Abstract
Stp1p and Stp2p are homologous and redundant transcription factors that are synthesized as latent cytoplasmic proteins with N-terminal regulatory domains. In response to extracellular amino acids, the plasma membrane-localized Ssy1p-Ptr3p-Ssy5p (SPS) sensor induces an endoproteolytic processing event that cleaves away the N-terminal regulatory domains. The shorter forms of Stp1p and Stp2p are targeted to the nucleus, where they bind and activate the transcription of amino acid permease genes. A novel genetic screen, specifically designed to search for rare mutations that affect the SPS-sensing pathway, identified the F-box protein Grr1p as an obligatory factor required for Stp1p/Stp2p processing. Additionally, we have found that a null mutation in the ASI1 (amino acid sensor-independent) gene enables full-length unprocessed Stp1p/Stp2p to enter the nucleus and derepress SPS sensor-dependent genes. The N-terminal domains of Stp1p/Stp2p contain two conserved motifs that are required for proper nuclear exclusion and proteolytic processing. These motifs function in parallel; mutations that abolish processing inhibit signaling, whereas mutations that interfere with cytoplasmic retention result in constitutive derepression of SPS sensor-regulated genes independently of processing. The N-terminal domain of Stp1p is functionally autonomous and transferable to other transcription factors, where its presence confers ASI1-dependent nuclear exclusion and SPS sensor-induced proteolytic processing.
All cells have the capacity to sense and respond to extracellular environmental cues by changing patterns of gene expression. Eukaryotic cells can regulate patterns of gene expression simply by controlling the movement of transcription factors across the nuclear envelope. For example, cells can synthesize transcription factors as latent forms that are excluded from entering the nucleus and, in response to discrete environmental cues, activate processes that induce their nuclear targeting. Several latent transcription factors that transfer regulatory information from the plasma membrane to the nucleus in metazoan cells have been described (9). Regulated latent transcription factors involved in signal transduction include the well-studied NF-κB/Relish, Cubitus interruptus, and Notch proteins (4, 5, 20). In such instances, nuclear targeting not only is used to provide signals necessary for the correct timing of gene regulation but also provides the means to physically transfer signals from nonnuclear compartments to specific promoter sequences.
Fundamental to understanding regulated latent transcription factors is the elucidation of the mechanisms that direct these proteins to change compartments in a controlled fashion. This change may be achieved through a protein modification that regulates the activity of intrinsic nuclear localization determinants; nuclear localization sequences (NLS) are sequence motifs recognized by the nuclear import machinery and, in the opposing manner, nuclear export sequences (NES) are sequence motifs that mediate nuclear export. Although NLS and NES motifs are required for directed nuclear import and export, respectively, perhaps the simplest way to regulate nuclear targeting is through physically tethering or anchoring latent precursor forms of transcription factors outside the nucleus. The first example of such a mechanism is the sterol regulatory element-binding protein (SREBP) (10). SREBP is an integral membrane protein that is anchored in the membranes of the early secretory pathway. The cytoplasmically oriented domain possessing transactivation activity is released from membranes in two successive rounds of proteolytic processing by site-specific membrane-bound proteases (38).
The unicellular budding yeast Saccharomyces cerevisiae was recently found to assess the availability of extracellular nutrients through sensors in the plasma membrane. Sensors, presumably functioning as receptors, that recognize small molecules, such as sugars or amino acids, have been identified (for a review, see reference 18). Ssy1p is a nutrient receptor that functions together with the two peripheral membrane-associated proteins, Ptr3p and Ssy5p, as a sensor of extracellular amino acids. This function was initially demonstrated by the observation that amino acid permease promoters are derepressed upon the addition of small amounts of amino acids (13). Amino acid induction requires all three sensor components, Ssy1p-Ptr3p-Ssy5p (SPS), and the F-box protein encoded by GRR1 (7, 13, 17, 25, 26, 29). The participation of Grr1p in signal transduction is not well understood, but its role as a substrate specificity factor in the Skp1p-Cullin-F-box ubiquitin protein-ligase (SCF) complex suggests a role for ubiquitylation in SPS sensor signal transduction. Consistent with this suggestion, mutants of the SCF complex and ubiquitin have been shown to affect SPS sensor-dependent promoter derepression (7).
Based on several lines of evidence, the homologous zinc finger transcription factors Stp1p and Stp2p are redundant downstream effector components of the SPS sensor pathway. Single deletions of either STP1 or STP2 partially impair SPS sensor signaling, whereas deletions of both fully abolish signaling (1, 12). Stp1p and Stp2p bind to specific upstream activating sequences (UAS) present within SPS sensor-regulated promoters (12, 33). Both Stp1p and Stp2p are synthesized as latent cytoplasmic factors that are activated by receptor-mediated processing (1). In response to the addition of amino acids and in a strictly SPS sensor-dependent manner, Stp1p and Stp2p are endoproteolytically cleaved. This event liberates the DNA-binding and transactivation domains from an approximately 10-kDa N-terminal fragment. The shorter forms of Stp1p and Stp2p accumulate in the nucleus, where they function to transactivate SPS sensor-regulated genes.
In this study, we directly tested the requirement of the proteolytic processing of Stp1p/Stp2p. We found that the N-terminal domains of Stp1p/Stp2p contain two conserved motifs that are required for SPS sensor-dependent regulation. These motifs are functionally distinct and function in parallel to ensure proper nuclear exclusion and proteolytic processing. We also showed that Grr1p is required for the proteolytic processing of Stp1p/Stp2p and that Asi1p, an integral membrane protein, functions to restrict the entry of full-length unprocessed forms of Stp1p and Stp2p into the nucleus. Finally, the N-terminal regulatory domain of Stp1p is transferable and confers full SPS sensor pathway and Asi1p control when fused to an unrelated synthetic transcription factor. Our data clearly indicate that Stp1p and Stp2p are maintained as latent cytoplasmic transcription factors merely because of motifs within their N-terminal regulatory regions.
MATERIALS AND METHODS
Media.
Standard media, including yeast extract-peptone-dextrose (YPD) medium and ammonia-based synthetic minimal dextrose (SD) medium, supplemented as required to enable the growth of auxotrophic strains, were prepared as described previously (11). Ammonia-based synthetic complex dextrose (SC) medium was prepared as described previously (1). When needed, l-leucine was added at a concentration of 1.3 mM to induce the SPS sensor, and l-glutamate was added at a final concentration of 1 mM to SD medium. When needed, 5-fluoroortic acid (5-FOA) was added to SC medium (1 g/liter). Media were made solid with 2% (wt/vol) Bacto Agar (Difco). Antibiotic selections were made on solid YPD medium supplemented with 200 mg of G418 (Invitrogen, Carlsbad, Calif.)/liter, 100 mg of clonNAT (Werner Bioagents, Jena, Germany)/liter, or 300 mg of hygromycin B (Duchefa, Haarlem, The Netherlands)/liter. Sensitivity to azetidine-2-carboxylic acid (AzC) was tested at a final concentration of AzC of 1 mM on solid leucine-supplemented SD medium.
Yeast strains.
All yeast strains used in this work (Table 1) are isogenic descendants of the S288c-derived strain AA255/PLY115 (3). Generation of the deletion alleles asi1Δ8::kanMX, grr1Δ50::hphMX4, stp1Δ51::Agleu2, stp2Δ50::hphMX4, and gap1Δ::PAGP-lacZ has been described elsewhere (1, 16). CAY224 and CAY225 are meiotic segregants obtained from a cross between CAY161 and PLY126. CAY228 was constructed by transforming CAY224 with two PCR products, one containing a 3′ portion of kanMX (23) fused to SSY1 flanked by 3′ ADE2 genomic sequences and one containing 5′ ADE2 genomic sequences fused to PTR3 flanked by a 5′ portion of kanMX. Homologous sequences within the region of overlap between the two amplified portions of kanMX facilitated the simultaneous targeting of both fragments to the ADE2 locus (see Fig. 3A). Transformants were selected on G418-supplemented medium, and ade2Δ (red) colonies carrying PTR3 and SSY1 sequences were identified. Similarly, CAY229 was obtained by transforming CAY225 with two PCR products, one encompassing SSY5 fused to a 5′ portion of natMX4 (22) and one containing a 3′ portion of natMX4 fused to SHR3. Overhangs in the appropriate primers facilitated targeting to the CAN1 locus (see Fig. 3A). Transformants were selected on clonNAT-supplemented medium and screened for canavanine resistance.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| CAY29 | MATaura3-52 | 1 |
| CAY91 | MATaura3-52 ssy1Δ13::hisG | 1 |
| CAY86 | MATaura3-52 grr1Δ50::hphMX4 | 1 |
| CAY103 | MATaura3-52 lys2Δ201 grr1Δ50::hphMX4 | This work |
| CAY107 | MATaura3-52 lys2Δ201 asilΔ8::kanMX | This work |
| CAY109 | MATaura3-52 grr1Δ50::hphMX4 asi1Δ8::kanMX | This work |
| CAY123 | MATaura3-52 stp1Δ51::Agleu2 stp2Δ50::hphMX4 | 1 |
| CAY144 | MATaura3-52 asilΔ8::kanMX | This work |
| CAY152 | MATaura3-52 stp1Δ51::Agleu2 stp2Δ50::hphMX4 asi1Δ8::kanMX | This work |
| CAY161 | MATαura3-52 ade2Δ51 stp2Δ50::hphMX4 gap1Δ::PAGP1-lacZ | 1 |
| CAY206 | MATaura3-52 ssy1Δ13::hisG asi1Δ8::kanMX | This work |
| CAY224 | MATaura3-52 gap1Δ::PAGP1-lacZ | This work |
| CAY225 | MATαura3-52 gap1Δ::PAGP1-lacZ | This work |
| CAY228 | MATaura3-52 gap1Δ::PAGP1-lacZ ade2Δ::PTR3-loxP-kanMX-loxP-SSY1 | This work |
| CAY229 | MATαura3-52 gap1Δ::PAGP1-lacZ can1Δ::SSY5-natMX4-SHR3 | This work |
| CAY233 | MATαura3-52 gap1Δ::PAGP1-lacZ ade2Δ::PTR3-loxP-kanMX-loxP-SSY1 can1Δ::SSY5-natMX4-SHR3 | This work |
| CAY241 | MATαura3-52 his3Δ200 gap1Δ::PAGP1-lacZ ade2Δ::PTR3-loxP-kanMX-loxP-SSY1 can1Δ::SSY5-natMX4-SHR3 | This work |
| HKY20 | MATaura3-52 lys2Δ201 ssy1Δ13::hisG | 29 |
| HKY31 | MATaura3-52 lys2Δ201 ptr3Δ15::hisG | 29 |
| HKY77 | MATaura3-52 lys2Δ201 ssy5Δ2::hisG | 17 |
| PLY126 | MATaura3-52 lys2Δ201 | 29 |
| PLY861 | MATaura3-52 his3Δ200 | This work |
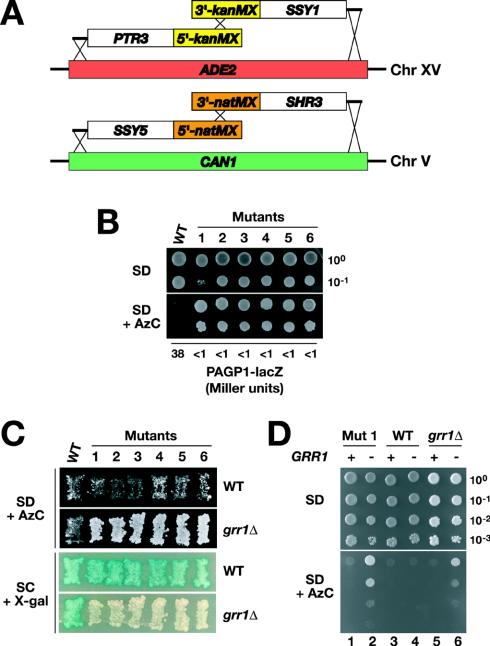
FIG. 3.
A genetic screen identifies GRR1 as an obligate component of the amino acid-induced SPS sensor pathway. (A) Merodiploid strains carrying duplicated genes encoding the known SPS sensor components and SHR3 were constructed by integrating PTR3 and SSY1 into the ADE2 locus on chromosome (Chr) XV and SSY5 and SHR3 into the CAN1 locus on chromosome V, creating the ade2Δ::PTR3-loxP-kanMX-loxP-SSY1 and can1Δ::SSY5-natMX-SHR3 alleles, respectively. (B) Tenfold dilution series of strain CAY241 (wild type [WT]) and six spontaneous AzC-resistant mutants lacking detectable β-galactosidase activity were applied as drops to solid SD medium (top) and SD medium containing leucine and AzC (bottom). The plates were incubated at 30°C for 7 days and photographed. β-Galactosidase activity (Miller units) present in each strain was quantified to monitor the expression of the integrated PAGP1-lacZ reporter construct. (C) Complementation analysis of diploid strains generated by mating CAY241 (WT) and AzC-resistant LacZ-negative mutants (mutants 1 to 6) with PLY126 (WT) and CAY103 (grr1Δ). The diploid strains were patched on SD medium supplemented with leucine, uracil, and AzC (top) and on SC medium (bottom). The plates were incubated at 30°C for 2 days, and β-galactosidase activity was visualized on SC medium by using an X-Gal overlay. (D) Tenfold dilution series of cell suspensions of mutant 1 (Mut 1), a WT strain (CAY241), or a grr1Δ strain (CAY86) transformed with pRS316 (−) or pCA212 (+) were spotted on plates containing SD medium (top) and SD medium supplemented with leucine and AzC (bottom). The plates were incubated at 30°C for 4 days and photographed.
The function of each of the targeted integration alleles was tested by complementation analysis. CAY228 and CAY229 were individually crossed to strains carrying URA3-marked deletions of SSY1, PTR3, SSY5, and SHR3. The resulting diploids were sporulated, and multiple meiotic segregants containing the integration alleles (Ade−/G418r or Canr/Natr) and the respective endogenous deletion alleles (Ura+) were analyzed. In all four cases, each integrated allele complemented the AzC resistance (AzC) exhibited by the individual ssy1Δ, ptr3Δ, ssy5Δ, and shr3Δ alleles. CAY233 is a meiotic segregant, containing both pairs of integration alleles, and was obtained by crossing CAY228 and CAY229. CAY241 is a his3 derivative of CAY233 and was obtained as a meiotic segregant from a cross between PLY861 and CAY233.
Plasmids.
The plasmids used in this study are listed in Table 2. The mutagenic oligonucleotides and PCR primers used are available on request. Plasmid pCA030 was created by ligating a 7.2-kb HindIII/SalI fragment obtained from YCpAGP1-LacZ (25) with similarly restricted pRS317. The insert of pCA047 was subcloned as an EcoRI/SmaI fragment into pRS317, creating pCA122. Plasmid pCA171 carrying HIS3 was isolated from yeast cells cotransformed with EagI/XhoI-restricted pRS313 and PvuI-restricted pCA047. Plasmid pCA120 was created by single-stranded mutagenesis of pCA047 with oligonucleotide 1 (30). Plasmids pCA127, pCA128, and pCA129 were created by single-stranded mutagenesis of pCA047 with oligonucleotides 2 to 4, respectively. Plasmid pCA135 was created by single-stranded mutagenesis of pCA047 with oligonucleotides 5 and 6. Plasmid pCA161 is a derivative of the commonly used two-hybrid bait plasmid pEG202 (24) with the following two modifications. First, the HIS3 marker was replaced by LYS2 amplified from pRS317 with primers 7 and 8. Second, the first 125 codons of STP1 were introduced in frame (with primers 9 and 10 and pCA029 as a template) (1) with the lexA gene fused to the activation domain (AD) of VP16 (with primers 11 and 12 and pCMXVP16 as a template) (a kind gift from T. Perlmann) (8). Plasmid pCA160 is a derivative of pCA161 created by deleting part of the ADH1 promoter and the entire lexA gene fusion by restriction with SphI and religation. Plasmid pCA212 was created by homologous recombination in yeast cells; a GRR1-containing PCR product (primers 13 and 14) amplified with S288c genomic DNA as a template was cotransformed with KpnI/SacI-restricted pRS316.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pCA027 | pRS316 (URA3) containing STP1Δ131 | 1 |
| pCA030 | pRS317 (LYS2) containing PAGP1-lacZ | This work |
| pCA047 | pRS316 (URA3) containing STP1-HA | 1 |
| pCA120 | pRS316 (URA3) containing STP1-133 | This work |
| pCA122 | pRS317 (LYS2) containing STP1-HA | This work |
| pCA127 | pRS316 (URA3) containing STP1-101-HA | This work |
| pCA128 | pRS316 (URA3) containing STP1-103-HA | This work |
| pCA129 | pRS316 (URA3) containing stp1-102-HA | This work |
| pCA135 | pRS316 (URA3) containing STP1-102,133 | This work |
| pCA160 | 2μm LYS2 PADH1 ΔSphI | This work |
| pCA161 | 2μm LYS2 PADH1 expressing Stp1(1-125)-LexA-AD | This work |
| pCA171 | pRS313 (HIS3) containing STP1-HA | This work |
| pCA212 | pRS316 (URA3) containing GRR1 | This work |
| pHK010 | pRS316 (URA3) containing SSY1 | 29 |
| pSH18-34 | 2μm URA3 containing OplexA-lacZ | 24 |
| pSSY1-102 | pRS316 (URA3) containing SSY1-102 | 19 |
Isolation of grr1 mutants.
Twenty independent colonies of CAY241 were inoculated into 5 ml of SD medium supplemented with adenine, glutamate, histidine, and uracil. The cultures were continuously shaken and grown to saturation for 36 h at 30°C. A 100-μl aliquot of each culture was plated on solid SD medium supplemented with adenine, glutamate, histidine, leucine, uracil, and AzC and incubated for 7 days at 30°C; between 50 and 200 AzC-resistant colonies formed on each of the 20 plates. The colonies were screened for β-galactosidase activity by using a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) overlay assay. Six AzC-resistant colonies with no detectable X-Gal staining were isolated.
β-Galactosidase activity assays.
β-Galactosidase activity was determined with N-lauroyl-sarcosine-permeabilized cells (28). After preincubation of cells in 800 μl of 0.2% (wt/vol) sodium N-lauroyl-sarcosine Z buffer at 30°C for 15 min, a 160-μl aliquot of o-nitrophenyl-β-d-galactopyranoside (ONPG) solution (4 mg/ml) was added and the tubes were mixed by inversion. The reaction was stopped by the addition of 400 μl of 1 M Na2CO3, and the tubes were centrifuged at 12,000 × g for 5 min. The absorbance of the supernatant was measured at 420 nm. Semiquantitative measurements of β-galactosidase activity were routinely made by using equally turbid cell suspensions (optical density at 600 nm, 1) diluted 1:1 in 0.4 M potassium phosphate buffer (pH 7) containing 0.2% (wt/vol) sodium N-lauroyl-sarcosine and 0.2 mg of X-Gal/ml. Cell suspensions were incubated at 30°C until blue color was detectable. A similar buffer containing 0.5% (wt/vol) low-melting-point agarose was used as an overlay to assess the β-galactosidase activity of whole colonies.
Immunoblot analysis.
Whole-cell extracts were prepared under denaturing conditions with NaOH and trichloroacetic acid treatment as described previously (35). Extracted proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and analyzed by immunoblotting. Immunoblots were incubated with primary antibody diluted in blocking buffer as follows: 12CA5 ascitic fluid (antihemagglutinin [HA] monoclonal antibody), 1:1,000; and anti-LexA polyclonal antibody (Invitrogen), 1:5,000. Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura extended-duration substrate; Pierce) of horseradish peroxidase conjugated to secondary antibodies (anti-mouse immunoglobulin from sheep and anti-rabbit immunoglobulin from donkey; Amersham) and quantified by using an LAS1000 system (Fuji Photo Film Co. Ltd).
RESULTS
The N-terminal domains of Stp1p and Stp2p contain conserved sequence elements.
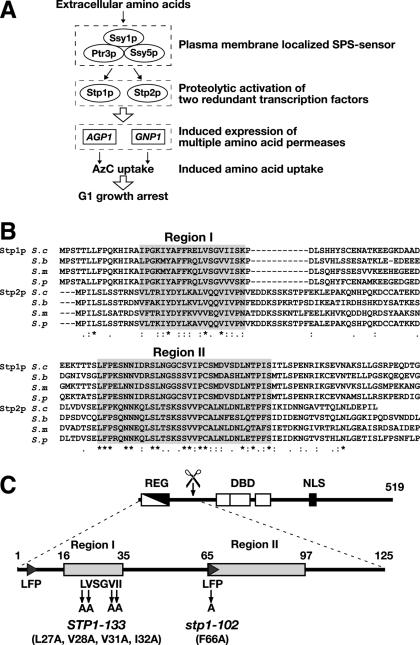
Based on current understanding of the SPS sensor signal transduction pathway (Fig. 1A), the N-terminal regulatory domains of Stp1p and Stp2p possess sequences that prevent their movement into the nucleus and protease recognition and cleavage sites (1). We sought to identify such elements by comparing the sequences of the first 125 N-terminal amino acids from the closely related orthologues of Stp1p and Stp2p from S. cerevisiae, S. bayanus, S. mikatae, and S. paradoxus (27). These domains are highly conserved between orthologues of Stp1p (60% identity) and Stp2p (55% identity). However, the degree of conservation is markedly reduced when Stp1p and Stp2p sequences are compared. Sequence alignments of all Stp1p and Stp2p homologues enabled two regions containing stretches of conserved amino acid residues to be identified (Fig. 1B). Region I is rich in branched-chain amino acids and is situated in the middle of the regulatory domain defined by the large in-frame deletion giving rise to the constitutive STP1Δ131 allele (Fig. 1C, REG) (1). Region II corresponds to the predicted site of proteolytic processing (Fig. 1C, scissor). The level of conservation within these regions and their locations suggest that they have important roles in the regulation of Stp1p and Stp2p nuclear exclusion (region I) and endoproteolytic processing (region II).
FIG. 1.
The N-terminal regulatory domains of the Stp1p and Stp2p orthologues contain two distinct regions of sequence conservation. (A) Schematic diagram of the SPS sensor pathway. Extracellular amino acids activate the SPS sensor, induce the endoproteolytic processing of transcription factors Stp1p and Stp2p, and thereby induce the expression of multiple amino acid permease genes. Consequently, cells grown in the presence of extracellular amino acids exhibit robust amino acid uptake and are sensitive to toxic amino acid analogues, e.g., AzC. (B) The first 125 amino acid residues from Stp1p/Stp2p (S.c.) and their orthologues in S. bayanus (S.b.), S. mikatae (S.m.), and S. paradoxus (S.p.) were aligned by using the CLUSTAL W algorithm (37). Conserved (asterisks) and similar (colons and periods) residues are marked. Regions I and II (gray boxes) are defined by stretches of enhanced sequence conservation. (C) Schematic representation of Stp1p. The locations of the inhibitory domain (REG), defined by the breakpoint of the STP1Δ133 allele (1), the DNA-binding domains (DBD), and the putative NLS are depicted in the full-length Stp1p (519 amino acids). Region I (amino acids 16 to 35), region II (amino acids 65 to 97), and the alanine substitution mutations of the STP1-133 and stp1-102 alleles are indicated in the enlargement of the N-terminal domain (amino acids 1 to 125).
A mutation that blocks proteolytic processing inhibits SPS signal transduction.
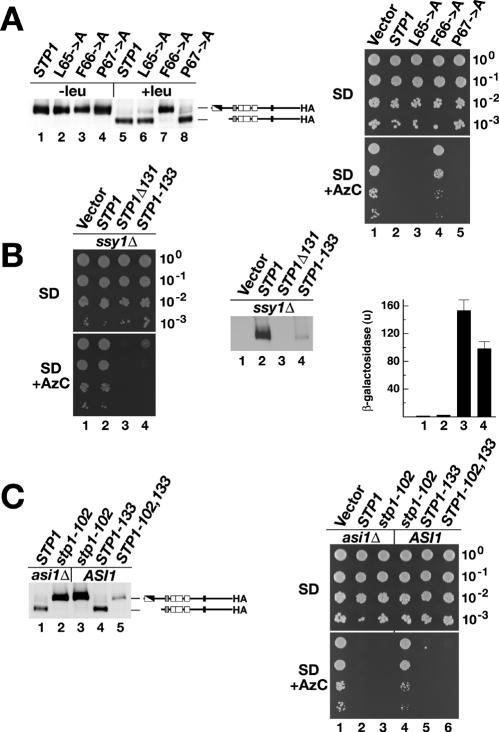
The first 3 amino acids of region II are conserved in all Stp1p and Stp2p homologues (Fig. 1B). To test the importance of these amino acids, residues L65, F66, and P67 were individually replaced with alanine in an initially functional protein carrying an HA tag at the extreme C terminus. An stp1Δ stp2Δ mutant strain was transformed with plasmids expressing wild-type Stp1p or alanine substitution L65A, F66A, and P67A mutant proteins. Cells were grown in amino-acid-free SD medium, and Stp1p cleavage in whole-cell extracts prepared before and 30 min after induction by leucine was monitored. Leucine induced the proteolytic processing of wild-type Stp1p (Fig. 2A, left panel, compare lanes 1 and 5). Alanine substitution mutations L65A and P67A had negligible effects on processing; the mutant proteins were proteolytically processed as effectively as wild-type Stp1p (Fig. 2A, left panel, compare lanes 2 and 6 and lanes 4 and 8). In contrast, leucine did not induce the proteolytic processing of the F66A mutant protein (Fig. 2A, left panel, compare lanes 3 and 7). The steady-state level of the F66A mutant protein was similar to that of wild-type Stp1p, suggesting that the alanine substitution does not grossly affect protein stability.
FIG. 2.
Mutations in regions I and II of Stp1p demonstrate the modular nature of the N-terminal inhibitory domain. (A) The stp1-102 allele encodes a mutant protein that is not endoproteolytically processed. (Left panel) Immunoblot analysis of protein extracts from strain CAY123 (stp1Δ stp2Δ) transformed with plasmids carrying wild-type STP1 (pCA047) or mutant allele L65→A (pCA127), F66→A (pCA129), or P67→A (pCA128). Cells were grown in SD medium (−leu) and, where indicated, leucine was added 30 min prior to harvest (+leu). Extracts were resolved by SDS-PAGE and immunoblotted with anti-HA. The immunoreactive forms of Stp1p present in the cell extracts are schematically represented at their corresponding positions of migration. (Right panel) Tenfold dilution series of cell suspensions of strain CAY123 (stp1Δ stp2Δ) transformed with pRS316 (Vector), pCA047 (STP1), pCA127 (L65→A), pCA129 (F66→A), or pCA128 (P67→A) were spotted on plates containing SD medium (top) and SD medium supplemented with leucine and AzC (bottom). (B) The STP1-133 allele encodes a constitutively active protein. (Left panel) Tenfold dilution series of cell suspensions of an ssy1Δ strain (HKY20) cotransformed with pCA030 (PAGP1-lacZ) and either pRS316 (Vector), pCA047 (STP1), pCA027 (STP1Δ131), or pCA120 (STP1-133) were spotted on plates containing SD medium (top) and SD medium supplemented with leucine and AzC (bottom). (Middle panel) Immunoblot analysis of protein extracts from HKY20 transformants grown in SD medium. Extracts were resolved by SDS-PAGE and immunoblotted with anti-HA. (Right panel) β-Galactosidase activity (Miller units, with standard deviations [n = 4]) present in each strain was quantified to monitor the expression of the PAGP1-lacZ reporter construct. (C) The STP1-133 mutation and the asi1Δ allele can suppress noncleavable stp1-102. (Left panel) Immunoblot analysis of protein extracts from strains CAY152 (asi1Δ) and CAY123 (ASI1) transformed with a plasmid carrying wild-type STP1 (pCA047) or mutant allele stp1-102 (pCA129), STP1-133 (pCA120), or STP1-102,133 (pCA135). Cells were grown in SD medium, and leucine was added 30 min priorto harvest. Extracts were resolved by SDS-PAGE and immunoblotted with anti-HA. (Right panel) Tenfold dilution series of cell suspensions of strains CAY152 (stp1Δ stp2Δ asi1Δ) and CAY123 (stp1Δ stp2Δ ASI1) transformed with pRS316 (Vector), pCA047 (STP1), pCA129 (stp1-102), pCA120 (STP1-133), or pCA135 (STP1-102,133) were spotted on plates containing SD medium (top) and SD medium supplemented with leucine and AzC (bottom).
The availability of a noncleavable mutant form of Stp1p enabled us to test whether proteolytic processing is obligatorily coupled to the transduction of SPS sensor-initiated signals. A growth assay based on the amino acid-induced uptake of the toxic proline analogue AzC was used to monitor Stp1p activity. AzC is transported into cells by Agp1p and Gnp1p (Fig. 1A); the genes encoding these amino acid permeases are transcribed in an Stp1p- or Stp2p-dependent manner (2). We examined the growth on SD medium and SD medium supplemented with AzC of an stp1Δ stp2Δ mutant strain transformed with a control vector or plasmids expressing wild-type Stp1p or L65A, F66A, and P67A mutant proteins. All strains exhibited robust growth on control SD medium (Fig. 2A, upper right panel). However, on SD medium supplemented with AzC, only cells carrying the control vector or the plasmid expressing the F66A mutant protein were able to grow (Fig. 2A, lower right panel, lanes 1 and 4). The growth of cells expressing the F66A mutant protein demonstrated that this protein is unable to transduce SPS sensor signals. We named the noncleavable and noninducible F66A mutant allele stp1-102 (Fig. 1C). The lack of growth on SD medium supplemented with AzC of cells carrying plasmids expressing wild-type Stp1p or L65A and P67A mutant proteins (Fig. 2A, right panel, lanes 2, 3, and 5) indicates that these proteins are functional, a finding consistent with their amino acid-induced processing.
Alanine mutations in conserved region I constitutively activate Stp1p.
We replaced multiple residues in the branched-chain amino acid cluster with alanine (Fig. 1B, region I). One allele with four alanine substitutions, STP1-133 (Fig. 1C), resulted in phenotypes consistent with constitutive signaling. It suppressed AzC resistance exhibited by an ssy1Δ strain (Fig. 2B, left panels, compare lanes 2 and 4) and activated the expression of β-galactosidase from a PAGP1-lacZ reporter plasmid almost to the extent of the constitutive STP1Δ131 allele (Fig. 2B, right panel, compare bar 2 with bars 3 and 4). The steady-state levels of full-length Stp1-133p present in SD medium-grown ssy1Δ cells were substantially lower than those in the same cells expressing wild-type Stp1p (Fig. 2B, middle panel, lanes 2 and 4), suggesting that the STP1-133 allele may result in misfolding of the N-terminal domain and destabilization of the protein. The fact that the STP1-133 allele phenocopies the characterized STP1Δ131 allele (1) indicates that residues affected by the mutations are important in the retention of unprocessed Stp1p outside the nucleus.
Proteolytic processing is not a prerequisite for transactivation, and full-length Stp1p activates transcription when inappropriately induced to enter the nucleus.
Loss-of-function mutations in the ASI1 gene enable the constitutive expression of Stp1p/Stp2p-dependent genes even in the absence of a functional SPS sensor (16). We previously showed that Stp1p/Stp2p processing does not occur in cells lacking a functional SPS sensor (1); thus, the constitutive expression of Stp1p/Stp2p-dependent genes in asi1Δ mutants without SPS sensor activity predicts that full-length Stp1p/Stp2p can activate transcription. To examine this possibility, we analyzed the status of Stp1p processing and AzC-resistant growth of asi1Δ mutant and ASI1 wild-type strains. An stp1Δ stp2Δ asi1Δ strain was transformed with a control plasmid (vector) or plasmids expressing either STP1 or the noncleavable stp1-102 allele. Immunoblot analysis revealed that Stp1p processing was not affected by the loss of ASI1; upon leucine induction, the shorter cleaved form was readily detected (Fig. 2C, left panel, lane 1). In constrast, all of the noncleavable Stp1-102p mutant protein migrated at the position of the full-length unprocessed form (Fig. 2C, left panel, lane 2). The activities of the expressed proteins were examined as AzC resistance. All transformed strains formed colonies of equal sizes on SD medium (Fig. 2C, upper right panel). Cells transformed with the empty plasmid (vector) readily formed colonies on AzC-containing medium (Fig. 2C, lower right panel, lane 1), whereas cells carrying a plasmid expressing wild-type STP1 (Fig. 2C, lower right panel, lane 2) or the stp1-102 allele did not form colonies (Fig. 2C, lower right panel, lane 3). The lack of growth of the STP1-expressing strain is consistent with the observed processing of Stp1p. The finding that stp1Δ stp2Δ asi1Δ cells expressing noncleavable Stp1-102p are unable to grow in the presence of AzC clearly demonstrates that unprocessed Stp1p is able to function as a transactivator if it is able to enter the nucleus.
The observation that the stp1-102 allele encodes a nonprocessed protein that is functional when expressed in an asi1Δ stp1Δ stp2Δ strain but not in an stp1Δ stp2Δ strain (Fig. 2C, right panels, lanes 4 and 3, respectively) enabled us to examine the epistasis of mutations in conserved regions I and II (Fig. 1B). We combined the recessive stp1-102 and the dominant STP1-133 mutations into a single gene, creating the double-mutant STP1-102,133 allele. An stp1Δ stp2Δ strain was transformed with plasmids expressing noncleavable Stp1-102p, constitutively active Stp1-133p, or double-mutant Stp1-102,133p. Immunoblot analysis showed that Stp1-133p was processed normally but that Stp1-102,133p was not (Fig. 2C, left panel, compare lanes 4 and 5). Cells transformed with the constitutively active STP1-133 allele exhibited AzC sensitivity and failed to form colonies on SD medium supplemented with AzC (Fig. 2C, right panels, lane 5). Also, cells carrying the double-mutant STP1-102,133 allele were unable to grow in the presence of AzC (Fig. 2C, right panels, lane 6). These results demonstrate that in the absence of a functional inhibitory domain (region I), cleavage is not required for unprocessed Stp1p to transactivate gene expression. Thus, it can be concluded that amino acid-induced cleavage is required merely to sever the inhibitory domain from the transactivating portion of Stp1p and that cytoplasmic retention and endoproteolytic activation are functionally separable and distinct processes.
A novel genetic screen identifies recessive mutations in GRR1 that inactivate the SPS sensor pathway.
Mutational analysis of the N-terminal region of Stp1p identified the endoproteolytic processing of Stp1p/Stp2p as the key regulatory step in the SPS sensor pathway and prompted us to search for additional factors that may be involved in the processing step. To enable the isolation of rare mutations affecting the SPS sensor pathway, we created a haploid strain harboring functional duplicated alleles of SSY1, PTR3, and SSY5 integrated into the genome (see Materials and Methods) (Fig. 3A). We also chose to duplicate SHR3, a gene encoding a membrane-localized chaperone that is specifically required for the exit of Ssy1p from the endoplasmic reticulum (21, 29). By making our starting strain merodiploid for these genes, our aim was to eliminate the reisolation of recessive loss-of-function mutations in previously identified SPS sensor components. The starting strain also carried an AGP1-promoted lacZ reporter gene integrated into the genome, leading to the expression of β-galactosidase when the SPS sensor pathway is intact. In contrast, cells with a mutant SPS sensor pathway will lack the expression of detectable β-galactosidase activity.
Signaling through the SPS sensor pathway leads to the transcription of AGP1 and GNP1, resulting in AzC import and cell cycle arrest (Fig. 1A). Consequently, mutations abolishing SPS sensor signaling make cells AzC resistant. Spontaneously arising AzC-resistant colonies were obtained by plating saturated cultures of the merodiploid starting strain on solid SD medium supplemented with AzC. Six slowly growing AzC-resistant colonies that were small and flat and that remained white after incubation with an X-Gal overlay were identified (Fig. 3B). Under all growth conditions, these six mutants consistently grew at greatly reduced rates compared to the wild-type starting strain. Microscopic analysis revealed that in all instances, the mutant cells were elongate and remained attached to one another, forming branched chains of cells. Consistent with these latter observations, the mutants exhibited a flocculent growth phenotype in liquid cultures.
The growth phenotypes of our newly identified mutant strains closely resemble those exhibited by grr1 mutant strains (14). Complementation analysis was used to address whether the mutations in our strains were indeed allelic with GRR1. All six strains were found to carry recessive mutations; diploids resulting from crosses with the wild-type strain were AzC sensitive and expressed robust β-galactosidase activity (Fig. 3C). In contrast, diploids from crosses with the grr1Δ tester strain were AzC resistant and lacked β-galactosidase activity (Fig. 3C). The lack of complementation of both phenotypes is consistent with the mutations being allelic with GRR1. Genetic linkage was established by sporulating the two diploids obtained from crosses with mutant 1; strict 2:2 segregation was observed in tetrads derived from the cross with the wild-type strain, and no AzC-sensitive spores were recovered from the cross with the grr1Δ strain. Additionally, plasmid-borne GRR1 complemented the AzC resistance exhibited by mutant 1 (Fig. 3D, compare lanes 1 and 2 with lanes 5 and 6). We conclude that the mutants isolated carry recessive mutations in GRR1.
GRR1 is required for the amino acid-induced endoproteolytic processing of Stp1p.
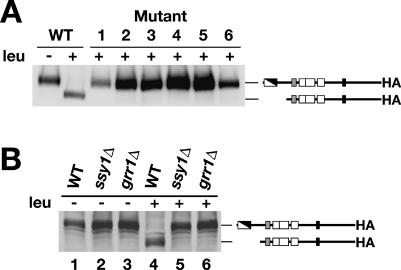
We previously reported that grr1Δ mutations do not impair the proteolytic processing of Stp1p, suggesting that Grr1p functions downstream of this regulatory event (1). We investigated whether our newly isolated grr1 mutant alleles affected the proteolytic processing of Stp1p. For wild-type extracts, the full-length form of Stp1p-HA was present in the noninduced control, and the cleaved 10-kDa lower-molecular-mass form was observed after leucine induction (Fig. 4A). In contrast, only the nonprocessed form of Stp1p-HA was found in leucine-induced extracts from the newly isolated grr1 mutants (Fig. 4A).
FIG. 4.
Loss-of-function mutations in GRR1 inhibit the endoproteolytic processing of Stp1p. (A) Immunoblot analysis of whole-cell extracts from a wild-type (WT) strain (CAY241) and AzC-resistant LacZ-negative mutant strains (Fig. 3) transformed with pCA171 (STP1-HA). Cells were grown in SD medium supplemented with adenine and uracil. Leucine (leu) was added 30 min prior to harvest. (B) Immunoblot analysis of whole-cell extracts from a WT strain (CAY29), an ssy1Δ strain (CAY91), and a grr1Δ strain (CAY86) transformed with pCA047 (STP1-HA). Cells were grown in SD medium. Leucine (leu) was added 30 min prior to harvest.
These unexpected results prompted us to repeat the analysis with a grr1Δ mutant strain. The cleavage of Stp1p-HA in a wild-type strain with an intact SPS sensor and that in an ssy1Δ mutant strain lacking the receptor component of the SPS sensor were monitored and served as positive and negative controls, respectively. Leucine-induced processing of Stp1p-HA was apparent in the wild-type strain (Fig. 4B, compares lane 1 and 4), whereas no processing was detected in the ssy1Δ strain (Fig. 4B, compare lanes 2 and 5). Consistent with our analysis of newly isolated grr1 mutant strains, no processing was observed in the grr1Δ mutant strain (Fig. 4B, compare lanes 3 and 6). Together, these results indicate, contrary to what we previously reported, that GRR1 is required for the proteolytic processing of Stp1p/Stp2p.
Constitutive activation of the Ssy1p receptor does not bypass the GRR1 requirement.
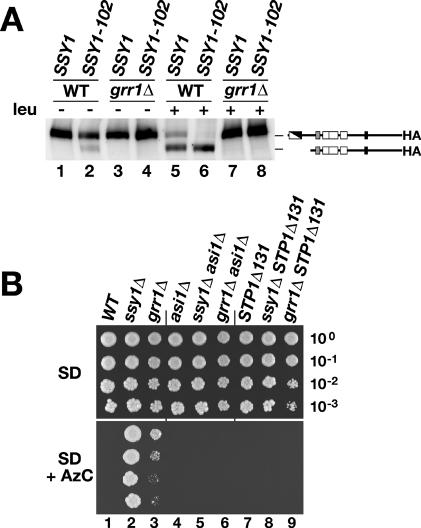
SSY1-102 encodes a dominant mutant form of Ssy1p that constitutively activates the expression of SPS sensor-regulated genes even in the absence of inducing amino acids (19). We tested whether SSY1-102 could suppress the Stp1p-processing defect in grr1Δ cells. Control wild-type and grr1Δ strains carrying a plasmid expressing Stp1p-HA were transformed with SSY1- or SSY1-102-containing plasmids and analyzed by immunoblotting. As previously shown, leucine induced Stp1p processing in wild-type cells (Fig. 5A, compare lanes 1 and 5) but not in grr1Δ cells (Fig. 5A, compare lanes 3 and 7). In uninduced wild-type cells expressing SSY1-102, a portion of immunodetectable Stp1p was found in its processed form (Fig. 5A, lane 2). Interestingly, leucine induction increased the amount of processed Stp1p to a level similar to that observed in SSY1-expressing cells (Fig. 5A, compare lanes 5 and 6). These results account for the constitutive amino acid-independent activation of SPS sensor-regulated genes in SSY1-102-expressing cells. Furthermore, these findings confirm that the proteolytic processing of Stp1p is a direct consequence of the amino acid-induced activation of Ssy1p and is not due to an indirect effect related to amino acid uptake (19). Thus, the constitutive nature of Ssy1-102p signaling appears to be mechanistically similar to amino acid-induced wild-type Ssy1p signaling. The Stp1p-processing defect in the grr1Δ strain could not be suppressed by the introduction of the SSY1-102 allele even in the presence of leucine (Fig. 5A, compare lanes 4 and 8).
FIG. 5.
Epistasis analysis of grr1Δ and SPS-sensing pathway mutations. (A) Immunoblot analysis of whole-cell extracts from a wild-type (WT) strain (PLY126) and a grr1Δ strain (CAY103) cotransformed with pCA122 (STP1-HA) and either pHK010 (SSY1) or pSSY1-102 (SSY1-102). Cells were grown in SD medium, and leucine (leu) was added 30 min prior to harvest. (B) Tenfold dilution series of cell suspensions of WT (CAY29), ssy1Δ (CAY91), grr1Δ (CAY86), asi1Δ (CAY144), ssy1Δ asi1Δ (CAY206), and grr1Δ asi1Δ (CAY109) strains carrying plasmid pRS316 and STP1Δ131 (CAY29), ssy1Δ STP1Δ131 (CAY91), and grr1Δ STP1Δ131 (CAY86) strains carrying plasmid pCA027 were spotted on plates containing SD medium (top) and SD medium supplemented with AzC (bottom). The plates were incubated at 30°C.
The dominant activated allele of STP1 (STP1Δ133) and the recessive asi1Δ mutation bypass the requirement for GRR1.
If the lack of Stp1p/Stp2p processing is the underlying reason for the signaling defect in grr1 mutant strains, mutations activating the pathway downstream of the processing step should exert a suppressing effect. To test this possibility, we examined whether the recessive asi1Δ and dominant activated STP1Δ133 alleles could bypass the need for GRR1. Wild-type, ssy1Δ, and grr1Δ cells, with and without the asi1Δ and STP1Δ133 alleles, were spotted on solid SD medium and SD medium supplemented with AzC. All strains grew on the control plate containing SD medium (Fig. 5B, upper panel). In the presence of AzC, the wild-type strain exhibited no growth, whereas the ssy1Δ and grr1Δ strains formed colonies, indicating that these strains are defective in SPS sensor signaling (Fig. 5B, lower panel, lanes 2 and 3). The introduction of either asi1Δ or STP1Δ133 mutations suppressed the loss of SPS sensor activity, as evidenced by the lack of growth of ssy1Δ asi1Δ and ssy1Δ STP1Δ133 strains (Fig. 5B, lower panel, compare lane 2 with lanes 5 and 8). The asi1Δ and STP1Δ133 mutations also suppressed the grr1Δ allele (Fig. 5B, lower panel, compare lane 3 with lanes 6 and 9). The epistasis analysis indicates that the defect in SPS sensor signaling exhibited by grr1Δ cells is identical to that observed in cells carrying loss-of-function mutations in any of SPS sensor component genes (SSY1, PTR3, or SSY5).
The N terminus of Stp1p is modular and can be transferred to regulate the activity of an artificial transcription factor.
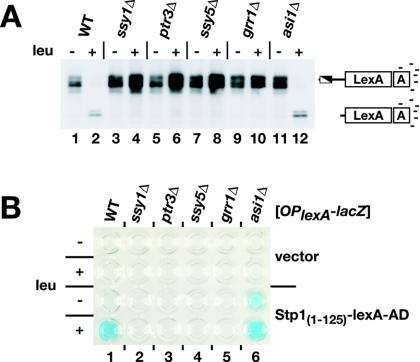
We examined the possibility that the first 125 residues of Stp1p [Stp1(1-125)], including the inhibitory motif (region I) and the cleavage sites (region II), could be transferred to confer amino acid-induced regulation of an artificial transcription factor. The N terminus of Stp1p was fused to a well-characterized synthetic transcription factor comprised of the bacterial LexA DNA-binding protein fused to the strong viral VP16 transcriptional AD. The LexA DNA-binding domain possesses an intrinsic NLS and is constitutively nuclear when expressed in yeast cells (34). A wild-type strain and strains carrying deletions of the SPS sensor components GRR1 and ASI1 were transformed with a plasmid expressing the Stp1(1-125)-LexA-AD fusion protein to assess the full spectrum of SPS sensor pathway regulation.
First, we examined whether the Stp1(1-125)-LexA-AD fusion protein was subject to amino acid-induced SPS sensor-dependent processing. Extracts were prepared from transformed strains and analyzed with anti-LexA antibodies by immunoblotting. In cells grown in SD medium, in the absence of an inducing amino acid, bands corresponding to the full-length Stp1(1-125)-LexA-AD fusion protein were readily detected (Fig. 6A, lane 1). When cells were grown in the presence of leucine, processing was observed in the wild-type and asi1Δ mutant cells (Fig. 6A, lanes 2 and 12). In these cells, the full-length Stp1(1-125)-LexA-AD fusion protein was detected only weakly, and more intense, faster-migrating bands were observed. No processing was observed in leucine-induced ssy1Δ, ptr3Δ, ssy5Δ, or grr1Δ mutant cells (Fig. 6A, lanes 3 to 10).
FIG. 6.
The N-terminal regulatory domain of Stp1p is transferable and confers full SPS sensor control of an artificial transcription factor. (A) Immunoblot analysis of protein extracts from wild-type (WT) (PLY126), ssy1Δ (HKY20), ptr3Δ (HKY31), ssy5Δ (HKY77), grr1Δ (CAY103), and asi1Δ (CAY107) strains transformed with pCA161 [Stp1(1-125)-LexA-AD] and grown in SD medium with or without leucine (leu). Extracts were resolved by SDS-PAGE and immunoblotted with anti-LexA antibodies. (B) Strains from panel A carrying pSH18-34 (OplexA-lacZ) and either pCA160 (vector) or pCA161 [Stp1(1-125)-LexA-AD] were grown in SD medium with or without leucine (leu). The levels of X-Gal staining resulting from the expression of lexA operator-promoted β-galactosidase in permeabilized cells were assessed.
Next, the ability of the Stp1(1-125)-LexA-AD fusion protein to activate a lexA operator-controlled lacZ reporter gene (OplexA-lacZ) was assessed. A plasmid-borne OplexA-lacZ reporter gene was introduced into strains carrying a vector without an insert or a plasmid expressing the Stp1(1-125)-LexA-AD fusion protein. Leucine-induced promoter activation could be detected readily in wild-type cells transformed with Stp1(1-125)-LexA-AD; a strong blue X-Gal precipitate formed when cells were grown in leucine-containing SD medium (Fig. 6B, lane 1). Derepression of the promoter was dependent on SPS sensor signaling; no blue precipitate was detected when Stp1(1-125)-LexA-AD was expressed in ssy1Δ, ptr3Δ, ssy5Δ, or grr1Δ mutant cells (Fig. 6B, lanes 2 to 5). Importantly, the regulation of Stp1(1-125)-LexA-AD derepressed the reporter even in the absence of amino acids in an asi1Δ strain (Fig. 6B, lane 6). These results indicate that Stp1(1-125) functions as a modular domain that can be transferred to an artificial transcription factor to faithfully confer full amino acid-induced SPS sensor-dependent regulation. Thus, the N-terminal domain is sufficient for both nuclear exclusion and protease recognition.
DISCUSSION
Prior to this report, the only known mechanism directly regulating the activity of Stp1p/Stp2p was the amino acid-induced SPS sensor-dependent endoproteolytic removal of their N-terminal domains (1). By comparing the N-terminal sequences of Stp1p/Stp2p from senso stricto strains of Saccharomyces, we identified two conserved sequence motifs, designated region I and region II (Fig. 1B). The functions of these motifs were assessed by using targeted alanine substitutions in Stp1p. A mutant protein (Stp1-102p) with a single alanine substitution at the start of homology region II (F66A) was not processed in response to amino acid induction (Fig. 2A). stp1-102 did not complement an stp1Δ stp2Δ double mutant, demonstrating that proteolytic removal of the inhibitory domain is strictly required for Stp1p/Stp2p activity. We could exclude the possibility that the lack of transcriptional induction by Stp1-102p was due to a folding defect or a general loss of function, since second site-suppressing mutations placed in cis (STP1-133) or in trans (asi1Δ) enabled the unprocessed mutant form of Stp1p to enter the nucleus and transactivate gene expression (Fig. 2C). The dominant nature and constitutive activity of the STP1-133 allele, which carries mutations within region I, suggest that this motif normally facilitates the cytoplasmic retention of unprocessed Stp1p. The observation that Stp1-133p is proteolytically processed (Fig. 2C), despite the presence of a nonfunctional inhibitory domain, indicates that the N-terminal regulatory domain of Stp1p is modular.
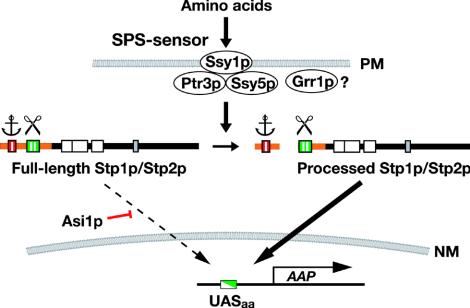
These results confirm that the processing of Stp1p/Stp2p is indeed the major regulatory event that transduces SPS sensor-initiated signals, generated at the plasma membrane, to the nucleus and to UASaa-containing promoters. Our analysis revealed that specific and functionally separable motifs within the N-terminal domain provide the molecular basis for the latent nuclear exclusion and amino acid-induced processing of Stp1p/Stp2p. Our current model for Stp1p/Stp2p regulation is presented in Fig. 7. Newly translated Stp1p/Stp2p is excluded from the nucleus by two parallel activities. The primary mechanism is linked to the inhibitory motif designated region I, which appears to anchor the unprocessed full-length forms to an as-yet-undefined cytoplasmic determinant. The secondary mechanism is dependent on the action of Asi1p.
FIG. 7.
Model of the amino acid-induced SPS-sensing pathway. The latent nature of Stp1p/Stp2p is determined by conserved sequence motifs region I (red) and region II (green) present in their N-terminal domains (orange). Region I appears to function as a cytoplasmic retention signal (anchor) that prevents the unprocessed full-length forms from efficiently entering the nucleus. Region II is required for amino acid-induced endoproteolytic processing (scissor). In the absence of Asi1p, full-length Stp1p/Stp2p can enter the nucleus at rates sufficient to partially derepress amino acid permease (AAP) gene expression. Extracellular amino acids induce the proteolytic processing of Stp1p/Stp2p in an SPS sensor- and Grr1p-dependent manner. The precise function of Grr1p remains to be defined (?); Grr1p either functions concomitantly with the previously characterized SPS sensor components (Ssy1p-Ptr3p-Ssy5p) or is required for SPS sensor assembly. The processed shorter forms of Stp1p and Stp2p containing DNA-binding domains (white boxes) accumulate in the nucleus, where they bind UAS present within the promoters of amino acid permease genes (UASaa). PM, plasma membrane; NM, nuclear membrane.
Forsberg et al. previously showed that recessive mutations in ASI1 constitutively derepress the transcription of SPS sensor-regulated genes in the absence of a functional SPS sensor (16). Remarkably, the lack of Asi1p function enables full-length Stp1-102p to enter the nucleus, indicating that the N-terminal regulatory domains in the native forms of Stp1p/Stp2p do not impair promoter binding or transactivation potential. These findings indicate that Asi1p normally functions to negatively regulate the transactivation potential of unprocessed Stp1p/Stp2p. Asi1p is a polytopic membrane protein with five membrane-spanning segments and a RING-like domain at its extreme C terminus (A. Zargari, C. Andréasson, and P. O. Ljungdahl, unpublished data). We anticipate that knowledge regarding the precise intracellular localization of Asi1p will dramatically influence the basis of the understanding of how this novel protein functions to modulate the activity of Stp1p/Stp2p.
The processing defect exhibited by grr1 mutant strains indicates that GRR1 encodes a component that functions in signaling events leading to Stp1p/Stp2p processing. This assignment is supported by our analysis of epistasis relationships. The constitutive activating form of Stp1p, encoded by the dominant STP1Δ131 allele, and mutations in ASI1 were found to efficiently suppress grr1Δ mutant phenotypes. However, the actual mechanistic role of Grr1p in the SPS sensor signal transduction pathway is difficult to predict based on these newly established genetic relationships. We are currently considering two possibilities; Grr1p either is directly involved in the signal transduction events within the SPS sensor or is a factor required for the activity of one of the SPS sensor components. The latter possibility defines a role for Grr1p without direct involvement in a dynamic signaling process. Based on the previously characterized functions of Grr1p in glucose sensing and cell cycle control (6, 15, 31, 32, 36), it is compelling to imagine a direct role for ubiquitin in the SPS sensor pathway, as previously suggested (7).
The modular nature of the N-terminal region of Stp1p motivated us to examine whether it could be transferred to the well characterized synthetic transactivator LexA-AD. The presence of the N terminus of Stp1p converted this constitutively nucleus-localized factor into a latent fully SPS sensor-regulated factor. This conclusion is based on several observations. First, immunoblot analysis demonstrated that Stp1(1-125)-LexA-AD is endoproteolytically processed in a manner similar to that of full-length Stp1p/Stp2p (Fig. 6A). Furthermore, lexA operator-driven promoter activation was shown to depend on amino acid induction and a functional SPS sensor (Fig. 6B). The OPlexA-lacZ reporter gene was fully repressed in strains carrying mutations in the upstream components of the SPS sensor pathway (ssy1Δ, ptr3Δ, ssy5Δ, and grr1Δ). Finally, constitutive derepression of the OPlexA-lacZ reporter construct was observed in asi1Δ cells. These data indicate that Stp1(1-125) is able to override the intrinsic NLS within LexA and to confer full SPS sensor pathway control.
The ability of the N-terminal domain of Stp1p to confer the entire known spectrum of SPS sensor pathway control to an unrelated protein supports the notion that all of the regulatory circuits converge on the N-terminal domain. This finding limits the number of feasible mechanisms that may regulate the latency of Stp1p/Stp2p. For example, it is unlikely that the N terminus masks or blocks an intrinsic NLS within Stp1p/Stp2p. Instead, the autonomous function of the N-terminal domain suggests that the inhibitory motif (region I) interacts with a protein that functions as a nuclear exclusion determinant, e.g., a cytoplasmic protein or a component of the nuclear export machinery. If this model proves correct, then the regulatory mechanism exhibits striking similarity with NF-κB/Rel signaling; IF-κB sequesters NF-κB in the cytoplasm and prevents nuclear translocation by binding the actin cytoskeleton via ankyrin repeats (9, 20).
Finally, it has not escaped our attention that the components described here comprise the basic building blocks of a highly specific system for the regulated expression of genes in yeast cells. The exploitation of such a system has some obvious advantages over existing expression systems. Presumably, the activities of other transcription factors (either heterologous, homologous, or completely artificial) can be placed under the control of the SPS sensor by appending the N-terminal domain of Stp1p. Consequently, amino acid-induced gene expression can be restricted to a single synthetic promoter, as we have accomplished with OPlexA-lacZ. Additionally, the transferability of the N-terminal inhibitory domain of Stp1p suggests the possibility of specifically restricting the access of individual proteins to the nucleus in a manner that is readily reversible. This scenario may be useful for testing the physiologic consequences of redirecting and sequestering presumed nuclear factors in the cytoplasm.
Acknowledgments
We thank the other members of the laboratory of P. O. Ljungdahl for constructive comments throughout the course of this work and Morten Kielland-Brandt for the kind gift of the plasmid carrying the dominant activated SSY1-102 allele.
This research was supported by the Ludwig Institute for Cancer Research.
REFERENCES
- 1.Andréasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson, C., E. P. A. Neve, and P. O. Ljungdahl. 2004. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 21:193-199. [DOI] [PubMed] [Google Scholar]
- 3.Antebi, A., and G. R. Fink. 1992. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3:633-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aza-Blanc, P., and T. B. Kornberg. 1999. Ci: a complex transducer of the hedgehog signal. Trends Genet. 15:458-462. [DOI] [PubMed] [Google Scholar]
- 5.Baron, M. 2003. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 14:113-119. [DOI] [PubMed] [Google Scholar]
- 6.Barral, Y., and C. Mann. 1995. G1 cyclin degradation and cell differentiation in Saccharomyces cerevisiae. C. R. Acad. Sci. Ser. III 318:43-50. [PubMed] [Google Scholar]
- 7.Bernard, F., and B. André. 2001. Ubiquitin and the SCF(Grr1) ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496:81-85. [DOI] [PubMed] [Google Scholar]
- 8.Botling, J., D. S. Castro, F. Oberg, K. Nilsson, and T. Perlmann. 1997. Retinoic acid receptor/retinoid X receptor heterodimers can be activated through both subunits providing a basis for synergistic transactivation and cellular differentiation. J. Biol. Chem. 272:9443-9449. [DOI] [PubMed] [Google Scholar]
- 9.Brivanlou, A. H., and J. E. Darnell, Jr. 2002. Signal transduction and the control of gene expression. Science 295:813-818. [DOI] [PubMed] [Google Scholar]
- 10.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 11.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Sping Harbor, N.Y.
- 12.de Boer, M., P. S. Nielsen, J. P. Bebelman, H. Heerikhuizen, H. A. Andersen, and R. J. Planta. 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28:974-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didion, T., B. Regenberg, M. U. Jørgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643-650. [DOI] [PubMed] [Google Scholar]
- 14.Flick, J. S., and M. Johnston. 1991. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol. Cell. Biol. 11:5101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flick, K. M., N. Spielewoy, T. I. Kalashnikova, M. Guaderrama, Q. Zhu, H. C. Chang, and C. Wittenberg. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14:3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg, H., M. Hammar, C. Andréasson, A. Moliner, and P. O. Ljungdahl. 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158:973-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91-109. [DOI] [PubMed] [Google Scholar]
- 19.Gaber, R. F., K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell 2:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilmore, T. D. 1999. The Rel/NF-kappaB signal transduction pathway: introduction. Oncogene 18:6842-6844. [DOI] [PubMed] [Google Scholar]
- 21.Gilstring, C. F., M. Melin-Larsson, and P. O. Ljungdahl. 1999. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into endoplasmic reticulum-derived transport vesicles. Mol. Biol. Cell 10:3549-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 23.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 25.Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jøorgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 27.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 28.Kippert, F. 1995. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128:201-206. [DOI] [PubMed] [Google Scholar]
- 29.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 31.Li, F. N., and M. Johnston. 1997. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16:5629-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosley, A. L., J. Lakshmanan, B. K. Aryal, and S. Özcan. 2003. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278:10322-10327. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen, R. J. Planta, M. C. Kielland-Brandt, and H. A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613-622. [DOI] [PubMed] [Google Scholar]
- 34.Rhee, Y., F. Gurel, Y. Gafni, C. Dingwall, and V. Citovsky. 2000. A genetic system for detection of protein nuclear import and export. Nat. Biotechnol. 18:433-437. [DOI] [PubMed] [Google Scholar]
- 35.Silve, S., C. Volland, C. Garnier, R. Jund, M. R. Chevallier, and R. Haguenauer-Tsapis. 1991. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell. Biol. 11:1114-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53-62. [DOI] [PubMed] [Google Scholar]