Enhanced chemiluminescence (ECL detection kit; Amersham) was used for immunoblotting detection.
To study the kinase activities of CDK2 and CDK4, protein lysates were obtained as described above, but the homogenate was frozen on powdered dry ice, thawed in ice water, incubated on ice for 15 min, and centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was collected and used for a kinase assay. Protein lysates (250 μg) were immunoprecipitated with antibodies against CDK2 or CDK4. Antibody-precoated beads (30 μl) were incubated with the lysates for 1 h at 4°C. The beads were washed twice with NP-40 buffer (Tris [pH 7.5], 150 mM NaCl, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride) and twice with kinase buffer (50 mM HEPES [pH 7], 10 mM MgCl2, 5 mM MbCl2). Then, 30 μl of kinase buffer, 1 μg of pRb (Santa Cruz Biotechnology) or histone H1 (Upstate Biotechnology Inc., Charlottesville, Va.) substrate, 5 μCi of [γ-32P]ATP (6,000 Ci/mmol), 1 mM DTT, and 5 μM ATP were added to the bead pellet and incubated for 30 min at 30°C. Sodium dodecyl sulfate sample buffer was added, and each sample was boiled for 5 min and electrophoresed through acrylamide gels.
Statistical analysis.
A one-way analysis of variance with a Tukey-Kramer multiple-comparison test was performed by using GraphPad InStat version 3.00 for Windows 95 (GraphPad Software, San Diego, Calif.).
RESULTS
Lack of CDK4 expression results in complete inhibition of tumor development.
Overexpression of the murine c-myc gene in the basal cell layer of the stratified epithelium (K5-Myc transgenic mice) resulted in epidermal hyperplasia and hypertrophy development (Fig. 1 and 2) (61). In addition, a high incidence of spontaneous tumors was observed in the skin and oral mucosa of transgenic mice (61). These results clearly showed that c-myc acts as an oncogene in the stratified epithelium, but the mechanisms leading to the malignant phenotype are not fully understood. Interestingly, the forced expression of CDK4 in the basal cell layer of stratified epithelium (K5-CDK4 transgenic mice) resulted in a skin phenotype similar to that of K5-Myc animals (46). Furthermore, chemically induced skin papillomas from both K5-CDK4 and K5-Myc transgenic mice showed an increase in the rate of malignant progression to squamous cell carcinomas (47, 61).
FIG. 1.
K5-Myc/CDK4−/− epidermis phenotype. Representative paraffin-embedded sections of skin stained with hematoxylin and eosin are shown. (A) K5-Myc transgenic mice. (B) K5-Myc/CDK4−/− mice. (C) CDK4−/− mice. Arrows indicate the hyperkeratosis that developed in K5-Myc transgenic mice and K5-Myc/CDK4−/− littermates.
Several other reports have demonstrated that Myc induces the transcription of CDK4 in cell cultures and colorectal tumor samples (6, 23, 36). In order to investigate whether CDK4 mediates the oncogenic activities of c-myc, we developed K5-Myc transgenic mice that lack the expression of CDK4 (K5-Myc/CDK4−/− mice). These mice, along with K5-Myc/CDK4+/−, K5-Myc/CDK4+/+, CDK4+/−, CDK4−/−, and wild-type siblings, were analyzed for the development of spontaneous tumors.
Histological analysis of mice bearing the c-myc transgene with one or two functional CDK4 alleles revealed a wide spectrum of tumors of the oral mucosa (Fig. 3 and Table 1) (61). The tumors were observed in mice as young as 8 weeks old with an incidence of 100%. These tumors were classified as keratinizing odontogenic tumors, neuroblastomas, and olfactory tumors derived from the oral mucosa, odontogenic tissues, and the olfactory epithelium (Table 1). In sharp contrast, CDK4−/− animals carrying the K5-Myc transgene did not develop any malignancies during the 3-month observation period (Table 1). Thus, genetic inhibition of CDK4 renders animals resistant to Myc-driven oncogenesis of the oral mucosa. No further observation was possible because the CDK4−/− mice died at 12 to 16 weeks of age. It is likely that these mice developed diabetes mellitus, as previously reported (56, 67). Thus, quantification and histological analysis of oral cavity tumors were performed until 12 weeks of age for all genotypes. Nontransgenic CDK4+/− and CDK4+/+ littermates did not show any signs of spontaneous tumor development, even when they were 12 months old.
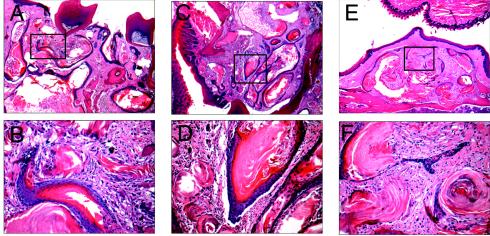
FIG. 3.
Odontogenic tumors in K5-Myc mice. Representative paraffin-embedded sections of odontogenic tumors obtained from K5-Myc mice and stained with hematoxylin and eosin are shown. (A, C, and E) Odontogenic tumors derived from the oral mucosa and classified as keratinizing odontogenic tumors (magnification, ×10). (B, D, and F) Magnifications (×40) of the insets from panels A, C, and E, respectively. (A to D) K5-Myc/CDK4+/+ mice. (E and F) K5-Myc/CDK4+/− mice.
TABLE 1.
Histopathologic analysis of K5-Myc mice, K5-Myc/CDK4+/− or K5-Myc/CDK4−/− siblings, and K5-Myc/cyclin D2−/− micea
| Mice | No. of mice/group | No. (%) of mice with the following tumor:
|
||
|---|---|---|---|---|
| Odontogenic | Neuroblastoma | Olfactoryb | ||
| K5-Myc | 15 | 8 (53) | 4 (27) | 3 (20) |
| K5-Myc/CDK4+/− | 15 | 6 (40) | 6 (40) | 3 (20) |
| K5-Myc/CDK4−/− | 10 | 0 (0) | 0 (0) | 0 (0) |
| K5-Myc/cyclin D2−/− | 14 | 9 (64) | 3 (21) | 2 (15) |
All of the K5-Myc, K5-Myc/CDK4+/−, and K5-Myc/cyclin D2−/− mice showed at least one of the tumors described above. None of the K5-Myc/CDK4−/−, CDK4−/−, K5-Myc CDK4+/−, or wild-type mice developed tumors.
Derived from olfactory epithelium.
It has also been shown that cyclin D2 expression is regulated by the c-myc-encoded transcription factor (14, 53), but the specific role as a mediator of the tumorigenic effects of c-myc in vivo has yet to be defined. To test the requirement for cyclin D2 in Myc-driven oncogenesis of the epithelium, we crossed K5-Myc mice with cyclin D2−/− mice (64) and generated K5-Myc/cyclin D2−/− animals. Our analysis revealed that the cyclin D2−/− mice remained fully susceptible to Myc-driven tumorigenesis of the oral mucosa (Table 1). Hence, these data suggest that cyclin D2 does not play a relevant role in the development of the spontaneous tumors observed in K5-Myc mice.
Collectively, these results indicate that CDK4, but not cyclin D2, plays a critical role in Myc-mediated tumor development.
Inactivation of CDK4 results in reduced epidermal thickness and proliferation.
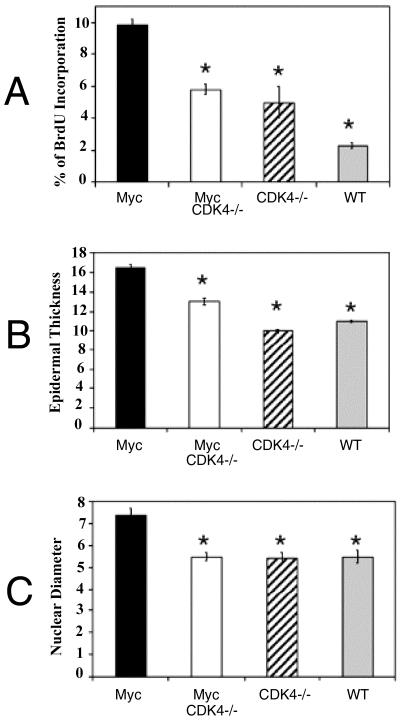
In order to investigate whether CDK4 also mediates the epidermal hyperproliferative phenotype triggered by Myc overexpression, we analyzed the epidermis of K5-Myc and K5-Myc/CDK4−/− mice. The skin of K5-Myc mice showed hyperplasia (increased cell number) and hypertrophy (increased cell size), two features that contributed to the increase in the epidermal thickness observed in these mice (Fig. 1 and 2) (61). A lack of CDK4 expression (K5-Myc/CDK4−/− mice) resulted in reversion of the increased epidermal thickness observed in K5-Myc mice, although the hyperkeratosis (accumulation of keratinized cells in the epidermal surface) characteristic of Myc overexpression still persisted (Fig. 1B). Rodriguez-Puebla et al. previously demonstrated that a lack of CDK4 expression does not affect the mouse epidermal architecture, which is similar to that in wild-type siblings (Fig. 1C) (58). Quantification of the epidermal thickness revealed a significant reduction in K5-Myc/CDK4−/− mice (13 μm) compared with K5-Myc siblings (16.6 μm), although the epidermis was still thicker than that in wild-type (11.1 μm) and CDK4−/− (10 μm) littermates (the P value, determined by the Tukey-Kramer multiple-comparison test, was <0.001) (Fig. 2B). However, the most significant difference was observed in the number of proliferative cells. The rate of keratinocyte proliferation was determined by BrdU incorporation. We found that the inhibition of CDK4 in K5-Myc mice resulted in a 40% reduction in the rate of proliferation compared to the rate in K5-Myc mice (the P value, as determined by the Tukey-Kramer multiple-comparison test, was <0.001) (Fig. 2A). In fact, the BrdU labeling index for K5-Myc/CDK4−/− mice was similar to that for CDK4−/− mice (Fig. 2A), showing again that a lack of CDK4 reversed the K5-Myc epidermal hyperproliferative phenotype. It is worth mentioning that the epidermis of CDK4−/− mice showed an increased level of proliferation compared with that of wild-type mice (58).
FIG. 2.
Quantification of keratinocyte proliferation, epidermal thickness, and nuclear diameter in mouse epidermis. (A) Epidermal proliferation in K5-Myc, K5-Myc/CDK4−/−, CDK4−/−, and wild-type (WT) littermates. The columns indicate the percentages of BrdU-positive cells in the basal cell layer of interfollicular epithelia. (B) Epidermal thickness, in micrometers. (C) Nuclei diameter in the interfollicular epidermis, in micrometers. The error bars indicate the standard error of the mean. An asterisk indicates a P value of <0.001 for differences between K5-Myc and K5-Myc/CDK4−/−, CDK4−/−, or wild-type littermates, as determined by the Tukey-Kramer multiple-comparison test.
To elucidate whether the reduced epidermal thickness observed in CDK4−/− epidermis was also a consequence of a decrease in cell size, we measured the nuclear diameters of K5-Myc, K5-Myc/CDK4−/−, CDK4−/−, and CDK4+/+ keratinocytes. The nuclear diameter of K5-Myc mice (7.4 μm) was 1.4-fold larger than that of wild-type siblings (the P value, as determined by the Tukey-Kramer multiple-comparison test, was <0.001) (Fig. 2C). Interestingly, the nuclear diameter of K5-Myc/CDK4−/− mice (5.5 μm) was similar to that of normal siblings. These data suggest that the reduction in epidermal thickness was probably due to a combination of the decreased cell size and the decreased number of cells observed in the epidermis of K5-Myc/CDK4−/− mice. Several laboratories have also reported that deregulated CDK4 influences cell size (19, 45). In fact, previous results showed that CDK4 overexpression in K5-CDK4 transgenic mice leads to an increase in epidermal thickness associated with increases in both cell number and nuclear diameter (46).
Together, these results suggest that CDK4 plays an important role in the deregulation of keratinocyte proliferation triggered by c-Myc overexpression. It is likely that this phenomenon is partially responsible for the inhibition of tumor development observed in K5-Myc/CDK4−/− mice, although the participation of other mechanisms, such as the regulation of apoptosis and terminal differentiation, cannot be ruled out.
Expression of cell cycle proteins.
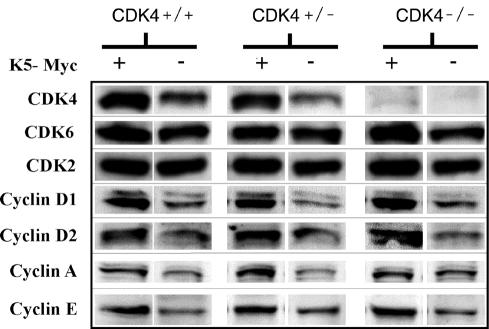
To elucidate whether the increased epidermal thickness and the increased epidermal proliferation in K5-Myc-overexpressing mice were due to changes in the patterns of expression of G1/S-phase regulators, Western blot analysis of epidermis from K5-Myc, K5-Myc/CDK4−/−, K5-Myc/CDK4+/−, CDK4−/−, CDK4+/−, and wild-type siblings was performed. The level of CDK4 protein was twofold higher in K5-Myc mice than in wild-type siblings (Fig. 4, lines 1 and 2). CDK4+/− mice showed half the level of CDK4 protein seen in wild-type littermates (Fig. 4, lines 2 and 4); however, when Myc was overexpressed in the epidermis of CDK4+/− mice, the expression of the CDK4 protein increased to a level as high as that observed in K5-Myc mice (Fig. 4, lines 1 and 3). These data suggest that a single functional allele of CDK4 is sufficient to produce aberrant levels of this protein in response to Myc overexpression. CDK2 and CDK6 proteins remained at the same levels in all of the mice regardless of the CDK4 status (Fig. 4, lines 2, 4, and 6). On the other hand, the levels of the cyclin D1 and cyclin D2 proteins were mildly increased in K5-Myc mice regardless of the presence or absence of CDK4, whereas cyclin D3 protein remain at the same level (data not shown). Deregulated c-myc expression is also linked to increased levels of cyclin A and cyclin E (16, 33, 37, 38, 54) and to the downregulation of p27Kip1 (8, 49, 50, 68). In agreement with those reports, we found a mild increase in the levels of both cyclin A and cyclin E in Myc-overexpressing animals compared to nontransgenic animals (Fig. 4). On the other hand, we did not observe changes in the p27Kip1 protein level in K5-Myc mice compared to wild-type siblings (data not shown). Together, these data demonstrate that the in vivo overexpression of c-myc in epithelial tissues results in the upregulation of several regulators of the cell cycle, including CDK4, albeit the regulation of other proteins, such as p27Kip1, appears to be tissue specific.
FIG. 4.
Western blot analysis of cell cycle proteins from mouse epidermis. Protein lysates of epidermis samples from K5-Myc, K5-Myc/CDK4−/−, K5-Myc/CDK4+/−, CDK4−/−, CDK4+/−, and wild-type siblings were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane. Primary antibodies against CDK4, CDK6, CDK2, cyclin D1, cyclin D2, cyclin A, and cyclin E were used for immunoblot analysis. Protein levels were quantified with a densitometer.
CDK4 upregulation results in the titration of p21Cip1 and p27Kip1.
It was previously established that CDK4 has dual functions, first as a kinase phosphorylating pRb and second as a noncatalytic protein that binds and sequesters the cell cycle inhibitors p21Cip1 and p27Kip1 (14, 53). The release of these CKIs from CDK2 complexes results in the activation of CDK2 and further progression through S phase (14, 53). The data presented here show that CDK4 is a critical mediator of the proliferative and tumorigenic activities of Myc. Therefore, in order to investigate the prevalent mechanisms for Myc-induced epidermal proliferation, we first analyzed complex formation between CDK4 and cyclin D1 in K5-Myc keratinocytes. We found that regardless of the increased levels of cyclin D1 and CDK4 observed in K5-Myc mice (Fig. 4 and 5A), there was not a proportional increase in CDK4/cyclin D1 complex formation in the skin of K5-Myc mice compared to wild-type siblings (Fig. 5A).
FIG. 5.
CDK4 complex formation and kinase assays of K5-Myc epidermis. (A) Complex formation between cyclin D1 and CDK4. Fresh epidermal proteins from K5-Myc and wild-type mice were immunoprecipitated (IP) with a polyclonal antibody against cyclin D1 and immunoblotted with polyclonal antibodies against CDK4 and cyclin D1. Lanes A and B, protein lysates from K5-Myc and wild-type epidermis, respectively. (B) Kinase activity of CDK4 from K5-Myc transgenic and wild-type siblings. Fresh epidermal proteins from two transgenic and two wild-type mice were immunoprecipitated with an anti-CDK4 (IP CDK4) antibody, and in vitro kinase assays were carried out with a pRb peptide as a substrate. (C) Kinase activity of CDK2 from K5-Myc/CDK4+/+, K5-Myc/CDK4+/−, K5-Myc/CDK4−/−, and normal siblings. Fresh epidermal proteins were immunoprecipitated with an anti-CDK2 (IP CDK2) antibody and normal rabbit immunoglobulin G (NR), and in vitro kinase assays were carried out with histone H1 as a substrate.
It was previously demonstrated that transgenic mice overexpressing CDK4 show hyperproliferation through a mechanism that involves the increased binding of CDK4 to p27Kip1 and a concomitant increase in CDK2 kinase activity (46, 47). To address the question of whether this mechanism is also valid for K5-Myc epidermis, CDK4 and CDK2 kinase assays were performed. We found that CDK4 kinase activities were equivalent in both K5-Myc and wild-type littermates (Fig. 5B). However, CDK2 kinase activity was fourfold higher in the epidermis of transgenic animals than in that of normal siblings (Fig. 5C, lanes 1 and 4). In contrast, K5-Myc/CDK4−/− mice showed diminished CDK2 kinase activity compared to K5-Myc mice (Fig. 5C, lanes 1 and 3). This effect was manifested only in the transgenic epidermis, as nontransgenic CDK4−/− mice showed the same level of CDK2 activity as wild-type littermates (58).
To elucidate whether the increased CDK4 protein level results in increased levels of CDK4/p27Kip1 and/or CDK4/p21Cip1 complex formation, epidermal lysates from K5-Myc and wild-type mice were immunoprecipitated with antibodies against p21Cip1 and p27Kip1, followed by Western blotting with anti-CDK4 antibody. Consistent with the role of CDK4 in titrating CKIs, we observed increases in CDK4/p21Cip1 and CDK4/p27Kip1 complex formation (3.12- and 1.5-fold increases, respectively) in K5-Myc mice compared to normal siblings (Fig. 6). It is worth mentioning that this effect was due to the elevated level of CDK4 observed in K5-Myc epidermis because p21Cip1 and p27Kip1 protein levels were not increased in K5-Myc epidermis (data not shown). These data suggest that CDK4 mediates the proliferative activity of Myc at least partly by binding to the CKIs p21Cip1 and p27Kip1, releasing them from CDK2 complexes and in turn activating CDK2 kinase activity.
FIG. 6.
p27Kip1/CDK4 and p21Cip1/CDK4 complex formation in K5-Myc epidermis. (A) Fresh protein lysates of epidermis from transgenic (K5-Myc) and wild-type (WT) samples were sequentially immunoprecipitated with anti-p21Cip1 or anti-p27Kip1 antibodies. Epidermal proteins were incubated with either anti-p21Cip1 or anti-p27Kip1 antibodies (1st), and then the supernatants were incubated again with the same antibodies (2nd). Proteins immunoprecipitated in each step were analyzed by Western blotting with antibodies against CDK4. CDK4/p21 (B) and CDK4/p27 (C) complex formation levels were quantified with a densitometer.
DISCUSSION
In the last few years, several groups have studied the role of Myc in both cell cycle progression and tumorigenesis. However, how these two events are linked is still poorly understood. In this work, we present direct evidence that Myc overexpression results in increased CDK4 protein expression, which stimulates cell proliferation and mediates tumor development. Transgenic mice that overexpress Myc in the basal cell layer of stratified epithelium developed severe epidermal hyperplasia and hypertrophy (61). This phenotype is consistent with previous reports that demonstrated the effects of Myc overexpression on cell proliferation in vivo (69, 70). Two other groups have also reported that the deregulation of Myc in the epidermis results in spontaneous tumor development (52, 61). Here we show that genetic inhibition of CDK4 severely cripples the ability of the c-myc oncogene to drive cell hyperproliferation and fully inhibits tumor development. In addition, we found that a single functional allele of CDK4 is sufficient to support tumor development. It can be hypothesized that metabolic alterations in CDK4−/− mice are the cause of the inhibition of tumorigenesis. However, Martin et al. (43) have shown that most, if not all, of the alterations observed in CDK4−/− mice are not due to metabolic alterations associated with insulin-deficient diabetes. Genetic rescue of CDK4−/− in the pancreas restored β-cell proliferation without changes in the sizes of the mice, which remained small. These results demonstrate that the reduced number of cells in CDK4−/− mice is attributable to the lack of proliferative capacity due to the absence of CDK4 (43) rather than a consequence of metabolic alterations in other tissues. Supporting this conclusion, a previous study showed normal architecture and proliferation in CDK4−/− epidermis as well as strong inhibition of chemically induced skin tumors (58).
Consistent with our results, Baudino et al. have shown that the loss of E2F1 in Eμ-Myc mice impairs proliferation and Myc-induced lymphomagenesis (8). However, whereas Myc overexpression in B cells results in E2F1-mediated downregulation of p27Kip1 (8), forced expression of Myc in epithelial tissues did not affect p27Kip1 protein levels. Thus, if p27Kip1 downregulation is a critical step in E2F1-mediated activity, it is plausible that the lack of E2F1 in K5-Myc epithelial cells will not result in the inhibition of tumorigenesis because p27Kip1 levels are not affected in these cells. Consistent with this idea, E2F1 inactivation in the K5-Myc transgenic model did not reduce tumorigenesis but enhanced it (60). It is also possible that reduced p27Kip1 levels in B cells overexpressing Myc can result in reduced binding of p27Kip1 to CDK2 and a further elevation of its kinase activity. We cannot easily reconcile the differences observed in B cells and epithelial cells, but it is conceivable that the ability of E2F1 to suppress carcinogenesis is part of a tissue-specific mechanism (61). Whatever the explanation, our studies support the idea that the lack of another positive regulator of the cell cycle, CDK4, results in the complete inhibition of carcinogenesis through a pathway that also involves p27Kip1. In this scenario, sequestration of p27Kip1 and p21Cip1 appears to play a significant role in CDK2 activation. Importantly, p27Kip1 also plays a key role in a setting where the lack of CDK4 results in delay entry into S phase in mouse embryo fibroblasts (67).
It is worth mentioning that odontogenic tumors are uncommon in mice (20) and in most other animals but can be induced through a variety of means, including viruses and aflatoxins (15, 28, 71). Gibson et al. observed a high incidence of odontogenic tumors in albumin-Myc/albumin-Ras transgenic mice (30). Thus, K5-Myc/CDK4−/− mice provide an excellent model for studying the role of CDK4 in this process, since the lack of CDK4 results in the inhibition of odontogenic tumors.
Two groups have reported that Myc induces cyclin D2 expression, contributing to cell cycle progression due to an increase in the formation of cyclin D2/CDK4/p27Kip1 ternary complexes (14, 53). The sequestration of p27Kip1, a specific inhibitor of cyclin E/CDK2, results in the activation of cyclin E-associated kinase activity. In our in vivo model, the overexpression of Myc in mouse epidermis also results in a mild increase in cyclin D1 and D2 expression. However, cyclin D2 appears to be dispensable for Myc-induced tumor development and hyperproliferation. The absence of cyclin D2 in K5-Myc mice did not change the outcome of tumor formation, which was as high as that in K5-Myc littermates (Table 1). In addition, paraffin-embedded sections of mouse skin showed no difference in the rates of epidermal proliferation between K5-Myc and K5-Myc/cyclin D2−/− mice (unpublished data). Also, Yu et al. showed that the lack of cyclin D2 does not result in protection against breast cancer in mouse mammary tumor virus Myc mice (74). Thus, contrary to the in vitro evidence implicating cyclin D2 as a mediator of Myc proliferation, here we show in an in vivo setting that cyclin D2 is not required for cell proliferation and does not modulate the oncogenic effects of Myc overexpression. However, Bouchard et al. (14) and Perez-Roger et al. (54) concluded that the c-myc oncogene signals through both cyclin D1 and cyclin D2. Thus, it is possible that in the oral epithelium, Myc-driven oncogenic proliferation is mediated by cyclin D1/CDK4 and cyclin D2/CDK4 complexes. According to this scenario, the inhibition of both cyclin D1 and cyclin D2 would be required to block the oncogenic action of Myc in the oral mucosa.
Together, these results show that CDK4 plays a critical role in tumor development and epidermal proliferation, although the role of its regulatory subunit, cyclin D1, in K5-Myc-mediated tumorigenesis has not yet been investigated. Emerging evidence indicates that the wiring of the oncogenic pathways to the core cell cycle machinery is cell type specific. For instance, mammary epithelial cells critically require cyclin D1 for Ras-driven tumorigenesis (74). In contrast, Ras can cause oncogenic transformation of fibroblasts in the absence of cyclin D1 (74), while cyclin D1-deficient skin keratinocytes show reduced susceptibility to chemical tumorigenesis (57, 59). The results of our current study reveal that CDK4 is the critical target of the c-myc oncogene in the oral mucosa.
It is possible that in other cell types, the c-myc oncogene impinges on the core cell cycle machinery through other targets. In addition, CDK4−/− mouse embryo fibroblasts are resistant to transformation in response to Ras activation with dominant-negative p53 expression or in an Ink4a/Arf−/− background (75). The resistance to transformation of CDK4−/− mouse embryo fibroblasts has been associated with the elevated expression of p21Cip1; however, we did not observe increased p21Cip1 expression in CDK4−/− keratinocytes. It is also worth mentioning that the lack of CDK4 in mouse epidermis results in elevated CDK6 kinase activity (58). We have hypothesized that CDK6 activity can compensate for the lack of CDK4 and is partly responsible for the mild increase in keratinocyte proliferation observed in CDK4−/− mice (Fig. 2A) (58); however, it is clear that it cannot compensate for the lack of CDK4 in tumor development. However, it is feasible to hypothesize that increased CDK6 activity is the result of increased CDK6/cyclin complex formation and further titration of p27Kip1 and/or p21Cip1. If this is the case, then elevated levels of CDK6/cyclin complexes in CDK4−/− mice will result in at least steady levels of CDK2 activity, as we observed in CDK4−/− epidermis (58). Thus, the implications of CDK6 activity for the homeostasis of mouse epidermal tissue warrant further investigation.
Biochemical studies demonstrated that there is a significant increase in the binding of p21Cip1 and p27Kip1 to CDK4 in K5-Myc mouse epidermis, supporting a model in which an increased level of CDK4 indirectly activates CDK2 through its noncatalytic function (Fig. 6). In contrast, we did not detect changes in the catalytic activities of CDK4, which were similar in K5-Myc and wild-type animals (Fig. 5B). Together, these results show that CDK4 plays an important role in Myc-induced proliferation by indirectly activating CDK2. The role of CDK2 in cell proliferation was recently challenged by the fact that CDK2−/− mice developed normally with minor phenotypic effects, mainly present during the meiotic cell cycle (11, 51). It has also been shown that some tumor cell lines can proliferate independently of CDK2 (66). However, the role of CDK2 in an in vivo tumorigenesis model has not been studied yet. It will be interesting to determine the role of CDK2 in Myc-induced tumorigenesis through the generation of transgenic mice overexpressing c-myc in a CDK2−/− background.
It is worth mentioning that transgenic mice which overexpress a kinase-dead mutant form of CDK4 (K5-CDK4dn) that can bind to CKIs also showed epidermal hyperplasia and increased epidermal thickness similar to those in previously reported K5-CDK4 mice (46; unpublished data). Thus, the hyperproliferation observed in the epidermis of K5-CDK4 dominant-negative mice is dependent not on CDK4 kinase activity but on CDK4 sequestration activity. These results support the idea that the noncatalytic function of CDK4 is critically involved in cell proliferation. Together, these data suggest that in epithelial tissues, deregulated Myc can induce the overexpression of several G1/S-phase regulators, including cyclin D1, cyclin D2, cyclin A, cyclin E, and CDK4, resulting in the sequestration of negative regulators, such as p21Cip1 and p27Kip1, and leading to the activation of CDK2 kinase activity. The requirement for both Myc and CDK4 in malignant transformation was first reported by Haas et al. (32), who described the oncogenic action of CDK4 when coexpressed with Ha-ras and Myc. In fact, they proposed that the ability of CDK4 to bind to p16Ink4 and not its kinase activity was important for its transforming potential (32). Rodriguez-Puebla et al. previously demonstrated that the lack of CDK4 expression also inhibits tumorigenesis in a setting in which mutations in the Ha-ras gene result in the initiation of keratinocytes (58). Thus, these results demonstrate the potential use of CDK4 as a therapeutic target not only in Ras- but also in Myc-mediated tumorigenesis.
Acknowledgments
This work was supported by PHS grants CA 42157 and CA 90864 from NCI.
We especially thank April Weiss for helping with the mouse experiments, the CVM-NCSU and Science Park animal facility personnel, and the Science Park histologic service for assistance with the immunohistochemical staining.
REFERENCES
- 1.Adams, J. M., A. W. Harris, C. A. Pinkert, L. M. Corcoran, W. S. Alexander, S. Cory, R. D. Palmiter, and R. L. Brinster. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318:533-538. [DOI] [PubMed] [Google Scholar]
- 2.Akervall, J., U. Bockmuhl, I. Petersen, K. Yang, T. E. Carey, and D. M. Kurnit. 2003. The gene ratios c-MYC:cyclin-dependent kinase (CDK)N2A and CCND1:CDKN2A correlate with poor prognosis in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 9:1750-1755. [PubMed] [Google Scholar]
- 3.Amati, B., and H. Land. 1994. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 4:102-108. [DOI] [PubMed] [Google Scholar]
- 4.An, H., M. W. Beckmann, G. Reifenger, H. G. Bender, and D. Niederacher. 1999. Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation. Am. J. Pathol. 154:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Askew, D., R. Ashmun, B. Simmons, and J. Cleveland. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6:1915-1922. [PubMed] [Google Scholar]
- 6.Augenlicht, L. H., S. Wadler, G. Corner, C. Richards, L. Ryan, A. S. Multani, S. Pathak, A. Benson, D. Haller, and B. G. Heerdt. 1997. Low-level c-myc amplification in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 57:1769-1775. [PubMed] [Google Scholar]
- 7.Baudino, T. A., and J. L. Cleveland. 2001. The Max network gone mad. Mol. Cell. Biol. 21:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baudino, T. A., K. H. Maclean, J. Brennan, E. Parganas, C. Yang, A. Aslanian, J. A. Lees, C. J. Sherr, M. F. Roussel, and J. L. Cleveland. 2003. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2F1 loss. Mol. Cell 11:905-914. [DOI] [PubMed] [Google Scholar]
- 9.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bemark, M., and M. Neuberger. 2000. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt's lymphoma line. Oncogene 13:3404-3410. [DOI] [PubMed] [Google Scholar]
- 11.Berthet, C., E. Aleem, V. Coppola, L. Tassarollo, and P. Kaldis. 2003. Cdk2 knockout mice are viable. Curr. Biol. 13:1775-1785. [DOI] [PubMed] [Google Scholar]
- 12.Bishop, J. M., M. Eilers, A. L. Katzen, T. Kornberg, G. Ramsay, and S. Schirm. 1991. MYB and MYC in the cell cycle. Cold Spring Harbor Symp. Quant. Biol. 56:99-107. [DOI] [PubMed] [Google Scholar]
- 13.Bitzer, M., M. Stahl, J. Arjumand, M. Rees, B. Klump, H. Heep, H. E. Gabbert, and M. Sarbia. 2003. c-Myc gene amplification in different stages of oesophageal squamous cell carcinoma: prognostic value in relation to treatment modality. Anticancer Res. 23:1489-1493. [PubMed] [Google Scholar]
- 14.Bouchard, C., K. Thieke, A. Maier, R. Saffrich, J. Hanley-Hyde, W. Ansorge, S. Reed, P. Sicinski, J. Bartek, and M. Eilers. 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18:5321-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen, J. M., B. H. Ruebner, D. P. Hsieh, and E. J. Burkes, Jr. 1987. Odontogenic tumors in Fischer rats. J. Oral Pathol. 16:469-473. [DOI] [PubMed] [Google Scholar]
- 16.Daksis, J. I., R. Y. Lu, L. M. Facchini, W. W. Marhin, and L. J. Penn. 1994. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene 9:3635-3645. [PubMed] [Google Scholar]
- 17.Dang, C. V. 1999. c-myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 253:63-77. [DOI] [PubMed] [Google Scholar]
- 19.Datar, S. A., H. W. Jacobs, A. F. de la Cruz, C. F. Lehner, and B. A. Edgar. 2000. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 19:4543-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dayan, D., T. Waner, A. Harmelin, and A. Nyska. 1984. Bilateral complex odontoma in a Swiss (CD-1) male mouse. Lab. Anim. 28:90-92. [DOI] [PubMed] [Google Scholar]
- 21.Downs, K. M., G. R. Martin, and J. M. Bishop. 1989. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 3:860-869. [DOI] [PubMed] [Google Scholar]
- 22.Eilers, M., S. Schirm, and J. M. Bishop. 1991. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 10:133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erisman, M. D., P. G. Rothberg, R. E. Diehl, C. C. Morse, J. M. Spandorfer, and S. M. Astrin. 1985. Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol. Cell. Biol. 5:1969-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evan, G., A. Wyllie, C. Gilbert, T. Littlewood, H. Land, M. Brooks, C. Waters, L. Penn, and D. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 25.Facchini, L. M., and L. Penn. 1998. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 12:633-651. [PubMed] [Google Scholar]
- 26.Field, J. K., D. A. Spandidos, P. M. Stell, E. D. Vaughan, G. I. Evan, and J. P. Moore. 1989. Elevated expression of the c-myc oncoprotein correlates with poor prognosis in head and neck squamous cell carcinoma. Oncogene 4:1463-1468. [PubMed] [Google Scholar]
- 27.Galaktionov, K., X. Chen, and D. Beach. 1996. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382:511-517. [DOI] [PubMed] [Google Scholar]
- 28.Gardner, D. G. 1992. An orderly approach to the study of odontogenic tumors in animals. J. Comp. Pathol. 107:427-438. [DOI] [PubMed] [Google Scholar]
- 29.Gaubatz, S., A. Meichle, and M. Eilers. 1994. An E-box element localized in the first intron mediates regulation of the prothymosin alpha gene by c-myc. Mol. Cell. Biol. 14:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson, C. W., E. Lally, R. C. Herold, S. Decker, R. L. Brinster, and E. P. Sandgren. 1992. Odontogenic tumors in mice carrying albumin-myc and albumin-rats transgenes. Calcif. Tissue Int. 51:162-167. [DOI] [PubMed] [Google Scholar]
- 31.Grandori, C., and R. N. Eisenman. 1997. Myc target genes. Trends Biochem. Sci. 22:177-181. [DOI] [PubMed] [Google Scholar]
- 32.Haas, K., P. Staller, C. Geisen, J. Bartek, M. Eilers, and T. Moroy. 1997. Mutual requirement of CDK4 and Myc in malignant transformation: evidence for cyclin D1/CDK4 and p16INK4A as upstream regulators of Myc. Oncogene 15:179-192. [DOI] [PubMed] [Google Scholar]
- 33.Hanson, K. D., M. Shichiri, M. R. Follansbee, and J. M. Sedivy. 1994. Effects of c-myc expression on cell cycle progression. Mol. Cell. Biol. 14:5748-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He, J., J. R. Allen, V. P. Collins, M. J. Allalunis-Turner, R. Godbout, R. S. Day, and C. D. James. 1994. CDK4 amplification is an alternative mechanism to p16 homozygous deletion in glioma cell lines. Cancer Res. 54:5804-5807. [PubMed] [Google Scholar]
- 35.Henriksson, M., B. Lüscher, and A. Pardee. 1996. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res. 68:109-182. [DOI] [PubMed] [Google Scholar]
- 36.Hermeking, H., C. Rago, M. Schuhmacher, Q. Li, J. Barret, A. Obaya, B. O'Connell, M. Mateyak, W. Tam, F. Kolhlhuber, C. Dang, J. Sedivy, D. Eick, B. Vogelstein, and K. Kinzler. 2000. Identification of CDK4 as a target of c-Myc. Proc. Natl. Acad. Sci. USA 97:2229-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoang, A. T., K. J. Cohen, J. F. Barrett, D. A. Bergstrom, and C. V. Dang. 1994. Participation of cyclin A in Myc-induced apoptosis. Proc. Natl. Acad. Sci. USA 91:6875-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansen-Durr, P., A. Meichle, P. Steiner, M. Pagano, K. Finke, J. Botz, J. Wessbecher, G. Draetta, and M. Eilers. 1993. Differential modulation of cyclin gene expression by MYC. Proc. Natl. Acad. Sci. USA 90:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanoe, H., T. Nakayama, H. Murakami, T. Hosaka, H. Yamamoto, Y. Nakashima, T. Tsuboyama, T. Nakamura, M. Sasaki, and J. Toguchida. 1998. Amplification of CDK4 gene in sarcomas: tumor specificity and relationship with the Rb mutation. Anticancer Res. 18:2317-2321. [PubMed] [Google Scholar]
- 40.Little, C. D., M. M. Nau, D. N. Carney, A. F. Gazdar, and J. D. Minna. 1983. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 306:194-196. [DOI] [PubMed] [Google Scholar]
- 41.Marcu, K. B., S. A. Bossone, and A. J. Patel. 1992. Myc function and regulation. Annu. Rev. Biochem. 61:809-860. [DOI] [PubMed] [Google Scholar]
- 42.Mariani-Costantini, R., C. Escot, C. Theillet, A. Gentile, G. Merlo, R. Lidereau, and R. Callahan. 1988. In situ c-myc expression and genomic status of the c-myc locus in infiltrating ductal carcinomas of the breast. Cancer Res. 48:199-205. [PubMed] [Google Scholar]
- 43.Martin, J., S. L. Hunt, P. Dubus, R. Sotillo, F. Nehme-Pelluard, M. A. Magnuson, A. F. Parlow, M. Malumbres, S. Ortega, and M. Barbacid. 2003. Genetic rescue of Cdk4 null mice restores pancreatic beta-cell proliferation but not homeostatic cell number. Oncogene 22:5261-5269. [DOI] [PubMed] [Google Scholar]
- 44.Meichle, A., A. Philipp, and M. Eilers. 1992. The functions of Myc proteins. Biochim. Biophys. Acta 1114:129-146. [DOI] [PubMed] [Google Scholar]
- 45.Meyer, C. A., H. W. Jacobs, S. A. Datar, W. Du, B. A. Edgar, and C. F. Lehner. 2000. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 19:4533-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miliani de Marval, P., I. Gimenez-Conti, M. LaCava, L. Martinez, C. Conti, and M. Rodriguez-Puebla. 2001. Transgenic expression of CDK4 results in epidermal hyperplasia and severe dermal fibrosis. Am. J. Pathol. 159:369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miliani de Marval, P. L., E. Macias, C. J. Conti, and M. L. Rodriguez-Puebla. 2004. Enhanced malignant tumorigenesis in Cdk4 transgenic mice. Oncogene 23:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreno de Alboran, I., R. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, and R. DePinho. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14:45-55. [DOI] [PubMed] [Google Scholar]
- 49.Muller, D., C. Bouchard, B. Rudolph, P. Steiner, I. Stuckmann, R. Saffrich, W. Ansorge, W. Huttner, and M. Eilers. 1997. Cdk2-dependent phosphorylation of p27 facilitates its Myc-induced release from cyclin E/cdk2 complexes. Oncogene 15:2561-2576. [DOI] [PubMed] [Google Scholar]
- 50.O'Hagan, R. C., M. Ohh, G. David, I. M. de Alboran, F. W. Alt, W. G. Kaelin, Jr., and R. A. DePinho. 2000. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 14:2185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ortega, S., I. Prieto, J. Odajima, A. Martin, P. Dubus, R. Sotillo, J. L. Barbero, M. Malumbres, and M. Barbacid. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35:25-31. [DOI] [PubMed] [Google Scholar]
- 52.Pelengaris, S., T. Littlewood, M. Khan, G. Elia, and G. Evan. 1999. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell 3:565-577. [DOI] [PubMed] [Google Scholar]
- 53.Perez-Roger, I., S. H. Kim, B. Griffiths, A. Sewing, and H. Land. 1999. Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1). EMBO J. 18:5310-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Roger, I., D. L. Solomon, A. Sewing, and H. Land. 1997. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene 14:2373-2381. [DOI] [PubMed] [Google Scholar]
- 55.Rane, S. G., S. Cosenza, R. V. Mettus, and E. P. Reddy. 2002. Germ line transmission of the Cdk4R24C mutation facilitates tumorigenesis and escape from cellular senescence. Mol. Cell. Biol. 22:644-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rane, S. G., P. Dubus, R. V. Mettus, E. J. Galbreath, G. Boden, E. Premkumar Reddy, and M. Barbacid. 1999. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in B-islet cell hyperplasia. Nat. Genet. 22:44-52. [DOI] [PubMed] [Google Scholar]
- 57.Robles, A., M. Rodriguez-Puebla, A. Glick, C. Trempus, L. Hansen, P. Sicinski, R. Tennant, R. Weinberg, S. Yuspa, and C. Conti. 1998. Reduced skin tumor development in cyclin D1 deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 12:2469-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez-Puebla, M. L., P. L. Miliani de Marval, M. LaCava, D. S. Moons, H. Kiyokawa, and C. J. Conti. 2002. Cdk4 deficiency inhibits skin tumor development but does not affect keratinocyte proliferation. Am. J. Pathol. 161:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Puebla, M. L., A. I. Robles, and C. J. Conti. 1999. Ras activity and cyclin D1 expression: an essential mechanism of mouse skin tumor development. Mol. Carcinogenesis 24:1-6. [PubMed] [Google Scholar]
- 60.Rounbehler, R. J., P. M. Rogers, C. J. Conti, and D. G. Johnson. 2002. Inactivation of E2f1 enhances tumorigenesis in a Myc transgenic model. Cancer Res. 62:3276-3281. [PubMed] [Google Scholar]
- 61.Rounbehler, R. J., R. Schneider-Broussard, C. J. Conti, and D. G. Johnson. 2001. Myc lacks E2F1's ability to suppress skin carcinogenesis. Oncogene 20:5341-5349. [DOI] [PubMed] [Google Scholar]
- 62.Sears, R., G. Leone, J. DeGregori, and J. R. Nevins. 1999. Ras enhances Myc protein stability. Mol. Cell 3:169-179. [DOI] [PubMed] [Google Scholar]
- 63.Sears, R., F. Nuckolls, E. Haura, Y. Taya, K. Tamai, and J. R. Nevins. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sicinski, P., J. Donaher, Y. geneg, S. Parker, H. Garder, M. Park, R. Robker, J. Richards, L. McGinnis, J. Biggers, J. Epping, R. Bronson, S. Elledege, and R. Weinberg. 1996. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 384:470-474. [DOI] [PubMed] [Google Scholar]
- 65.Sotillo, R., P. Dubus, J. Martin, E. de la Cueva, S. Ortega, M. Malumbres, and M. Barbacid. 2001. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 20:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tetsu, O., and F. McCormick. 2003. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cells 3:233-245. [DOI] [PubMed] [Google Scholar]
- 67.Tsutsui, T., B. Hesabi, D. S. Moons, P. Pandolfi, K. Hansel, A. Koff, and H. Kiyokawa. 1999. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27Kip1 activity. Mol. Cell. Biol. 19:7011-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlach, J., S. Hennecke, K. Alevizopoulos, D. Conti, and B. Amati. 1996. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 15:6595-6604. [PMC free article] [PubMed] [Google Scholar]
- 69.Waikel, R. L., Y. Kawachi, P. A. Waikel, X. J. Wang, and D. R. Roop. 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 28:165-168. [DOI] [PubMed] [Google Scholar]
- 70.Waikel, R. L., X. J. Wang, and D. R. Roop. 1999. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene 18:4870-4878. [DOI] [PubMed] [Google Scholar]
- 71.Walsh, K. M., L. J. Denholm, and B. J. Cooper. 1987. Epithelial odontogenic tumours in domestic animals. J. Comp. Pathol. 97:503-521. [DOI] [PubMed] [Google Scholar]
- 72.Waters, C., T. Littlewood, D. Hancock, J. Moore, and G. J. Evan. 1991. c-Myc protein expression in untransformed fibroblasts. Oncogene 6:797-805. [PubMed] [Google Scholar]
- 73.Wölfel, T., M. Hauer, J. Schneider, M. Serrano, C. Wölfel, E. Klehmann-Hieb, E. De Plaen, T. Hankeln, K. H. Meyer zum Büschenfelde, and D. Beach. 1995. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269:1281-1284. [DOI] [PubMed] [Google Scholar]
- 74.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 75.Zou, X., D. Ray, A. Aziyu, K. Christov, A. D. Boiko, A. V. Gudkov, and H. Kiyokawa. 2002. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 16:2923-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zuo, L. 1996. Germline mutation in the p16Ink4a binding domain of cdk4 in familial melanoma. Nat. Genet. 12:97-99. [DOI] [PubMed] [Google Scholar]