Abstract
The development of the epidermis of Caenorhabditis elegans involves cell fusion, migration, and differentiation events. To understand the mechanisms underlying these processes, we characterized the roles of NHR-25, a member of the nuclear receptor family of transcription factors. The NHR-25 homologs Ftz-F1 in Drosophila and SF-1 in mammals are involved in various biological processes, including regulation of patterning during development, reproduction, metabolism, metamorphosis, and homeostasis. Impairment of nhr-25 activity leads to severe phenotypes in embryos and many postembryonic tissues. Further analysis has indicated that nhr-25 activity is required for the proper development, including cell-cell fusion, of several epidermal cell types, such as the epidermal syncytial, seam, and Pn.p cells. Our results also suggest that nhr-25 is likely to regulate cell-cell junctions and/or fusion. In a subset of Pn.p cells, called vulval precursor cells, nhr-25 acts collaboratively with the lin-39 Hox gene in regulating vulval cell differentiation. Additionally, our data suggest that nhr-25 may also function with another Hox gene, nob-1, during embryogenesis. Overall, our results indicate that nhr-25 plays an integral role in regulating cellular processes of epidermal cells.
Members of the nuclear receptor (NR) superfamily of transcription factors (also known as nuclear hormone receptors [NHRs]) share some highly conserved peptide motifs, including DNA-binding, ligand-binding, and transactivation domains (for a review, see reference 44). However, these proteins carry out diverse physiological functions (for a review, see references 8, 44, and 60). As more NRs have been identified, the features of the ligands for these receptors have been shown to be more chemically diverse, and not exclusively endocrine (for a review, see references 8 and 15). For many of the identified NRs, no apparent ligands have been found, and these proteins are therefore referred to as orphan nuclear receptors (for a review, see references 8 and 43).
Members of one subfamily of the orphan receptors bind to nonrepetitive DNA sequences as monomers and are constitutively localized to the nucleus; these members include Drosophila αFtz-F1 (fushi tarazu factor 1) and mammalian SF-1 (steroidogenic factor 1) (42). Previous studies of Drosophila have shown that αFtz-F1 functions collaboratively with a homeodomain protein, fushi tarazu (Ftz), to promote cell fate specification in fly embryos (25, 70). Mutations in ftz or αftz-f1 cause a common pair rule phenotype, which is associated with defects in alternate segments of the embryos. The alternatively spliced β isoform of Ftz-F1 has been found to regulate ecdysone-induced gene expression and therefore is involved in the metamorphosis of flies (4). Mammalian homologues of ftz-f1 include the SF-1 and LRH-1 (liver receptor homologue 1) proteins. SF-1 functions in regulating the transcription of some steroidogenic enzymes and plays an essential role in the development of the adrenal gland and gonad (28, 30, 37). LRH-1 has been shown to play a key role in regulating bile acid synthesis and cholesterol homeostasis (22, 39). In addition to two highly conserved zinc finger motifs found in all NRs (43), the DNA-binding domains (DBDs) of αFtz-F1 and SF-1 contain a basic amino acid-rich region called the Ftz-F1 box (63). The only Caenorhabditis elegans homologue of the Ftz-F1/SF-1 subfamily is encoded by the nhr-25 locus. NHR-25 contains a putative DBD that is ∼75% identical to those of Drosophila Ftz-F1 and human SF-1. NHR-25 also shares a less conserved ligand-binding domain and a transcriptional activation domain with the other members of this subfamily (31).
Previous studies of C. elegans have shown that eliminating nhr-25 gene activity with double-stranded RNA (dsRNA) interference (RNAi) or a deletion mutation results in gross defects in the animals, including embryonic lethality, defects in molting between larval stages, sterility, and vulval abnormality, indicating that this C. elegans NR homologue plays multiple roles throughout C. elegans development (1, 21). However, the cellular function of nhr-25 in these developmental events remains largely unknown. Here, our study focuses on the requirement for nhr-25 for the normal functions of the epidermal cells. In C. elegans, most epidermal (also known as hypodermal) cells fuse to form multinucleate syncytia, which play essential roles in protecting the animal's body by secreting collagens that make up the cuticle surrounding the body (36); in regulating cell fate specification of the neighboring cells, including the seam cells and the Pn.p cells (see below) (23, 46); and in taking up dietary sterols (69). A major epidermal syncytium, hyp7, is formed by the fusion of >100 cells, and it covers the majority of the animal's body (55). Our study revealed that nhr-25 mutants display defects in the ventral epidermal cells in embryos and in the epidermal syncytial cells, the seam cells, and the Pn.p cells in larvae. The seam cells and Pn.p cells are specialized epidermal cells. Cell fusion defects were often observed in the epidermal cells in nhr-25 mutants, which might reflect the involvement of nhr-25 in cell-cell junction or fusion.
MATERIALS AND METHODS
C. elegans strains.
C. elegans strains were maintained according to the standard protocol (3). All genetic analyses were performed at 20°C, unless otherwise noted. The ku217 allele was isolated in a screen for temperature-sensitive mutants defective in vulval morphogenesis following ethyl methanesulfonate mutagenesis (26).
The alleles used in this study were as follows (52): LGIII unc-119(ed3), nob-1(ct230), lin-39(n709), and lin-39(n1760) (11); LGX dpy-6(e14) and unc-9(e101). An integrated ajm-1::gfp {pJS191 [ajm-1::gfp] plus pDP#MM016B [unc-119(+)]} was used to visualize adherens junctions (40, 50; our unpublished results). An integrated seam cell marker-green fluorescent protein (SCM::GFP) strain, JR 672, was used to score the presence of seam cell nuclei. The SCM construct in this strain contains GFP with a nuclear localization signal driven by an undefined fragment of C. elegans genomic DNA (59; J. Rothman, personal communication). This reporter specifically marks the seam cells from the twofold stage through adulthood.
Molecular cloning of the gene defined by ku217.
Standard three-point mapping techniques were used to locate the mutation in the ku217 allele. From dpy-6 unc-9/ku217 heterozygotes, 2 out of 38 Unc non-Dpy recombinant progeny segregated in the Egl (egg-laying-defective) phenotype of ku217, and 33 out of 35 Dpy non-Unc progeny segregated in the Egl phenotype of ku217. We thus mapped ku217 to a genetic location of 10.9 map units on chromosome X.
In DNA-mediated germ line transformation experiments (45), cosmids that contained genomic sequences in the corresponding regions were microinjected into ku217 hermaphrodites, together with a sur-5::gfp reporter, pTG96 (24). The concentrations used were 15 ng/μl for each cosmid and 40 ng/μl for the reporter construct, and at least three independent lines were examined for all experiments. A single cosmid, F11C1, rescued the egg-laying defects associated with ku217 mutants (7% Egl; n = 44). A deletion subclone of F11C1 generated by XbaI digestion that removed all but one predicted open reading frame, F11C1.6, retained the ability to rescue the ku217 mutant egg-laying defects.
Two cDNA clones that represent the coding sequence of F11C1.6, yk175f2 and yk342d8, were obtained from Y. Kohara (National Institute of Genetics, Mishima, Japan). Both of them contained a stop codon and a poly(A) tail at their 3′ ends but lacked an ATG at their 5′ ends. We PCR amplified the upstream cDNA sequence from a C. elegans mixed-staged cDNA library (a gift from R. Barstead) using an SL1 (a spliced leader sequence in C. elegans) primer and a unique sequence found in both of the cDNA clones (5′-CAGTGCGTTTGAAGAAGCC-3′). A single PCR product that corresponds to the coding sequence of nhr-25 was isolated, which contained the predicted start codon and an in-frame stop codon immediately in front of the ATG. This clone was ligated with the cDNA clones from Y. Kohara to generate a full-length cDNA for F11C1.6. The resulting cDNA sequence, together with 3 kb of upstream sequence, rescued the sterility, egg-laying defects, larval lethality, and embryonic lethality for >80% of ku217 transgenic animals from three independent lines. To determine the molecular lesion in ku217 animals, mutant genomic DNA corresponding to all of the exons and intron-exon boundaries was amplified. In addition, ∼3 kb of sequence upstream of the start codon and 1 kb of sequence downstream of the stop codon were examined. Only one mutation was found in two independent PCR and sequencing experiments, and the antisense strand of DNA was further sequenced to confirm the result.
EMSA.
Electrophoresis mobility shift assays (EMSAs) were carried out as previously described (2) with oligonucleotides purchased from Operon (Alameda, Calif.). The wild-type DNA sequence contained the in vivo target nucleotides of Drosophila Ftz-F1 (5′-TTGCAGCACCGTCTCAAGGTCGCCGAGTAGGAG-3′). A single-nucleotide switch (G to T; in italics below) was introduced into the wild-type DNA sequence to generate the mutant DNA sequence (5′-TTGCAGCACCGTCTCAAGTTCGCCGAGTAGGAG-3′). Oligonucleotides wereend labeled with [α-32P]dATP (Perkin-Elmer Life Sciences Inc., Boston, Mass.) by fill-in reactions using Klenow DNA polymerase (Gibco BRL, Carlsbad, Calif.). nhr-25 full-length cDNA sequence (either wild type or mutant) was inserted into a pGSTag2 vector (a gift from D. Ron and H. Dressler) for expression of a glutathione S-transferase (GST)::NHR-25 fusion protein. This fusion protein was expressed in the Escherichia coli BL21 strain and was purified according to the protocol of Ausubel et al. with some modifications. Briefly, glutathione-Sepharose resin (Amersham Biosciences, Piscataway, N.J.) was loaded into a Poly-Prep chromatography column from Bio-Rad (Hercules, Calif.). Bacterial lysate containing GST::NHR-25 fusion protein was added to the column and was washed on the column. The purified protein was eluted off the column by adding 50 mM reduced glutathione. The eluate was run on a sodium dodecyl sulfate-10% polyacrylamide gel and was visualized by staining it with Coomassie blue. The protein concentration in the eluate was determined by comparing it with bovine serum albumin proteins at various concentrations in Coomassie blue staining. The purified GST::NHR-25 protein was then incubated with 0.05 pmol of labeled oligonucleotides in 20 μl of incubation buffer (2) for 30 min at 20°C, and 10 μl of the reaction mixture was loaded onto a 0.5% polyacrylamide gel containing 0.5 mM MgCl2. The gel was then dried and exposed to a phosphorimager screen (Amersham Biosciences).
GFP reporters.
Full-length cDNA sequence of nhr-25, together with 3 kb of upstream regulatory sequence, was inserted into the pPD95.77 vector (a gift from A. Fire), which contains the GFP coding sequence (7) and the 3′ untranslated region of unc-54 (16) at the 3′ end. A second GFP reporter construct was generated by inserting the gfp coding sequence, which was excised from the plasmid pPD103.87 (a gift from A. Fire), in front of the stop codon of nhr-25. This construct also contained the native 3′ untranslated region of nhr-25. The two constructs were injected into unc-119(ed3) animals at 30 ng/μl, together with an unc-119(+) plasmid, pDP#MM016B (40), at 40 ng/μl.
RNAi.
PCR primers that each contained a T7 promoter sequence and a unique nhr-25 cDNA sequence were used to amplify the first 0.9 kb of the nhr-25 coding sequence. dsRNA was generated from the 0.9-kb template using a large-scale T7 transcription kit (Novagen, Madison, Wis.). RNAi by dsRNA injection was carried out as described previously (17). The injected animals were transferred to individual fresh plates after 16 h, and their progeny were scored for the mutant phenotype.
For RNAi feeding, the first 1.4 kb of nhr-25 coding sequence was inserted into the vector pPD129.36 (a gift from A. Fire). The resulting plasmids were transformed into an E. coli strain, HT115, and the bacteria were streaked on agar plates containing 10 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 50 μg of ampicillin/ml to induce the production of dsRNA (32, 61). Worms were raised on these plates for at least two generations before the phenotype was scored to ensure the effectiveness of RNAi.
For tissue-specific RNAi, the first 0.9 kb of nhr-25 coding sequence was fused in the sense and antisense orientations to the col-10 promoter (a gift from V. Ambros). The col-10 promoter contains 1.2 kb of col-10 genomic DNA upstream of its ATG. Both constructs were injected at 50 ng/μl, and a sur-5::gfp transcriptional fusion construct was used as a coinjection marker (24).
MH27 antibody staining.
Large-scale embryo preparations were fixed and stained according to the method of Miller and Shakes (47). The monoclonal antibody MH27, which recognizes adherens junctions, was a generous gift of R. Waterston and was used at a dilution of 1:250 (19).
GST pull down.
Both wild-type and mutant NHR-25 and GST proteins were expressed and purified on glutathione-Sepharose resin as described above. Full-length LIN-39, NOB-1, and luciferase were in vitro translated using a TNT-coupled reticulocyte lysate system from Promega (Madison, Wis.) and labeled with [35S]methionine (Promega). Thirty microliters of a 1:1 slurry of Sepharose resin was incubated with 5 μl of the labeled protein in the incubation buffer (250 mM NaCl, 50 mM Tris · HCl [pH 7.5], 0.5 mM sucrose, 1 mM dithiothreitol, 5% glycerol) for 2 h at 4°C. Subsequently, the reaction mixture was washed five times in the incubation buffer at 4°C, sodium dodecyl sulfate sample buffer was added, and the Sepharose resin was spun down. The supernatant was run on a 10% polyacrylamide gel to separate proteins retained on the resin. The resulting gel was dried and exposed to film.
RESULTS
ku217 mutants were associated with embryonic and larval lethality, sterility, egg-laying defects, and male tail abnormalities.
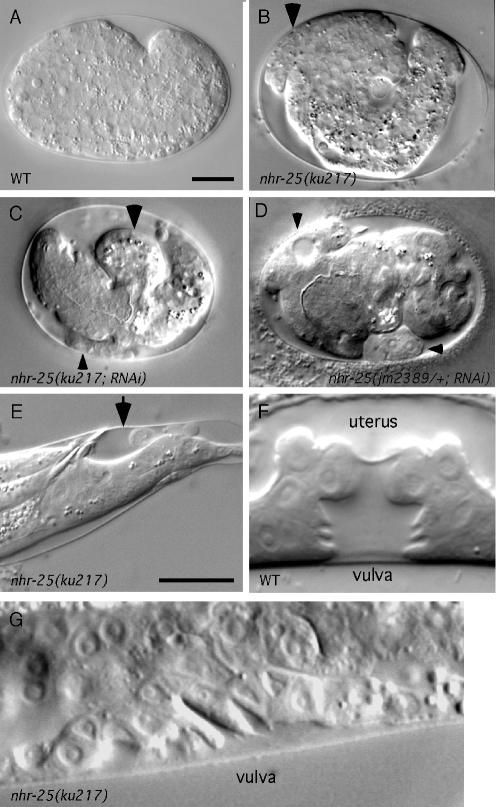
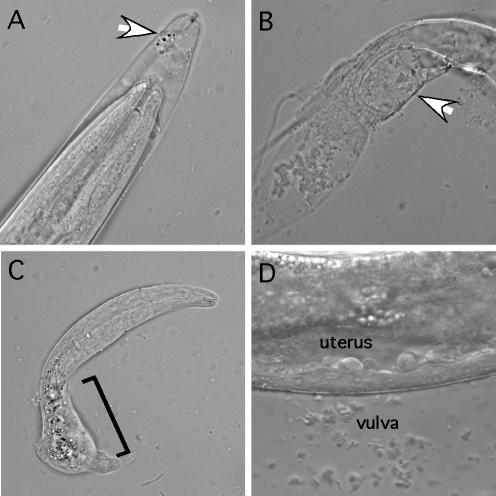
We initiated our study of the nhr-25 gene by isolating the ku217 allele in a screen for temperature-sensitive mutants defective in vulval development (9, 26). At 20°C, 74% of adult ku217 homozygous hermaphrodites were egg-laying defective, and some of them were sterile (Table 1). Lethality at either embryonic or larval stages was also observed (Table 1 and Fig. 1B and E). The severity of the above-mentioned phenotypes increased at higher temperatures (Table 1), suggesting that ku217 is a temperature-sensitive allele. nhr-25(ku217) mutant males were sterile at all temperatures; several structures located at the tail that are required for copulation, including spicules, sensory rays, and the hook, were aberrant (data not shown). Arrested ku217 larvae were observed with an apparent molting defect; the mutants were unable to shed the old cuticles even when the new ones had fully formed (Fig. 1E). ku217 animals that survived further to adulthood were sometimes sterile. In these sterile animals, gonadal cells appeared to undergo excess proliferation. The resulting tumorous gonad often failed to elongate and migrate correctly to form functional tissue (reference 21 and data not shown). Vulval development in 9% (n = 22) of ku217 animals was completely absent, possibly due to the lack of differentiation of the anchor cell in the gonad, which is required for signaling the vulval precursor cells to adopt the vulval fate (23). However, all ku217 animals that were examined appeared to have a normal anchor cell. In the majority of ku217 animals where vulval differentiation did initiate, cell division sometimes prematurely stopped. Vulval precursor cells underwent fewer rounds of cell division and therefore generated fewer progeny than in wild-type animals (see below). Furthermore, vulval cells in ku217 hermaphrodites sometimes did not migrate to the right positions so that the invagination formed by these cells was abnormally wide (Fig. 1G).
TABLE 1.
Phenotypes of nhr-25 mutants
| Genotype | Temp (°C) | n | % Phenotypea:
|
|||
|---|---|---|---|---|---|---|
| Emb let | Lar let | Egl | Sterility | |||
| N2 | 20 | 196 | <1 | 0 | 0 | 0 |
| 15 | 215 | 13 | 2 | 30 | 5 | |
| nhr-25(ku217) | 20 | 228 | 14 | 2 | 74 | 9 |
| 25 | 118 | 29 | 6 | 39 | 26 | |
| nhr-25(jm2389/+) | 20 | 176 | 22 | 2 | 0 | 0 |
| nhr-25(RNAi injection) | 20 | 372 | 5 | 6 | ND | ND |
| nhr-25(ku217 RNAi injection) | 20 | 67 | 100 | 0 | ND | ND |
| nhr-25(jm2389/+ RNAi injection) | 20 | 53 | 100 | 0 | ND | ND |
| nhr-25(ku217 RNAi feeding) | 20 | 149 | 32 | 31 | 13 | 24 |
| nhr-25(jm2389/+ RNAi feeding) | 20 | 96 | 33 | 30 | 16 | 21 |
| nhr-25 (ku217) unc-22(RNAi) | 20 | 177 | 10 | 3 | ND | ND |
Emb let, embryonic lethality; Lar let, larval lethality; Egl, egg-laying defective; ND, no data.
FIG. 1.
Phenotypes of nhr-25 mutant animals. (A to D) Wild-type, nhr-25(ku217), nhr-25(ku217 RNAi), and nhr-25(jm2389/+ RNAi) embryos, respectively, during elongation. The arrowheads point to the positions where the animals ruptured. (E) Molting defect of an nhr-25(ku217) larva during the L2 stage. The arrow indicates the old cuticle that remained surrounding the tail of the animal. (F and G) Vulva morphology of wild-type and nhr-25(ku217) animals, respectively, during L4. Anterior is to the left. Scale bars, 10 μm.
ku217 is a reduction-of-function mutation in the nhr-25 nuclear receptor locus.
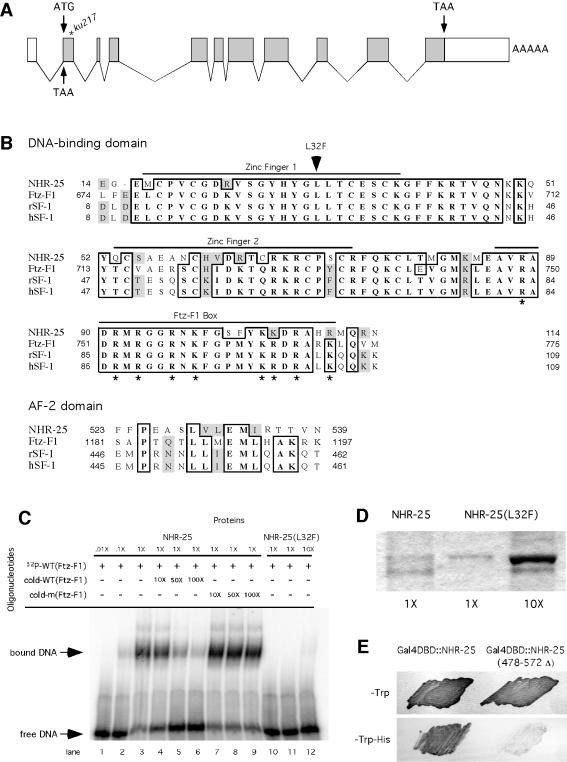
A single open reading frame that includes the nhr-25 gene, F11C1.6, fully rescued the mutant phenotypes of ku217 (see Materials and Methods). This locus is predicted by the genome-sequencing consortium to encode a 572-amino-acid-long peptide (21). A full-length cDNA clone that correlates with the predicted coding sequence of nhr-25 was isolated, and it retained the ability to rescue the ku217 mutant phenotypes (Fig. 2A) (see Materials and Methods). The nhr-25 gene product is homologous to Drosophila Ftz-F1 and mammalian SF-1 in a putative DBD at the N terminus and a transcriptional activation domain at the C terminus. The DBD in these proteins comprises two C2C2-type zinc fingers and a basic amino acid-rich domain called the Ftz-F1 box (Fig. 2B) (63).
FIG. 2.
Gene structure of the nhr-25 locus and biochemical properties of the NHR-25 protein. (A) nhr-25 cDNA contains an in-frame stop codon in front of ATG and a poly(A) tail at the 3′ end, suggesting that the cDNA is full length. The ku217 mutation was a C-to-T transition in the first exon and caused a Leu-to-Phe change in the protein sequence. (B) Alignment of peptide sequences of the DBD and transcriptional activation domain (AF-2) from C. elegans NHR-25, Drosophila Ftz-F1, and rat and human SF-1 (also see reference 21). Identical residues are boxed, and similar residues are shaded. The DBD consists of two C2C2-type zinc fingers and a basic amino acid-rich domain (Ftz-F1 box). ku217 was associated with a missense mutation (L32F) in the first zinc finger. The AF-2 domain contains an invariant glutamic acid residue and is flanked by several other conserved amino acids. (C) DNA-binding activities of wild-type NHR-25 and mutant NHR-25 [NHR-25(L32F)] generated by the ku217 mutation. In an electrophoresis motility shift assay, wild-type NHR-25 specifically bound to a DNA sequence containing the highly conserved core recognition sequence (PyCAAGGPyCPu) of Ftz-F1. NHR-25(L32F) bound poorly to this DNA sequence. A 1× amount of GST::NHR-25or GST::NHR-25(L32F) is equal to ∼50 ng of protein. (D) Coomassie blue staining of the NHR-25 proteins used in panel C. Note that NHR-25(L32F) was used in 10-fold excess compared to wild-type NHR-25 protein for the binding assay in panel C. (E) Transactivation activities of NHR-25 and NHR-25(478-572Δ). NHR-25(478-572Δ), which lacks the AF-2 domain, only weakly activated transcription of the HIS3 reporter gene, and therefore, the yeast strain (Y190) carrying this truncated form of NHR-25 grew poorly in media lacking histidine (−His). Yeast strains expressing NHR-25 or NHR-25(478-572Δ) grew equally well in media supplemented with histidine. The growth media lacked tryptophan in order to select strains expressing the NHR-25 proteins.
Previously, it was shown that deletion of the nhr-25 locus results in completely penetrant embryonic lethality (1), suggesting that this locus is essential for early embryonic development. To investigate the genetic nature of the ku217 allele, we constructed transheterozygous animals bearing the ku217 mutation and the jm2389 deletion allele (1). These transheterozygous animals arrested during embryogenesis, and only a few could survive to late larval stages, indicating that ku217 is a partial loss-of-function mutation in the nhr-25 locus. Animals subjected to RNAi against nhr-25 by injection (see Materials and Methods) in a wild-type background displayed less severe embryonic and larval phenotypes (Table 1 and data not shown). However, in combination with molecular lesions in nhr-25, either ku217 or jm2389/+ nhr-25(RNAi) resulted in increased lethality (Fig. 1C and D and Table 1), similar to what has been observed in nhr-25(jm2389) deletion mutants. RNAi of an unrelated gene, unc-22, had little effect on lethality (Table 1). Taken together, impairment of nhr-25 function, either by RNAi or by ku217 and jm2389 lesions, had a common effect during embryonic and larval development, although with different severities.
The ku217 mutation disrupted the DNA-binding ability of NHR-25.
A single-nucleotide switch (C to T) was found in the first exon of the nhr-25 coding sequence in ku217 mutants, and it was predicted to change a highly conserved leucine residue to phenylalanine in the putative DBD of NHR-25 (Fig. 2A and B). We investigated the DNA-binding abilities of the resulting mutant protein and wild-type NHR-25 using an EMSA (Fig. 2C). As the downstream targets of nhr-25 in C. elegans have not been identified, in this assay we made use of a 33-bp oligonucleotide sequence taken from the upstream promoter of ftz, which has been shown to be an in vivo target of Drosophila Ftz-F1 (62). In vitro DNA-binding assays have suggested that Ftz-F1 and SF-1 recognize and bind to a consensus 9-bp DNA sequence (PyCAAGGPyCPu) as a monomer (62, 67). Wild-type NHR-25 was able to bind to the oligonucleotide containing the 9-bp sequence, generating a slowly migrating species in the gel (Fig. 2C, lanes 1 to 3). The binding between a 32P-labeled oligonucleotide and NHR-25 could be competed away by adding an excess amount of cold oligonucleotide (Fig. 2C, lanes 4 to 6). A mutant sequence bearing a single-nucleotide change (G to T) in the consensus sequence (see Materials and Methods) was unable to affect the binding between the wild-type DNA sequence and NHR-25 even when it was 100-fold in excess (Fig. 2C, lanes 7 to 9). On the other hand, the mutant NHR-25(L32F) protein interacted with the target DNA sequence at an extremely low affinity (Fig. 2C, lanes 10 to 12, and D). Therefore, the missense mutation (L32F) in the ku217 mutant protein, located in the highly conserved zinc finger motif, greatly compromised the DNA-binding ability of NHR-25. These data suggest that, like its homologues in flies and in mammals, NHR-25 binds to DNA in a sequence-specific manner and that disruption of the DNA-binding activity leads to the loss-of-function phenotypes observed in ku217 animals. However, the dramatic >100-fold loss of affinity for DNA by NHR-25(L32F) is somewhat surprising given the partial loss-of-function phenotype in ku217 animals. This discrepancy could simply be a result of the differences between in vitro and in vivo experiments or a consequence of the use of the Drosophila Ftz-F1 binding site in our assay, which might differ somewhat from a true high-affinity NHR-25 site in vivo. Alternatively, NHR-25 could have a DNA-independent function in vivo that is not disrupted in ku217 mutants.
NHR-25 is a nuclear protein that is likely to function as a transcriptional activator.
C. elegans NHR-25 also contains a putative transcriptional activation motif, called the AF-2 domain (31). When fused to the GAL4 DBD, full-length NHR-25 was able to activate the HIS3 reporter gene expression driven by a GAL4 gene promoter (Fig. 2E). Deletion of a peptide sequence (after amino acid 477) of NHR-25 that contains the AF-2 domain greatly compromised the reporter gene transcription (Fig. 2E). Therefore, NHR-25 is likely to function as a transcriptional activator.
In support of the idea of NHR-25 being a transcription factor, an nhr-25::gfp reporter was found to be expressed in the nuclei of many epidermal cells, including the epidermal syncytial cells, the seam cells, and all the Pn.p cells and their progeny. Many cells in the head and tail also showed nhr-25::gfp expression (reference 21 and data not shown). nhr-25 expression was first observed during embryogenesis around the 100-cell stage (data not shown) and continued to be expressed throughout development in the epidermal and seam cells. In the Pn.p-derived vulval cells, when vulval morphogenesis (including cell division, fusion, and migration) was almost complete during the L4-to-adult transition, nhr-25::gfp expression significantly decreased, whereas in other Pn.p cells that fused with hyp7, nhr-25::gfp expression remained largely unchanged (data not shown).
nhr-25 mutants display defects in a subset of embryonic-cell fusion events.
To find out why impairment of nhr-25 function led to embryonic lethality, we first examined cell division and differentiation during early embryogenesis. Using a four-dimensional Nomarski DIC microscope to record early development of embryos, we found no apparent defect in cell division and differentiation (data not shown). However, later during embryo morphogenesis, the mutant animals often ruptured at the onset of body elongation along the A-P axis (Fig. 1B to D). Such a phenotype often reflects a deficit or malfunction of the epidermal cells, since the external epithelium in the embryos is crucial in sustaining the pressure generated during elongation and keeping the proper shape of the body (for a review, see references 46 and 54). Before embryo elongation initiates, the worm body is covered by six longitudinal rows of epidermal cells, two on the dorsal side, and two on each of the lateral sides. The ventral-lateral epidermal cells elongate and migrate circumferentially to meet at the ventral midline and form adherens junctions at their adjoining plasma membrane. This developmental process is referred to as ventral enclosure (66). In wild-type animals, ventral enclosure occurs first in the anterior end and proceeds to the posterior. Two pairs of the most anterior epidermal cells fuse (Fig. 3A, pairs 2 and 4) when the posterior cells start making contact with each other. The normal junction and/or fusion of the anterior epidermal cells allows them to withstand the pressure when cells squeeze into a long tubular worm shape and may be necessary for the enclosure in the posterior end (66).
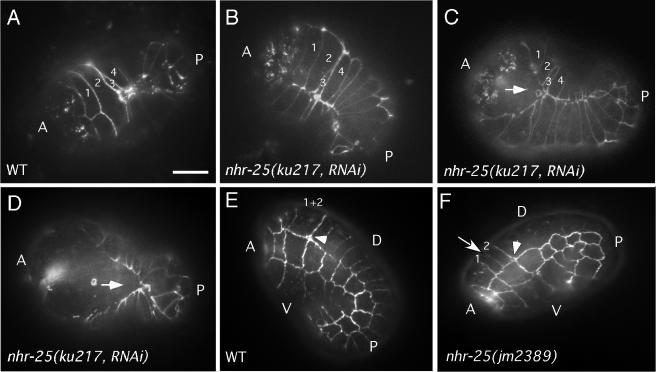
FIG. 3.
Fusion defects of nhr-25 mutant embryos. A and P, anterior and posterior, respectively. (A to D) Ventral side. (A) In wild-type (WT) embryos, ventral enclosure initiates at the anterior end and proceeds to the posterior end. Note that the pair 2 epidermal cells had already fused with one another when the posterior cells started to make contact. The pair 4 cells eventually fused. The other ventral epidermal cells remained unfused. (B) In an nhr-25(ku217 RNAi) animal, pair 2 and 4 cells did not fuse to each other, even after posterior cells had already formed adherens junctions at the adjoining membranes. (C) At a later time point, when the embryo shown in panel B attempted to elongate, the junction between anterior ventral epidermal cells ripped open, as indicated by the arrow. (D) Finally, the posterior epidermal cells could no longer maintain the adherens junctions between them, and the ventral side of the animal was no longer covered by any epidermal cells. (E and F) Fusion defect in the embryonic dorsal epidermis. The arrowheads indicate the positions of deirid sensillae. Dorsal epidermal cells 1 and 2 are located just anterior of the deirid sensillae. D and V, dorsal and ventral, respectively. (E) Lateral view of 1.5-fold MH27-stained wild-type embryo that has lost adherens junctions between dorsal epidermal cells 1 and 2 prior to any of the dorsal epidermal cells that are posterior to the deirid sensillae. (F) Lateral view of 1.5-fold MH27-stained embryo from an nhr-25(jm2389/+) parent that still retains the adherens junction between dorsal epidermal cells 1 and 2 (arrow) despite the fusion of most of the posterior dorsal epidermal cells. Scale bar, 10 μm.
To investigate cell differentiation, formation of adherens junction, and fusion of the epidermal cells in nhr-25 mutant embryos, we made use of an ajm-1::gfp reporter, which is present in the apical junctions between epithelial cells (50). We found that the right number of cells expressed AJM-1 at their peripheries at the proper stage (data not shown), suggesting that the defect seen in mutants was not due to epidermal cell proliferation. In addition, the ventral-lateral epidermal cells in nhr-25 mutant embryos were properly localized and were able to meet at the ventral midline and form adherens junctions as in wild-type animals (Fig. 3B to D), arguing against any possible defect in cell migration. However, the anterior ventral epidermal cells that normally fuse to each other, i.e., pairs 2 and 4, failed to fuse in some nhr-25 mutants. By the time posterior epidermal cells enclosed, the anterior cells detached from each other (Fig. 3B to D), and the embryos died shortly thereafter with a morphology identical to that of the embryos shown in Fig. 1B to D. The presence of a morphology associated with a failure in ventral enclosure was observed in 19% (n = 43) of the embryos from nhr-25(ku217) animals injected with nhr-25 dsRNA and 7% (n = 43) of the embryos from an nhr-25(jm2389/+) animal. A further 16% (n = 43) of the nhr-25(ku217 RNAi) embryos appeared to have ruptured during or shortly after the onset of embryonic elongation at a location distinct from the ventral midline.
Since some nhr-25 mutant embryos rupture on the dorsal side, we used the MH27 antibody that recognizes adherens junctions to investigate contacts and fusion events in the dorsal epidermal cells. In wild-type animals, the dorsal epidermal cells 1 and 2, lying just anterior to the deirid sensillae, usually fuse to each other shortly after the fusion of the pair 2 ventral epidermal cells involved in ventral enclosure (51). Further fusions of the dorsal epidermal cells occur in an anterior-to-posterior manner. Thirteen percent (n = 45) of wild-type embryos at the 1.5-fold stage had MH27 staining which indicated the presence of a cell boundary between the two cells (Fig. 3E). In contrast, 56% (n = 41) of 1.5-fold embryos from an nhr-25(jm2389/+) parent had a failure or delay in fusion between cells 1 and 2 (Fig. 3F). These two dorsal epidermal cells mirror the pair 2 cells of the ventral epidermis with respect to their position along the anterior-posterior axis and the time at which fusion occurs. It is possible that nhr-25 is required specifically for the differentiation and/or fusion of these two groups of cells. Another possibility is that these two groups of cells are the most sensitive to fusion defects in general and are therefore most easily assayed as being defective in an nhr-25 mutant background.
nhr-25(ku217) mutants had abnormal seam cell morphology and a greater number of adult seam cell nuclei.
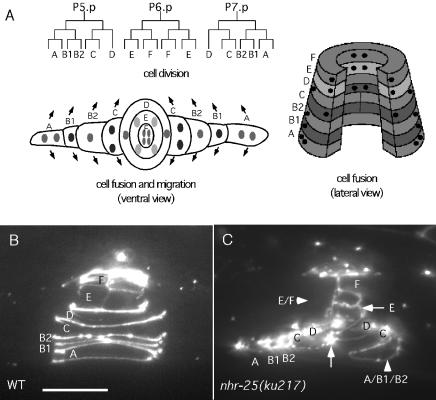
The two rows of dorsal epidermal cells interdigitate and fuse to generate the dorsal epidermal syncytium during embryogenesis (58). The ventral epidermal cells either fuse during embryogenesis (as described above) or, except for six pairs (referred to as the P cells [see below]), interdigitate and fuse to form the ventral epidermal syncytium during the L1 larval stage. Epidermal cells that lie on the lateral side of the body (10 on each lateral side) do not fuse and are referred to as the seam cells. The seam cells remain as stem cells during larval stages and divide asymmetrically to generate one stem cell and one daughter cell that differentiates into other cell types. Most of the differentiated daughter cells fuse to the surrounding epidermal syncytium; some, in males, give rise to the male-specific ray sensory neurons in the tail (14, 58). The seam cells connect to both dorsal and ventral syncytia and are therefore important for regulating the body shape of the worms. Additionally, the seam cells of L1 larvae and dauer worms are responsible for producing a specialized cuticle structure called the alae on the lateral surface of the animal. During the L4-to-adult transition, the seam cells eventually fuse together, and this syncytial cell is responsible for producing the adult alae (57). In nhr-25(ku217) ajm-1::gfp animals, the lateral epidermal cells appeared normal during embryogenesis. However, as the worm hatched and increased in size, particularly in length, the seam cells failed to elongate along the A-P axis (Fig. 4B). These seam cells had a round instead of a rectangular and elongated shape (Fig. 4A and B). As the worm body further elongated when the animal went through the four larval stages, junctions between neighboring seam cells were no longer maintained, so that gaps between seam cells were often observed even after seam cell fusion (Fig. 4C). Gaps in adult syncytial seam cells were seen in 76% (n = 38) of nhr-25(ku217) animals examined. Injection of nhr-25 dsRNA caused a seam cell phenotype similar to that seen in nhr-25(ku217) mutant animals. In the adult nhr-25(ku217) or nhr-25 dsRNA-injected animals, the lateral surface of the body was also characterized by patches of alae instead of a continuous one as in wild-type animals. The seam cell morphology and alar defects suggest that seam cell differentiation and/or execution of seam cell function is compromised in nhr-25 mutants.
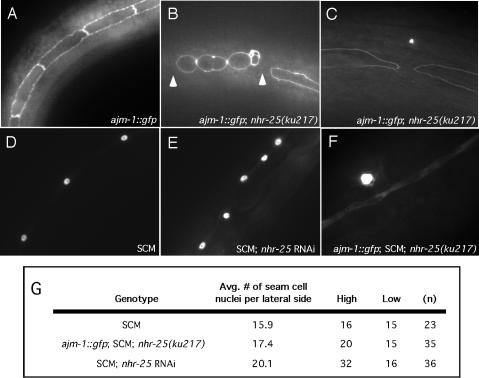
FIG. 4.
ajm-1::gfp and SCM expression in seam cells. (A) ajm-1::gfp expression in seam cells in a wild-type animal. (B) ajm1::gfp expression in seam cells of nhr-25(ku217) animals at L1. Note that the seam cells in the mutant had a round shape and were not always connected to other seam cells. The arrowheads indicate gaps between seam cells. (C) Adult nhr-25(ku217) animal that still had a gap in a lateral syncytial seam cell. (D) Evenly spaced SCM in a wild-type adult animal. (E) Presence of extra adult seam cell nuclei as indicated by the SCM following injection of nhr-25 dsRNA. (F) SCM was occasionally observed outside of the ajm-1::gfp-delineated boundary of adult syncytial seam cells in nhr-25(ku217) mutants. (G) On average, more SCM-positive nuclei were observed in adult syncytial seam cells of nhr-25 mutants than in wild-type animals.
We took advantage of a number of GFP markers for seam cell differentiation to better understand the nature of the seam cell defects in nhr-25 mutant animals. Using SCM (see Materials and Methods) (59), we determined that seam cells were able to differentiate properly in nhr-25(ku217) and nhr-25 RNAi animals (Fig. 4E), and the number of seam cells present at the time of hatching in nhr-25(ku217) animals was not noticeably different from that in wild-type animals (data not shown). However, when nhr-25(ku217) or nhr-25 dsRNA-injected young adults were scored for the number of SCM-expressing nuclei in their syncytial seam cells, it was observed that they had a higher number of SCM nuclei than did wild-type animals (Fig. 4E and G). Furthermore, SCM was occasionally expressed outside of the syncytial seam cell, as delineated by ajm-1::gfp (Fig. 4F). These defects could indicate a failure of the seam cell daughters to properly differentiate and fuse with the epidermal syncytia; hence, they abnormally maintained the seam cell fate. In support of the idea that seam cell differentiation did occur in nhr-25 compromised animals, the expression patterns of the seam cell markers elt-5::gfp, nhr-73::gfp, nhr-75::gfp, and nhr-77::gfp were not reproducibly altered when subjected to nhr-25 RNAi (references 34 and 48 and data not shown). It is unlikely that the GATA transcription factor ELT-5/EGL-18 regulates nhr-25 transcription, since injection of elt-5 dsRNA did not disrupt the expression of nhr-25::gfp in the embryo or in the larval stages of RNAi escapers (data not shown).
Pn.p and vulval cell differentiation was aberrant in nhr-25(ku217) mutants.
Six pairs of the ventral epidermal cells do not fuse during embryogenesis; they interdigitate and line up on the ventral midline of the animal at the end of the L1 stage (58). These cells are referred to as the P1 through P12 cells, anterior to posterior. Their posterior daughters (Pn.p cells) can adopt either the epidermal cell fate or the vulval cell fate (for a review, see reference 23). In wild-type animals, P(1, 2, 9 to 11).p cells fuse to hyp7 during L1 and lose their potential to adopt the vulval fate. The other six Pn.ps, P3.p through P8.p (also called vulval precursor cells [VPCs]), remain unfused with the surrounding hyp7 until L2-L3 and are competent to adopt the vulval fate. Using ajm-1::gfp to visualize the fusion of Pn.ps, we found that in nhr-25 mutants, some Pn.ps that normally fuse to hyp7 during L1 did not fuse as late as L2 (Table 2). Conversely, P4.p through P8.p, which should always remain unfused until late L2, sometimes fused to hyp7 during L1 or early L2 and therefore could no longer become vulval cells. In wild-type animals, three of the six VPCs, P(3, 4, 8).p, adopt the epidermal cell fate during the L3 stage; they divide only once and fuse to hyp7. The remaining three VPCs, P(5 to 7).p, are induced by an epidermal growth factor signal to become vulval cells and undergo a total of three rounds of cell division and a series of subsequent morphogenetic events (23, 53). In the ku217 mutants, fewer VPCs (an average of 2.7 cells; n = 39) were induced to become vulval cells at 20°C, consistent with the fact that some of the VPCs fused earlier and therefore lost their competence to acquire the vulval fate. Further lineage analysis revealed that some vulval cells prematurely stopped dividing in the ku217 mutants, particularly those that normally divide transversely, i.e., vulC, -E, and -F cells (Table 3). Using ajm-1::gfp, we investigated whether the defect in vulval cell division is accompanied by abnormal cell fusion. As shown in Fig. 5C, the boundary between vulE, vulF, and hyp7 was missing, indicative of abnormal fusion between these vulval cells and hyp7.
TABLE 2.
Pn.p cell fusion in nhr-25 mutants
| Genotype | % Unfused Pn.psa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | n | |
| Wild type | 0 | 0 | 38 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 56 |
| nhr-25(ku217 RNAi feeding) | 0 | 4 | 75 | 89 | 93 | 93 | 96 | 71 | 11 | 4 | 25 | 28 |
| nhr-25(ku217) | 0 | 0 | 70 | 100 | 100 | 100 | 100 | 97 | 3 | 0 | 3 | 31 |
| lin-39(n709) | 0 | 0 | 53 | 100 | 100 | 100 | 95 | 100 | 0 | 0 | 0 | 38 |
| lin-39(n709) nhr-25(ku217) | 0 | 0 | 50 | 91 | 91 | 91 | 82 | 65 | 0 | 3 | 6 | 34 |
1 to 11, P1.p to P11.p; n, number of animals scored.
TABLE 3.
nhr-25 and lin-39 mutations disrupt vulval cell induction and division
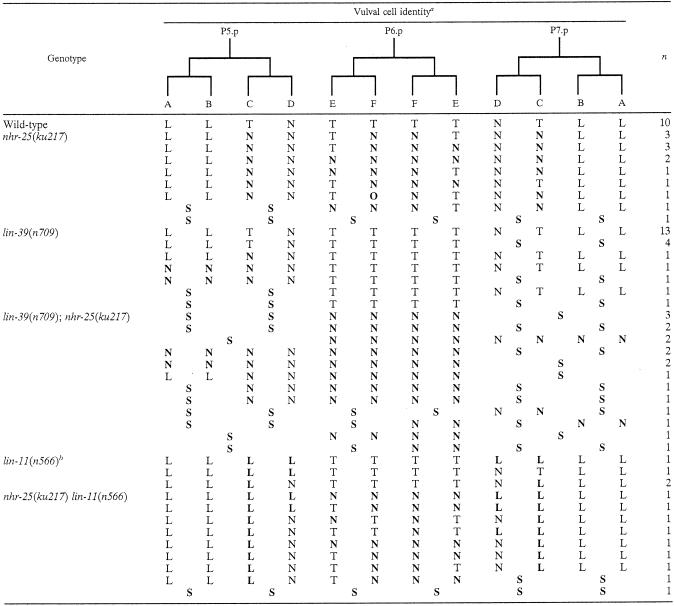
L, T, N, and O represent the division axes of the vulval cells during the third round of cell division. L, longitudinal division; T, transverse division; N, no division; O, oblique division axis; S, no vulval induction. Abnormal axes are highlighted in boldface. The number of letters in each cell indicates the number of progeny of each Pn.p cell.
See Ferguson et al. (15a) for lineage data for lin-11(382) and lin-11(n389).
FIG. 5.
Vulval cell division, fusion, and migration. (A) Twenty-two vulval cells are generated from three rounds of cell division. Except for vulB1 and vulB2, vulval cells that adopt the same cell fate, e.g., the two vulA cells, on either the anterior or posterior side of the vulva first fuse. These cells then send out cytoplasmic processes to the other side and fuse to the corresponding cells. The arrows indicate the directions of outgrowth of the cytoplasmic processes. A total of nine syncytial cells are generated, and they stack into seven toroidal rings (A through F). (B) ajm-1::gfp expression in a wild-type (WT) vulva (lateral view). The letters between rings of ajm-1::gfp-expressing membranes represent the identities of the syncytia. (C) ajm-1::gfp expression in an nhr-25(ku217) mutant (dorsal-lateral view). The arrowheads indicate where abnormal cell fusion occurred, and the arrows point to the junctions between cells that failed to fuse. Note that some vulC, -E, and -F cells are no longer enclosed by ajm-1::gfp-expressing membranes, suggesting that these cells have already fused to the surrounding hyp7. The posterior vulA, -B1, and -B2 abnormally fused together. In contrast, vulD cells and the posterior vulE cells failed to fuse. Anterior is to the left, and ventral is down. Scale bars, 10 μm.
In wild-type animals, as the vulval cells finish dividing, they fuse with each other in an invariant pattern to form nine multinucleate syncytial cells. During L4, sister cells from the third division (except for vulB1 and vulB2) on either the anterior or posterior side of the vulva first fuse with each other (Fig. 5B). These cells then send out cytoplasmic processes to the other side of the vulva and fuse with their counterparts. Although the division pattern of the vulA, vulB1, and vulB2 cells did not seem to be much affected by the nhr-25(ku217) mutation, they sometimes abnormally fused after division and formed a large syncytial cell (Fig. 5C). As mentioned above, migration of the vulval cells was also disrupted by the nhr-25(ku217) mutation so that the vulval cells on either the anterior or posterior side of the vulva often did not make contact and therefore did not fuse with each other (Fig. 5C).
Major adherens junction components appeared to be normal in nhr-25 mutants.
To test the possibility that the abnormalities in various epidermal cells are a result of disruption of adherens junctions that bring two neighboring cells together, we examined the expression level and localization of adherens junction components in nhr-25 mutant animals. At least two layers of physical contacts are present at the adherens junctions in C. elegans, one consisting of HMR-1, HMP-1, and HMP-2 (12) and the other consisting of AJM-1 (50). The complex formed by HMR-1 (E-cadherin), HMP-1 (α-catenin), and HMP-2 (β-catenin) has been found to connect the intracellular actin network to the extracellular matrix (12). A number of membrane-associated proteins have also been implicated in the proper assembly of the adherens junctions. For example, DLG-1 and LET-413 are required for the proper organization of the actin cytoskeleton, and they cooperatively control AJM-1 localization in the apical junction (18, 35, 38). Mutations in the above-mentioned genes often result in the rupture of the animal body during embryonic elongation, a phenotype similar to that observed in the nhr-25 mutant animals. Using antibodies against HMR-1, HMP-1, and HMP-2, we found that the expression levels, as well as the localizations, of all three proteins in ku217 animals injected with nhr-25 dsRNA, which caused fully penetrant embryonic lethality, were indistinguishable from those in wild-type animals (data not shown). Even in arrested embryos, adherens junctions, as marked by the antibodies of these three proteins, were still clearly visible. As mentioned above, the ajm-1 expression pattern was not affected in the embryonic epidermal cells, as well as the seam and vulval cells, of larvae (Fig. 3, 4, 5, and 6). Taken together, epidermal cells in nhr-25 mutants appeared to have properly differentiated and were able to establish the initial physical contact with each other. However, these contacts were often disturbed when the cells underwent extensive morphological changes.
FIG. 6.
Disruption of nhr-25 function through tissue-specific RNAi treatment. (A to D) Phenotypes caused by expression of partial nhr-25 sense and antisense sequence under the control of the col-10 promoter in a sid-1(qt2) background. (A and B) Arrows indicate old cuticle still attached to the anterior (A) and the posterior (B) of the worm. (C) L1 larvae had normal anterior structures but had a shortened posterior half, as indicated by the bracket. (D) Adult hermaphrodite with no recognizable vulval differentiation, very similar to nhr-25(ku217) mutant animals. Anterior is to the right.
nhr-25 likely functions cell autonomously in epidermal cells.
Although nhr-25 is expressed throughout the epidermis, we sought to further investigate where nhr-25 function is required during development. The gene sid-1 was shown to be essential for systemic, but not cell-autonomous, RNAi in C. elegans (68). We used a mutant sid-1(qt2) strain to perform tissue-specific RNAi by expressing the sense and antisense strands of partial nhr-25 coding sequence under the control of the col-10 promoter (see Materials and Methods). col-10 codes for a collagen protein produced by the epidermal cells during embryonic and larval stages (V. Ambros, personal communication). Larval-stage expression of a GFP reporter under the control of the col-10 promoter appeared to be restricted to hyp7, seam cells, and Pn.ps (data not shown). We found that, in the background of sid-1(qt2), animals expressing the pcol-10::nhr-25 sense and antisense constructs sometimes had a shorter body length and were more severely defective in the posterior end (Fig. 6C). Ten percent (n = 191) of progeny segregating from an animal carrying pcol-10::nhr-25(sense/antisense) constructs displayed a larval molting defect at various stages (Fig. 6A and B), suggesting that inhibition of nhr-25 activity in the epidermal cells interfered with the normal function of these cells. Additionally, 22% (n = 143) of animals positive for pcol-10::nhr-25(sense/antisense) constructs were Egl. Although these animals were not specifically examined for fusion defects using an adherens junction marker, a number of them had failures in Pn.p cell division and vulval morphogenesis similar to the fusion-defective nhr-25 RNAi or nhr-25(ku217) mutants (Fig. 6D). Control animals carrying empty pcol-10 vector did not display molting defects (0%; n = 301) or the Egl phenotype (<1%; n = 301). Overall, these results suggest that nhr-25 function is likely to be required cell autonomously in the epidermal cells and that the vulval defects seen in nhr-25 mutants are probably not the result of defective anchor cell signaling. In further support of this hypothesis, a recent study has shown that the anchor cell expression pattern of a lin-3::gfp reporter was unchanged in an nhr-25 RNAi background, where vulval differentiation was greatly compromised (29).
nhr-25 and lin-39 act collaboratively in regulating VPC fusion.
As discussed above, in the nhr-25 mutant, vulval precursor cells sometimes abnormally fused to hyp7; as a result, these cells could no longer acquire the vulval fate and failed to go through all three rounds of cell division. Such a phenotype was also observed in lin-39 Hox mutants (11, 65). We went on to test possible genetic and physical interactions between nhr-25 and lin-39. We first constructed and examined the phenotypes of double mutants for the two genes. When nhr-25 RNAi was carried out in the background of a lin-39(n1760) null mutant, which has not been found to be associated with any embryonic defects, only mild embryonic lethality was observed, as in nhr-25 dsRNA-treated animals alone, suggesting that these two genes are unlikely to have synergistic or redundant functions during embryogenesis. However, in lin-39(n709); nhr-25(ku217) double mutants, both alleles being partial loss-of-function mutations, a higher percentage of VPCs failed to adopt the vulval fate (Table 3). P5.p and P7.p sometimes fused to hyp7 before they underwent any division, and P6.p never divided more than twice (Table 3). We further characterized the fusion defects of P1.p to P11.p in a lin-39(n709); nhr-25(ku217) background by using the ajm-1::gfp marker. The VPC fusion defects seen in this double-mutant background were more severe than in any of the single weak loss-of-function mutants alone but no more severe than a strong loss-of-function in nhr-25 (i.e., RNA: in the ku217 background) (Table 2). These data suggest that lin-39 and nhr-25 may function collaboratively in preventing P(3 to 8).p cell fusion with the hyp7 cell and subsequently maintaining their competence for the vulval fate.
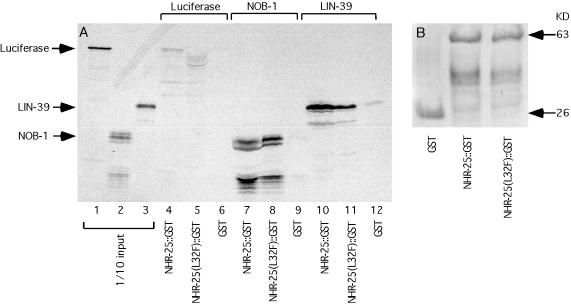
Secondly, a GST pull-down experiment was carried out to examine the direct physical interaction between NHR-25 and LIN-39. As shown in Fig. 7A, 35S-labeled LIN-39, but not luciferase, was retained on the glutathione-Sepharose resin that was conjugated with wild-type NHR-25, suggesting that NHR-25 specifically interacts with LIN-39 (Fig. 7A). The L32F mutation in the ku217 allele, which greatly compromised the DNA-binding activity of the NHR-25 protein, did not significantly affect the binding of LIN-39 (Fig. 7A). These data, in conjunction with the genetic evidence, suggest that NHR-25 and LIN-39 likely function together in a complex to regulate the postembryonic development of the VPCs.
FIG. 7.
NHR-25 physically interacts with LIN-39 and NOB-1. (A) Binding between LIN-39 and NHR-25. Lanes 1, 2, and 3, 1/10 total input of the in vitro-translated luciferase, NOB-1, and LIN-39, respectively; lanes 4, 5, and 6, amounts of luciferase protein retained on the Sepharose resin conjugated with NHR-25, NHR-25(L32F), and GST, respectively. None of the three proteins interacted with luciferase. Lanes 7, 8, and 9, amounts of NOB-1 retained on the Sepharose resin conjugated with NHR-25, NHR-25(L32F), and GST, respectively; lanes 10, 11, and 12, amounts of LIN-39 retained on the Sepharose resin conjugated with NHR-25, NHR-25(L32F), and GST, respectively. Only NHR-25 and NHR-25(L32F) (and not GST alone) were able to interact with NOB-1 and LIN-39. Note that the NHR-25(L32F) mutation resides in the DBD of the protein. (B) Similar amounts of GST, NHR-25::GST, and NHR-25(L32F)::GST were used for incubation with in vitro-translated proteins.
nhr-25 and nob-1 act together to promote embryogenesis.
Because nhr-25 mutant phenotypes were seen in a number of cell types throughout development, we sought to determine if NHR-25 was functioning with Hox proteins other than LIN-39. We chose to investigate the Hox genes ceh-13, nob-1, and mab-5 due to their phenotypes and/or expression patterns. The Hox protein CEH-13 is an attractive candidate for functioning with NHR-25 in the embryo and seam cells due to the ceh-13 loss-of-function phenotypes, which include rupture of the embryonic epidermis, fusion defects, and seam cell abnormalities (6). MAB-5, another Hox protein, is also known to regulate differentiation of the seam cells (33). However, we failed to observe genetic interactions between nhr-25 and the Hox genes ceh-13 and mab-5 (data not shown). The posterior-group Hox gene nob-1 was previously shown to have an important role in embryonic morphogenesis (64). nob-1(ct230), a weak loss-of-function allele, results in animals that are embryonic lethal and larval lethal (with missing posterior regions) and viable animals with variably misshapen tails. Because nhr-25 mutants sometimes have shortened posterior ends (data not shown and Fig. 6C), we looked for genetic interactions between nhr-25 RNAi and nob-1(ct230). We observed enhanced embryonic lethality in a nob-1(ct230); nhr-25 RNAi background that was considerably greater than the sum of the lethalities seen in the single mutants (Table 4). Dead embryos from this double-mutant background appeared to be similar to those resulting from a strong loss of function in nhr-25. A control RNAi experiment directed against unc-22 did not show increased lethality in nob-1(ct230) animals (Table 4). Thus, these results suggest the possibility that nhr-25 and nob-1 function together to promote embryogenesis; conversely, nhr-25 is not likely to function with ceh-13 or mab-5.
TABLE 4.
Genetic interaction between nhr-25 and nob-1 during embryogenesis
| Genotype | % Phenotypea
|
n | |
|---|---|---|---|
| Emb let | Nob | ||
| nhr-25 RNAi | 11 | <1 | 562 |
| nob-1(ct230) | 12 | 6 | 663 |
| nob-1(ct230) nhr-25 RNAi | 56 | 10 | 714 |
| nob-1(ct230) unc-22 RNAi | 14 | 5 | 145 |
Emb let, embryonic lethality; Nob, no back end.
To further support the genetic results discussed above, we performed GST pull-down experiments. 35S-labeled NOB-1, but not luciferase, was retained on the glutathione-Sepharose resin that was conjugated with wild-type NHR-25 (Fig. 7A). Again, the L32F mutation in the ku217 allele did not affect the binding of NOB-1 (Fig. 7A). Thus, NHR-25 may function in a complex with NOB-1 to promote some aspect of embryonic development. As expected, CEH-13 and MAB-5 displayed no physical interaction in the GST pull-down assay (data not shown).
DISCUSSION
Cell fusion defects were observed in the ventral and dorsal embryonic epidermal cells, Pn.p cells, and vulval cells of nhr-25 mutants. It is possible that the presence of extra SCM-positive nuclei in the adult syncytial seam cell is caused by a failure of the seam cell daughters to properly fuse to hyp7 during larval development, thus maintaining the seam cell fate. Mutations in other genes that disrupt cell fusion, such as eff-1, lead to the presence of extra seam cells (49). Alternatively, some seam cell daughters may not be able to properly adopt the cell fate in the first place and consequently are unable to fuse to hyp7. The mutant phenotypes in the male tail may also result from cell fusion defects, since cell fusion plays an important role in the morphogenesis of the male tail (14). However, outside of these defects, fusion in general does not appear to be affected, and no gross changes in eff-1::gfp expression were observed in nhr-25 mutant embryos (data not shown). Additionally, nhr-25 appears to both promote and inhibit Pn.p and vulval cell fusion. Taken together, these results suggest that there is a failure in the regulation of fusion and differentiation and not a specific defect in the fusion machinery. The altered morphology of seam cells, improper migration of Pn.ps, and alar and molting defects in nhr-25 mutants all support the idea that nhr-25 functions in additional aspects of normal epidermal cell function that remain to be understood. However, nhr-25 function is not likely to dramatically control cell fate determination, given the presence of most epidermal cell junction components and seam cell markers in mutant nhr-25 animals.
It has been suggested that syncytial cells formed by cell-cell fusion are in general in a nonproliferative state (for a review, see reference 55). Therefore, the fusion between a syncytial cell and its neighboring cells may play an important role in controlling cell differentiation of the neighboring cells. During L3, the epidermal growth factor/Ras/MAPK pathway, together with the activities of genes in several other regulatory pathways, including lin-39, lin-12/Notch, and components of the Wnt pathway, actively promote the vulval fate in P5.p to P7.p and repress the alternative epidermal fate by preventing cell fusion between Pn.ps and hyp7 (10, 13, 23, 27, 41). Reduction of nhr-25 function in the vulva often led to a failure in cell division during the last round of cell division; the undivided cells appeared to have fused with neighboring hyp7 syncytium. Therefore, nhr-25 is likely to repress cell fusion in order to maintain the proliferating capability of the vulval cells. We noticed that in the mutant vulC cells that stopped dividing, a vulva-specific gene, egl-17, was no longer expressed (data not shown). Since vulD cells, the nondividing sister cells of vulC, did express egl-17 in both mutant and wild-type animals, it suggests that the mutant vulC cells did not transform into a cell fate similar to their sisters' vulval cells. Instead, these mutant vulC cells might have adopted an epidermal cell fate and therefore fused to the hyp7 syncytial cell. A similar choice between fusion and division is also critical for seam cell differentiation: the elt-5 and -6 genes are known to be essential for repressing seam-hypodermis fusion and for promoting proper seam cell differentiation (34). However, it remains a question whether the fusion defect seen in the nhr-25 mutant vulval cells is a consequence of failure in cell division. Previous work by G. Shemer and colleagues has suggested that fusion is not a default state after failure in cell division in the Pn.ps (54). Therefore, we favor the model that the fusion defect is causal in the division defects seen in nhr-25 mutants.
lin-39 activity in P3.p to P8.p, during the L1 and L2 larval stages, prevents these six cells from fusing to hyp7 and maintains their ability to divide and differentiate into vulval cells. Without lin-39 function, such as in lin-39 mutants or in P(1, 2, 9 to 11).p, the Pn.p cells fuse to hyp7 and can no longer divide (11, 65). nhr-25 mutants reveal a similar function for nhr-25 in repressing fusion in the VPCs. Ftz-F1 and its mammalian homologues are known to act as cofactors for other transcriptional regulators (22, 25, 70). For example, Ftz-F1 interacts physically and acts collaboratively with Ftz, a homeodomain-containing protein, to regulate gene expression in every other segment of fly embryos (25, 70). Our results suggest that NHR-25 may function as a cofactor for a Hox protein, LIN-39, in the vulval lineage, based on the physical interaction between the two proteins and the genetic synergy of fusion defects seen with partial loss-of-function mutations in the two genes. However, unlike lin-39, nhr-25 appears to simultaneously promote fusion of the non-vulval-lineage Pn.p cells, P(1, 2, 9 to 11).p. Thus, an attractive model for nhr-25 regulation of Pn.p cell fusion consists of NHR-25 functioning with LIN-39 to repress fusion of P3.p to P8.p while acting with another transcription factor in the non-VPC Pn.p cells (or in other cell types, such as the seam cells) to promote their fusion to hyp7. The identification of downstream target genes for NHR-25 and LIN-39 is likely to be crucial in further investigating how they cooperate in controlling vulval cell fusion and differentiation. It has been shown that lin-39 acts to repress Pn.p cell fusion by repressing the expression of EFF-1, a membrane protein directly involved in cell fusion (49, 56). However, we did not observe significant changes in eff-1::gfp expression in an nhr-25 mutant background (due to the embryonic lethality, we were unable to use the null mutation to assay the effect of completely eliminating nhr-25 activity on eff-1 expression), although this does not exclude the possibility that nhr-25 acts on other targets that are involved in cell fusion. A recent study of a novel member of the angiotensin-converting enzyme family, acn-1, has suggested that NHR-25 positively regulates acn-1 expression (5). This study also demonstrated that loss of acn-1 function results in a set of molting, vulval, and seam cell defects similar to those seen in nhr-25 mutants. Therefore, the acn-1 gene might define at least one potentially important transcriptional target of NHR-25.
Given the fact that LIN-39 activity is restricted to postembryonic stages in the Pn.p cells, it is possible that NHR-25 functions together with other factors, likely other Hox proteins, during embryogenesis and in different tissues. Indeed, we observed a physical and genetic interaction with the Hox gene nob-1. A thorough analysis of nob-1 expression and function has yet to be completed; therefore, it is difficult to speculate how or in what cells NHR-25 and NOB-1 might function together to promote embryogenesis. NHR-25 complex formation with various Hox or homeodomain-containing proteins might be an important and conserved mechanism for the regulation of NHR-25 function throughout development and in various cell types. However, experiments to look for ceh-13 or mab-5 genetic interactions with nhr-25 mutants, as well as in vitro binding assays to look for the physical interactions of CEH-13 and MAB-5 with NHR-25, were both negative (data not shown).
It remains unknown why the vulC, -E, and -F cells, which normally switch their division axes from the longitudinal to the transverse orientation, were more affected than the vulA and -B cells that continue to divide longitudinally in nhr-25 mutants (Table 3 and Fig. 6). The difference may imply that nhr-25 activity is essential in cells that undergo extensive cytoskeleton reorganization during division. Such an activity of nhr-25 is no longer required once vulC transforms to become vulA/B, such as in the lin-11 mutant background (20), since the transformed vulC regained its ability to divide along the longitudinal axis even when nhr-25 function was impaired (i.e., vulC divided longitudinally in both lin-11 and lin-11 nhr-25 animals [Table 3]). nhr-25 activity might also be required for the extensive cytoskeleton reorganization during and after migration and elongation of the ventral epidermal cells, for the elongation of seam cells, and in the migration of induced Pn.p cells.
Acknowledgments
We thank W. Hanna-Rose for isolating ku217 mutants; J. Rothman for the elt-5::gfp strain; J. Priess for HMR-1, HMP-1, and HMP-2 antibodies; J. Hardin for the ajm-1::gfp construct; M. Stern for the egl-17 construct; V. Ambros for the col-10 construct; D. Ron and H. Dressler for pGSTag2; R. Bastead for a cDNA library; A. Fire for GFP vectors; W. Wood for the nob-1 allele; A. Coulson at the Sanger Center for cosmids; Y. Kohara for cDNA clones; and the C. elegans Genetic Center (University of Minnesota) for some strains used in this work. We thank M. Tucker, D. Starr, W. Wood, and other members of the Han and Wood laboratories for helpful discussions and comments.
This work was supported by an RO1 grant from NIH (GM47869) to M.H., who is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Asahina, M., T. Ishihara, M. Jindra, Y. Kohara, I. Katsura, and S. Hirose. 2000. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells 5:711-723. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. Current protocols in molecular biology, vol. 3. John Wiley & Sons, Inc., New York, N.Y.
- 3.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broadus, J., J. R. McCabe, B. Endrizzi, C. S. Thummel, and C. T. Woodard. 1999. The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell 3:143-149. [DOI] [PubMed] [Google Scholar]
- 5.Brooks, D. R., P. J. Appleford, L. Murray, and R. E. Isaac. 2003. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J. Biol. Chem. 278:52340-52346. [DOI] [PubMed] [Google Scholar]
- 6.Brunschwig, K., C. Wittmann, R. Schnabel, T. R. Burglin, H. Tobler, and F. Muller. 1999. Anterior organization of the Caenorhabditis elegans embryo by the labial-like Hox gene ceh-13. Development 126:1537-1546. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 8.Chawla, A., J. J. Repa, R. M. Evans, and D. J. Mangelsdorf. 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866-1870. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z., and M. Han. 2001. Role of C. elegans lin-40 MTA in vulval fate specification and morphogenesis. Development 128:4911-4921. [DOI] [PubMed] [Google Scholar]
- 10.Clandinin, T. R., W. S. Katz, and P. W. Sternberg. 1997. Caenorhabditis elegans HOM-C genes regulate the response of vulval precursor cells to inductive signal. Dev. Biol. 182:150-161. [DOI] [PubMed] [Google Scholar]
- 11.Clark, S. G., A. D. Chisholm, and H. R. Horvitz. 1993. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell 74:43-55. [DOI] [PubMed] [Google Scholar]
- 12.Costa, M., W. Raich, C. Agbunag, B. Leung, J. Hardin, and J. R. Priess. 1998. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141:297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenmann, D. M., J. N. Maloof, J. S. Simske, C. Kenyon, and S. K. Kim. 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125:3667-3680. [DOI] [PubMed] [Google Scholar]
- 14.Emmons, S. W., and P. W. Sternberg. 1997. Male development and mating behavior, p. 295-334. In D. L. Riddle, T. Blumental, B. J. Meyer, and J. R. Priess (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 15.Escriva, H., F. Delaunay, and V. Laudet. 2000. Ligand binding and nuclear receptor evolution. Bioessays 22:717-727. [DOI] [PubMed] [Google Scholar]
- 15a.Ferguson, E. L., P. W. Sternberg, and H. R. Harvitz. 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326:259-267. [DOI] [PubMed] [Google Scholar]
- 16.Fire, A., K. Kondo, and R. Waterston. 1990. Vectors for low copy transformation of C. elegans. Nucleic Acids Res. 18:4269-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 18.Firestein, B. L., and C. Rongo. 2001. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Mol. Biol. Cell 12:3465-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis, G. R., and R. H. Waterston. 1985. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101:1532-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freyd, G., S. K. Kim, and H. R. Horvitz. 1990. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344:876-879. [DOI] [PubMed] [Google Scholar]
- 21.Gissendanner, C. R., and A. E. Sluder. 2000. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev. Biol. 221:259-272. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 23.Greenwald, I. 1997. Development of the vulva, p. 519-541. In D. L. Riddle, T. Blumental, B. J. Meyer, and J. R. Priess (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 24.Gu, T., S. Orita, and M. Han. 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18:4556-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guichet, A., J. W. Copeland, M. Erdelyi, D. Hlousek, P. Zavorszky, J. Ho, S. Brown, A. Percival-Smith, H. M. Krause, and A. Ephrussi. 1997. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385:548-552. [DOI] [PubMed] [Google Scholar]
- 26.Hanna-Rose, W., and M. Han. 1999. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development 126:169-179. [DOI] [PubMed] [Google Scholar]
- 27.Hoier, E. F., W. A. Mohler, S. K. Kim, and A. Hajnal. 2000. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 14:874-886. [PMC free article] [PubMed] [Google Scholar]
- 28.Honda, S., K. Morohashi, M. Nomura, H. Takeya, M. Kitajima, and T. Omura. 1993. Ad4BP regulating steroidogenic P-450 gene is a member of steroid hormone receptor superfamily. J. Biol. Chem. 268:7494-7502. [PubMed] [Google Scholar]
- 29.Hwang, B. J., and P. W. Sternberg. 2004. A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development 131:143-151. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda, Y., D. S. Lala, X. Luo, E. Kim, M. P. Moisan, and K. L. Parker. 1993. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol. Endocrinol. 7:852-860. [DOI] [PubMed] [Google Scholar]
- 31.Ito, M., R. N. Yu, and J. L. Jameson. 1998. Steroidogenic factor-1 contains a carboxy-terminal transcriptional activation domain that interacts with steroid receptor coactivator-1. Mol. Endocrinol. 12:290-301. [DOI] [PubMed] [Google Scholar]
- 32.Kamath, R. S., M. Martinez-Campos, P. Zipperlen, A. G. Fraser, and J. Ahringer. 20 December 2000. posting date. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2. [Online.] http://genomebiology.com/2000/2/1/RESEARCH/0002. [DOI] [PMC free article] [PubMed]
- 33.Kenyon, C. J., J. Austin, M. Costa, D. W. Cowing, J. M. Harris, L. Honigberg, C. P. Hunter, J. N. Maloof, M. M. Muller-Immergluck, S. J. Salser, D. A. Waring, B. B. Wang, and L. A. Wrischnik. 1997. The dance of the Hox genes: patterning the anteroposterior body axis of Caenorhabditis elegans. Cold Spring Harbor Symp. Quant. Biol. 62:293-305. [PubMed] [Google Scholar]
- 34.Koh, K., and J. H. Rothman. 2001. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128:2867-2880. [DOI] [PubMed] [Google Scholar]
- 35.Koppen, M., J. S. Simske, P. A. Sims, B. L. Firestein, D. H. Hall, A. D. Radice, C. Rongo, and J. D. Hardin. 2001. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3:983-991. [DOI] [PubMed] [Google Scholar]
- 36.Kramer, J. 1997. Extracellular matrix, p. 471-500. In D. L. Riddle, T. Blumental, B. J. Meyer, and J. R. Priess (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 37.Lala, D. S., D. A. Rice, and K. L. Parker. 1992. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol. Endocrinol. 6:1249-1258. [DOI] [PubMed] [Google Scholar]
- 38.Legouis, R., A. Gansmuller, S. Sookhareea, J. M. Bosher, D. L. Baillie, and M. Labouesse. 2000. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat. Cell Biol. 2:415-422. [DOI] [PubMed] [Google Scholar]
- 39.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 40.Maduro, M., and D. Pilgrim. 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maloof, J. N., and C. Kenyon. 1998. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development 125:181-190. [DOI] [PubMed] [Google Scholar]
- 42.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 43.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna, N. J., and B. W. O'Malley. 2002. Minireview: nuclear receptor coactivators—an update. Endocrinology 143:2461-2465. [DOI] [PubMed] [Google Scholar]
- 45.Mello, C., and A. Fire. 1995. DNA transformation. Methods Cell Biol. 48:451-482. [PubMed] [Google Scholar]
- 46.Michaux, G., R. Legouis, and M. Labouesse. 2001. Epithelial biology: lessons from Caenorhabditis elegans. Gene 277:83-100. [DOI] [PubMed] [Google Scholar]
- 47.Miller, D. M., and D. C. Shakes. 1995. Immunofluorescence microscopy. Methods Cell Biol. 48:365-394. [PubMed] [Google Scholar]
- 48.Miyabayashi, T., M. T. Palfreyman, A. E. Sluder, F. Slack, and P. Sengupta. 1999. Expression and function of members of a divergent nuclear receptor family in Caenorhabditis elegans. Dev. Biol. 215:314-331. [DOI] [PubMed] [Google Scholar]
- 49.Mohler, W. A., G. Shemer, J. J. del Campo, C. Valansi, E. Opoku-Serebuoh, V. Scranton, N. Assaf, J. G. White, and B. Podbilewicz. 2002. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell 2:355-362. [DOI] [PubMed] [Google Scholar]
- 50.Mohler, W. A., J. S. Simske, E. M. Williams-Masson, J. D. Hardin, and J. G. White. 1998. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 8:1087-1090. [DOI] [PubMed] [Google Scholar]
- 51.Podbilewicz, B., and J. G. White. 1994. Cell fusions in the developing epithelial of C. elegans. Dev. Biol. 161:408-424. [DOI] [PubMed] [Google Scholar]
- 52.Riddle, D. L., T. Blumental, B. J. Meyer, and J. R. Priess. 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 53.Sharma-Kishore, R., J. G. White, E. Southgate, and B. Podbilewicz. 1999. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development 126:691-699. [DOI] [PubMed] [Google Scholar]
- 54.Shemer, G., R. Kishore, and B. Podbilewicz. 2000. Ring formation drives invagination of the vulva in Caenorhabditis elegans: Ras, cell fusion, and cell migration determine structural fates. Dev. Biol. 221:233-248. [DOI] [PubMed] [Google Scholar]
- 55.Shemer, G., and B. Podbilewicz. 2000. Fusomorphogenesis: cell fusion in organ formation. Dev. Dyn. 218:30-51. [DOI] [PubMed] [Google Scholar]
- 56.Shemer, G., and B. Podbilewicz. 2002. LIN-39/Hox triggers cell division and represses EFF-1/fusogen-dependent vulval cell fusion. Genes Dev. 16:3136-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh, R. N., and J. E. Sulston. 1978. Some observations on molting in C. elegans. Nematologica 24:63-71. [Google Scholar]
- 58.Sulston, J. E., and H. R. Horvitz. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110-156. [DOI] [PubMed] [Google Scholar]
- 59.Terns, R. M., P. Kroll-Conner, J. Zhu, S. Chung, and J. H. Rothman. 1997. A deficiency screen for zygotic loci required for establishment and patterning of the epidermis in Caenorhabditis elegans. Genetics 146:185-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thummel, C. S. 1995. From embryogenesis to metamorphosis: the regulation and function of Drosophila nuclear receptor superfamily members. Cell 83:871-877. [DOI] [PubMed] [Google Scholar]
- 61.Timmons, L., D. L. Court, and A. Fire. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263:103-112. [DOI] [PubMed] [Google Scholar]
- 62.Ueda, H., S. Sonoda, J. L. Brown, M. P. Scott, and C. Wu. 1990. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 4:624-635. [DOI] [PubMed] [Google Scholar]
- 63.Ueda, H., G. C. Sun, T. Murata, and S. Hirose. 1992. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long terminal repeat-binding protein. Mol. Cell. Biol. 12:5667-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Auken, K., D. C. Weaver, L. G. Edgar, and W. B. Wood. 2000. Caenorhabditis elegans embryonic axial patterning requires two recently discovered posterior-group Hox genes. Proc. Natl. Acad. Sci. USA 97:4499-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, B. B., M. M. Muller-Immergluck, J. Austin, N. T. Robinson, A. Chisholm, and C. Kenyon. 1993. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell 74:29-42. [DOI] [PubMed] [Google Scholar]
- 66.Williams-Masson, E. M., A. N. Malik, and J. Hardin. 1997. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development 124:2889-2901. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, T. E., T. J. Fahrner, and J. Milbrandt. 1993. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol. Cell. Biol. 13:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winston, W. M., C. Molodowitch, and C. P. Hunter. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456-2459. [DOI] [PubMed] [Google Scholar]
- 69.Yochem, J., S. Tuck, I. Greenwald, and M. Han. 1999. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development 126:597-606. [DOI] [PubMed] [Google Scholar]
- 70.Yu, Y., W. Li, K. Su, M. Yussa, W. Han, N. Perrimon, and L. Pick. 1997. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature 385:552-555. [DOI] [PubMed] [Google Scholar]