Abstract
The orphan receptor SHP interacts with many nuclear receptors and inhibits their transcriptional activities. SHP is central to feedback repression of cholesterol 7α hydroxylase gene (CYP7A1) expression by bile acids, which is critical for maintaining cholesterol homeostasis. Using CYP7A1 as a model system, we studied the molecular mechanisms of SHP repression at the level of native chromatin. Chromatin immunoprecipitation studies showed that mSin3A and a Swi/Snf complex containing Brm as a central ATPase were recruited to the promoter. This recruitment was associated with chromatin remodeling after bile acid treatment that was blunted by inhibition of the endogenous Swi/Snf function by dominant-negative ATPase mutants. Biochemical studies indicated that SHP was associated with the mSin3A-Swi/Snf complex by direct interaction with Brm and mSin3A through its repression domain. Expression of Brm, but not an ATPase mutant, inhibited CYP7A1 promoter activity and further enhanced SHP-mediated repression. Bile acid-induced recruitment of mSin3A/Brm, chromatin remodeling, and concomitant repression of endogenous CYP7A1 expression were impaired when SHP expression was inhibited by SHP small interfering RNA. Our results suggest that SHP mediates recruitment of mSin3A-Swi/Snf to the CYP7A1 promoter, resulting in chromatin remodeling and gene repression, which may also be a mechanism for the repression by SHP of genes activated by many nuclear receptors. Our study establishes the first link between a Swi/Snf complex and regulation of cholesterol metabolism.
The orphan nuclear receptor small heterodimer partner (SHP) protein is an atypical nuclear receptor that lacks a conventional DNA binding domain but contains a putative ligand binding domain (45). SHP has been reported to interact with a number of nuclear receptors, including both ligand-regulated receptors, such as ER (estrogen receptor), GR, TR, AR, RAR, and RXR (retinoid X receptor), and orphan receptors, such as LRH-1 (liver receptor homologue 1), HNF-4 (hepatic nuclear factor 4), ERR, CAR, LXR, and PPAR, and to inhibit their transcriptional activities (1-3, 19, 29, 30, 43, 45). SHP, therefore, has been implicated in diverse biological activities, including cholesterol/bile acid and glucose/energy metabolic pathways.
SHP has been reported to play a key role in the negative feedback regulation of cholesterol 7α hydroxylase gene (CYP7A1) expression in the liver (15, 32). This hepatic enzyme catalyzes the first and rate-limiting step of the neutral pathway for the conversion of cholesterol into bile acids and thus plays a crucial role in enterohepatic cholesterol-bile acid homeostasis (42). Previous studies showed that the bile acid-activated farnesoid X receptor (FXR) binds as a heterodimer with RXR to the promoter of the SHP gene and increases transcription. The bile acid-induced SHP then interacts with LRH-1 and/or HNF-4, bound to the bile acid response element (BARE) in the CYP7A1 promoter, which results in transcriptional repression (7, 15, 29, 32). Although the key regulatory factors, FXR, LRH-1, and SHP, have been identified, the precise molecular events that occur at the level of CYP7A1 promoter chromatin and mediate gene repression by bile acids and bile acid-induced SHP are still unknown.
Chemical and structural changes in chromatin play fundamental roles in eukaryotic gene regulation (21, 26, 51). The primary structural units of chromatin are nucleosomes, which suppress transcription by imposing a barrier to the access of transcription factors and basal transcriptional machinery to DNA. There are two classes of chromatin-modifying complexes that alter chromatin structure. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) (21, 27, 51) covalently modify histones by adding or removing acetyl groups on lysine residues in core histones. Transcriptionally active or suppressed chromatin is associated with histone hyperacetylation or hypoacetylation, respectively. The second class of chromatin-modifying complexes includes ATP-dependent chromatin remodeling complexes, such as Swi/Snf. Using the energy from ATP hydrolysis, these complexes weaken DNA-histone contacts in the nucleosome core, which may disrupt or alter nucleosome conformation (36, 51).
Swi/Snf chromatin remodeling complexes have been implicated in diverse cellular activities, including cell proliferation, reproduction, development, and differentiation (13, 14, 41). The Swi/Snf complexes contain either of two ATPases, Brm or Brg-1, and variable subunits of Brm- or Brg-1-associated factors (BAFs). In Brm-null mice, Brg-1 levels are increased and functionally compensate for Brm (41). However, these mice have defects in the regulation of cellular proliferation. A recent study also demonstrated that Brm and Brg-1 preferentially interact with different types of transcription factors, and they appear to target different promoters during cellular proliferation and differentiation (20). While remodeling complexes have been generally implicated in gene activation, recent studies suggest that these complexes are also involved in gene repression. The corepressors HDAC1 and HDAC2 were reported to be integral components of the ATP-dependent remodeling complex, NURD (53). The Swi/Snf-Brg-1 complex was shown to form a complex with HDAC and the retinoblastoma (Rb) protein that inhibits transcription of cyclin genes (55).
Using the CYP7A1 gene as a model system, we examined the mechanism of SHP repression at the level of native chromatin. We analyzed the effects of bile acid and bile acid-induced SHP on chromatin structure at the endogenous human CYP7A1 promoter in HepG2 cells and on the association with the native promoter of protein coregulator complexes involved in chromatin remodeling and histone acetylation. Here, we report the unexpected finding that bile acid-induced SHP recruits to the endogenous human CYP7A1 promoter a Swi/Snf-Brm chromatin remodeling complex that also contains an mSin3A/HDAC-1 corepressor complex, which is associated with chromatin remodeling and gene repression in vivo.
MATERIALS AND METHODS
Cell culture.
Human hepatoma cells (HepG2; ATCC HB8065) were grown in Dulbecco's modified Eagle's medium-F12 (1:1) phenol red-free medium. Human adrenal carcinoma cells (SW13; ATCC-CCL 105) were maintained in Dulbecco's modified Eagle's medium. The media were supplemented with 100 U of penicillin G-streptomycin sulfate/ml and 10% heat-inactivated charcoal-treated fetal bovine serum. The HepG2 cells were treated with 25 to 75 μM chenodeoxycholic acid (CDCA) (Sigma) for 8 to 16 h in serum-free medium. Frozen HepG2 cells were thawed every 6 to 8 weeks, and reverse transcription (RT)-PCR was regularly done to ensure that CYP7A1 gene expression was properly regulated by CDCA.
RT-PCR analysis.
Total RNA was isolated using Trizol, cDNA was synthesized, and semiquantitative PCR was carried out. The sequences for the primers are as follows: forward primer, 5′-CTAAGGAGGATTTCACTTGC-3′, and reverse primer, 5′-ACTGGTCCAAAGGTGGACAT-3′ for CYP7A1; forward primer, 5′-CAAGAAGATTCTGCTGGAGG-3′, and reverse primer, 5′-GGATGTCAACATCTCCAATG-3′ for SHP; and forward primer, 5′-GTCATCACCATTGGCAATGAG-3′, and reverse primer, 5′-CGTCATACTCCTGCTTGCTG-3′ for β-actin. Amplifications of 20 to 22, 25 to 27, and 29 to 32 cycles were performed for β-actin, SHP, and CYP7A1, respectively.
Low-resolution nucleosome mapping.
Nuclei were isolated in 300 mM sucrose, 60 mM KCl, 60 mM Tris-HCl, pH 8.0, 2 mM EDTA, 0.5% NP-40, 0.15 mM spermine, 0.5 mM spermidine, 1 mM dithiothreitol (DTT), and protease inhibitors. Either cells that were permeabilized by treatment with 0.05% lysolecithin or isolated nuclei were incubated with 15 to 75 U of micrococcal nuclease (MNase)/ml for 3 min at 37°C in digestion buffer (150 mM sucrose, 50 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 3 mM CaCl2, 0.15 mM spermine, 0.5 mM spermidine, protease inhibitors, and 1 mM DTT), and the stop solution (20 mM Tris-HCl, pH 8.0, 20 mM NaCl, 20 mM EDTA, 1% sodium dodecyl sulfate [SDS], 300 μg of proteinase K/ml) was added. The DNA was purified and subjected to genomic Southern analysis as described previously (25). Briefly, DNA was cleaved by HindIII, separated by electrophoresis, transferred to a membrane, and subjected to hybridization at 42°C overnight with the γ-32P-end-labeled CYP7A1 promoter-specific primer (−375 to −335) in the presence of hybridization solution (7% SDS, 0.5 M sodium phosphate, pH 7.2, 1 mM EDTA).
LMPCR-mediated nucleosome mapping.
HepG2 cells were treated with 1% formaldehyde at 25°C for 10 min. Nuclei were isolated and partially digested with MNase as described above. After reverse cross-linking at 65°C for 4 h, mono- and dinucleosomal DNA was isolated by gel purification and subjected to ligation-mediated PCR (LMPCR) as described previously (23-25). Since MNase digestion generates double-strand blunt ends without 5′ phosphates, phosphorylation of the 5′ ends by polynucleotide kinase is required before PCR. Primer set a includes SP2 (−267 to −241) (5′-CCAAACTCTTAATATTAGCTGTTGTCC-3′) and SP3 (−249 to −218) (5′-GCTGTTGTCCCCAGGTCCGAATGTTAAGTCAAC-3′). Primer set c includes NSP2 (−205 to −183) (5′-GACCTTCAACTTATCAAGTATTG-3′) and NSP3 (−195 to −168) (5′-TATCAAGTATTGCAGGTCTCTGATTGC-3′). Primer set b includes AP2 (+72 to +48) (5′-GGTCATCATTTTGCAAATCTAGGCC-3′) and AP3 (+60 to +32) (5′-GCAAATCTAGGCCAAAATCTCTGAGGA-3′). Primer set d includes AP2 (+72 to +48) (5′-GGTCATCATTTTGCAAATCTAGGCC-3′) and NAP3 (+42 to +11) (5′-CTCTGAGGAAGAAAATCTCTGATTAGAAAGGG-3′). Sequencing ladders from pcDNA3.1 served as size markers. The primer sequences for LMPCR 1 and 2 are 5′-GGTGACCCGGGAGATCTGAATTC-3′ and 5′-GAATTCAGATC-3′, respectively.
In vivo restriction endonuclease accessibility assays.
DNA in intact nuclei was partially digested with a restriction enzyme (10 to 25 U/100 μl) in 1× buffer (New England Biotechnology) at 37°C for 10 to 30 min (9, 14, 49). The DNA was isolated and digested to completion with AvaII (primer A set) or SphI (primer B set) in vitro. The DNA was purified and subjected to LMPCR using primer sets SP1 (−311 to −292) (5′-GCTATGCCCATCTTAAACAG-3′)-SP2-SP3 and AP1 (+87 to +66) (5′-CCAAATCAAAGATGTGGTCATC-3′)-AP2-AP3. The sequences for SP2, SP3, AP2, and AP3 are described above.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were carried out as described previously with minor modifications (4, 47, 49). HepG2 cells were incubated with 1% formaldehyde at 25°C for 10 min. Nuclei were isolated and sonicated to reduce the DNA length to 0.3 to 1.5 kb. Chromatin was precleared in the presence of 20 μl of normal serum, 2 μg of salmon sperm DNA, and 80 μl of a 25% protein A-agarose slurry. Precleared chromatin samples were subjected to immunoprecipitation at 4°C overnight in the presence of 3 to 10 μg of antisera for Brm (sc-4650), Brg-1 (sc-10768), BAF155 or BAF57 (kindly provided by W. Wang), BAF170 (sc-9744), mSin3A (sc-994), HDAC-1 (Upstate Biotechnology), p300 (sc-585), CBP (sc-369), SRC-1 (sc-6098), and Gal4DBD (sc-510), as well as AcH3 (antiserum against amino acids 1 to 21; Upstate Biotechnology), AcH4 (antiserum against amino acids 2 to 19; Upstate Biotechnology), or immunoglobulin G (IgG). The antisera with “sc” designations were obtained from Santa Cruz Biotechnology. After the complex was collected by incubation in 60 μl of a 25% protein A-Sepharose slurry and centrifugation, the beads were washed five times as described previously (4) and the chromatin immune complex was eluted. After the cross-links were reversed, the DNA was purified and used as a template in PCR. Semiquantitative PCR was performed using primer sets specific for the CYP7A1 promoter (forward, 5′-GATATCTATGCCCATCTTAAACAGG-3′; reverse, 5′-GAATTCGGGAAGGATGCCACTG-3′) and a control region from +860 to +1160 in CYP7A1 (forward, 5′-GAACCACCTCTAGAGAATG-3′; reverse, 5′-GAATCTCCACATAAGGATAAC-3′).
Plasmid construction.
SUPER vectors encoding SHP small interfering RNA (siRNA) were constructed by ligation of a 64-mer double-stranded oligonucleotide containing +76 to +94 of the human SHP cDNA sequence into the pSUPER vector (Oligoengine) digested with BglII and HindIII. Positive clones were confirmed by sequencing. The plasmids pcDNA3-Brm and pcDNA3-Brm mutant were constructed by EcoRI digestion of pBSK Brm constructs (provided by S. Sif and A. Imbalzano), followed by ligation into an EcoRI fragment of pcDNA3 (Invitrogen). The plasmids 4ERE-tk-luc (35), CMV-hERα (35), G4DBD-SHP (29), and human 371-CYP7A1-luc (6) have been described. Transfection was carried out using either electroporation for ChIP, coimmunoprecipitation (CoIP), and remodeling assays (Bio-Rad electroporator at 600 μF and 300 V) or Lipofectamine 2000 for transfection assays. β-Galactosidase (β-Gal) assays were performed to normalize for transfection efficiency.
GST pull-down assay.
Glutathione S-transferase (GST), GST-SHP, or GST-SHP deletion mutants were expressed in Escherichia coli BL21/DE3/RIL and immobilized on glutathione-Sepharose as described previously (1, 35). 35S-labeled Brm or mSin3A was synthesized by in vitro transcription and translation (Promega). One microgram of GST fusion proteins was incubated with 35S-labeled proteins in incubation buffer (20 mM K+-HEPES, pH 7.8, 0.1 mM EDTA, 10% glycerol, 200 mM KCl, 0.25% NP-40, protease inhibitors, 1 mM DTT) at 23°C for 40 min. Swi/Snf complexes were purified from HeLa cells stably producing Flag-Ini 1 (provided by R. Kingston and S. Sif) by preparation of nuclear extracts, followed by M2 agarose purification as described previously (52). One microgram of GST-SHP or a GST-SHP mutant was incubated with the purified Swi/Snf complex in incubation buffer at 4°C for 2 h. After being washed five times with incubation buffer, proteins associated with GST-SHP or GST-SHP mutants were eluted and analyzed by SDS-polyacrylamide gel electrophoresis. Interaction with Swi/Snf was detected by Western blotting using antiserum against Flag peptide, BAF57, or Brm. To detect the binding of endogenous proteins in HepG2 nuclear extracts to SHP, 5 μg of GST-SHP or a GST-SHP mutant was incubated with 500 μg of HepG2 nuclear extracts in incubation buffer at 4°C for 2 h. After being washed five times with incubation buffer (supplemented to 350 mM KCl and 0.5% NP-40), the protein complex was eluted and detected by Western blotting.
CoIP assay.
HepG2 cells were pelleted by centrifugation and resuspended in lysis buffer (20 mM K+-HEPES, pH 8.0, 0.2 mM EDTA, 5% glycerol, 250 mM NaCl, 1%NP-40, 0.25% sodium deoxycholate, 1 mM NaF, 1 mM sodium orthovanadate, 1 mM DTT, and protease inhibitors). After incubation on ice for 10 min, followed by sonication and centrifugation, the cell extracts were precleared by incubation with 60 μl of a 25% protein A-Sepharose slurry for 30 min at 4°C and incubated with 3 to 5 μg of either IgG or appropriate antisera for 4 h to overnight at 4°C. Immune complexes were collected by incubation with 30 μl of a 25% protein A-Sepharose slurry for 1 h. Immunoprecipitates were washed five times with lysis buffer and subjected to Western blotting.
RESULTS
Bile acid-induced in vivo chromatin remodeling at the human CYP7A1 promoter.
Previous reports showed that bile acid-activated FXR binds to the SHP promoter as a heterodimer with RXR and induces expression of SHP (15, 32). SHP interacts with a hepatic transcription factor, LRH-1, on the BARE at the CYP7A1 promoter and suppresses transcription of the CYP7A1 gene. Thus, we measured the endogenous levels of CYP7A1 and SHP mRNAs in HepG2 cells treated with a primary bile acid, CDCA, or dimethyl sulfoxide (DMSO) vehicle, using semiquantitative RT-PCR. Consistent with previous studies (6, 8, 33), CYP7A1 expression was decreased and SHP expression was increased by CDCA (Fig. 1A). These results indicate that the HepG2 cell line is a suitable system to study the negative feedback regulation of human CYP7A1 expression by bile acids.
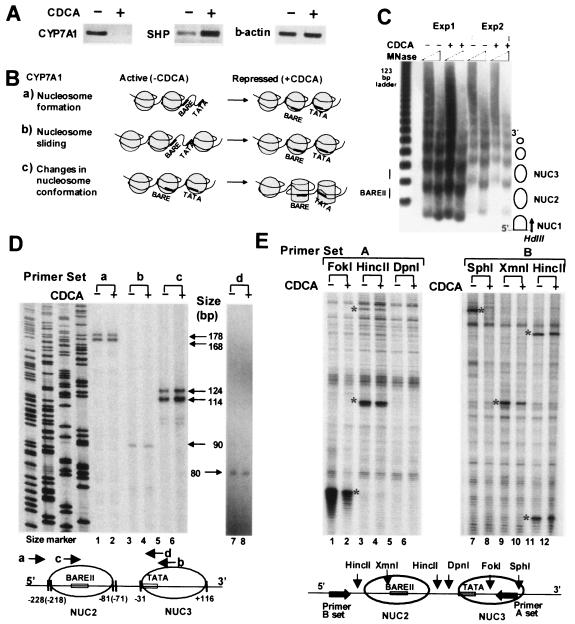
FIG. 1.
Bile acid-induced chromatin remodeling at the CYP7A1 promoter. (A) Effects of bile acid treatment on SHP and CYP7A1 expression. Total RNA was isolated from HepG2 cells and subjected to semiquantitative RT-PCR using primer sets specific for human CYP7A1, SHP, and β-actin. +, present; −, absent. (B) Three potential models of chromatin remodeling after CDCA treatment. (C) Low-resolution mapping of nucleosome positions. Permeabilized HepG2 cells (Exp1) or isolated nuclei (Exp2) were incubated with MNase, and purified DNA was cleaved with HindIII and subjected to indirect end-labeling genomic Southern analysis using the probe indicated by the arrow. Nucleosome positions are indicated as circles. (D) High-resolution mapping of nucleosome positions. HepG2 cells were incubated with formaldehyde to cross-link proteins and nucleic acids. Nuclei were isolated and partially digested with MNase. After the cross-links were reversed, nucleosomal DNA was isolated and subjected to LMPCR using the primer sets a, b, c, and d. Sequencing ladders served as size markers. The lengths of LMPCR products are indicated. A schematic diagram of nucleosome positioning at the promoter is shown below. (E) Accessibility of CYP7A1 chromatin to restriction endonucleases. Isolated nuclei were partially digested with restriction enzymes. DNA was purified and subjected to LMPCR using primer set A or B. The asterisks indicate the restriction enzyme cleavages detected by LMPCR. Background bands present in all lanes result from nonspecific cleavages by endogenous nucleases. Reproducible results were obtained in two to five independent studies.
Chromatin remodeling is defined as any detectable change in chromatin or mononucleosomal structure (21). Three potential remodeling events, nucleosome formation, sliding, and conformational change, that could occur at the suppressed CYP7A1 promoter after bile acid treatment are illustrated in Fig. 1B. The activation of genes is often associated with disruption of a nucleosome in a regulatory region (34). Analogous to this, suppression of an active gene could result from nucleosome formation. In this case, the nucleosome would be disrupted in the active CYP7A1 promoter and CDCA treatment would result in nucleosome formation, which could inhibit binding of transcription factors to the BARE and TATA regions (Fig. 1B, a). Therefore, we performed low-resolution nucleosome mapping by indirect end-labeling genomic Southern analyses using MNase that preferentially cleaves in the linker region of nucleosomes. A regular ladder of DNA fragments was present regardless of bile acid treatment (Fig. 1C), suggesting that the CYP7A1 promoter is constantly occupied by a regular array of nucleosomes, and thus, gene repression by CDCA cannot result from nucleosome formation.
A second possibility is that a nucleosome may be repositioned to block the binding sites of the transcription factors in the BARE and TATA regions in response to bile acid treatment (Fig. 1B, b). Such nucleosome sliding has been shown for transcriptional activation of the mouse mammary tumor virus promoter by GR or the β interferon gene in response to viral infection (31, 40). To examine whether bile acid treatment resulted in nucleosome sliding, we performed LMPCR high-resolution mapping. Two PCR products were detected for nucleosome 2 using primer set a or c, suggesting the existence of two populations of nucleosomes (Fig. 1D, lanes 1, 2, 5, and 6). Consistent results were observed with two additional primer sets (data not shown). The significance of the two populations is not known, but importantly, bile acid treatment did not result in changes in the intensities or positions of these bands. Using primer set b, a single PCR fragment was observed at the 5′ end of nucleosome 3 (Fig. 1D, lanes 3 and 4). The 5′ end mapped at −31, indicating that the TATA box is located at the 5′ edge of the core of nucleosome 3. An additional primer set (primer set d) confirmed the position of the nucleosome containing the TATA box (Fig. 1D, lanes 7 and 8). Again, no difference was observed between the CDCA- and vehicle-treated cells, indicating that nucleosomal sliding did not occur at the CYP7A1 promoter after CDCA treatment.
Instead of gross chromatin structural alterations, such as nucleosome formation or sliding, changes in nucleosome conformation could occur after bile acid treatment (Fig. 1B, c). Thus, we carried out restriction endonuclease accessibility assays which have been successfully utilized to monitor in vivo chromatin remodeling (14, 49). Isolated nuclei were partially digested with restriction enzymes that cleave either in the nucleosomal core or in the linker. The cleavage sites were then detected by LMPCR. Interestingly, FokI and SphI cleavages within the nucleosome 3 core were dramatically decreased by CDCA (Fig. 1E, lanes 1, 2, 7, and 8). XmnI cleavage in the nucleosome 2 core was modestly decreased by CDCA (lanes 9 and 10). In contrast, cleavage by HincII, which cleaves within the linkers, was not altered by CDCA (lanes 3, 4, 11, and 12). DpnI cleavage was not observed (lanes 5 and 6), indicating that the DpnI sites in this region are not methylated, which is required for cleavage. These in vivo chromatin remodeling studies demonstrate that accessibility of the core nucleosomes to restriction enzymes is substantially reduced after bile acid treatment, implying that the CYP7A1 promoter is remodeled into a more closed configuration.
Bile acid treatment causes dissociation of coactivator HATs and recruitment of the corepressors mSin3A and HDAC-1, which are associated with histone deacetylation at the CYP7A1 promoter.
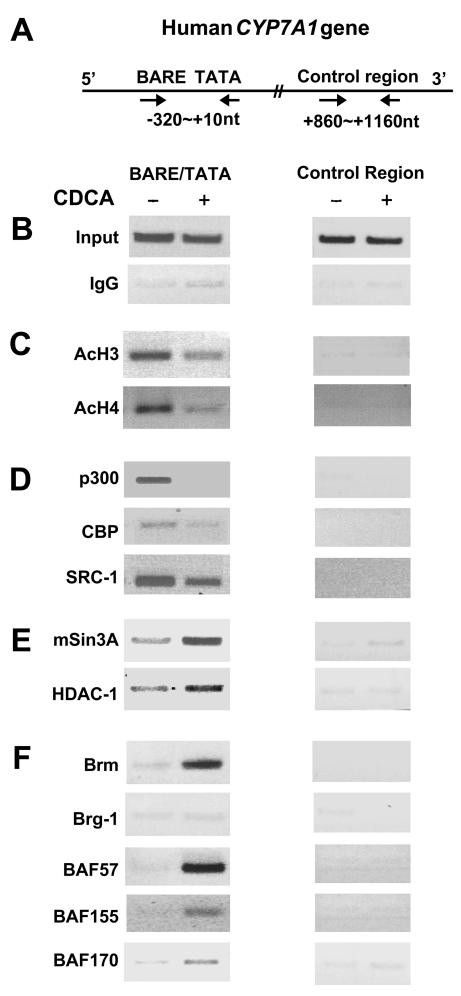
Since gene repression and closed chromatin conformation are often associated with decreased histone acetylation, we first determined whether bile acid treatment results in histone deacetylation at the promoter by using ChIP assays. The CYP7A1 promoter sequence precipitated by antiserum against either acetylated histone 3 (AcH3) or acetylated histone 4 (AcH4) was decreased in CDCA-treated samples (Fig. 2C). In contrast, little promoter DNA was present in precipitates with IgG (Fig. 2B). The control sequence from +860 to +1160 (Fig. 2A) was not enriched by antiserum against AcH3 or AcH4 (Fig. 2C). These results indicate that bile acid treatment results in decreased histone acetylation at the promoter. Decreased acetylation could result from dissociation of HATs and/or recruitment of HDACs. Thus, we examined the association of histone-modifying enzymes, such as HATs and HDAC, with the native CYP7A1 promoter. The CYP7A1 promoter sequence precipitated by antisera to the coactivator HATs, p300, CBP, and SRC-1, was reduced after bile acid treatment (Fig. 2D), indicating that bile acid treatment results in dissociation of coactivator HATs from the promoter. In contrast, the promoter sequences precipitated by antiserum against the corepressor mSin3A or HDAC-1 were increased in the CDCA-treated samples (Fig. 2E), suggesting that the mSin3A/HDAC-1 corepressor complex was recruited to the promoter. The dissociation of HATs and the recruitment of HDAC-1 are consistent with decreased histone acetylation at the CYP7A1 promoter after bile acid treatment.
FIG. 2.
Effects of bile acid treatment on association of HATs, corepressors, and Swi/Snf with the endogenous CYP7A1 promoter. (A) Schematic diagram of the CYP7A1 gene. (B to F) ChIP assays were performed as described in Materials and Methods. Chromatin DNA precipitated by the antibodies was analyzed by PCR using primer sets specific for the promoter or control regions (indicated by arrows in panel A). Consistent results from >3 ChIP assays in each case were obtained. +, present; −, absent.
Bile acid leads to recruitment of a Swi/Snf-Brm chromatin remodeling complex to the CYP7A1 promoter.
To determine whether the Swi/Snf remodeling complex is associated with the decreased accessibility at the CYP7A1 promoter chromatin to endonucleases after bile acid treatment, we carried out ChIP assays using antisera to components of the Swi/Snf complexes (Fig. 2F). Interestingly, the promoter sequence precipitated by Brm antiserum was increased in CDCA-treated samples. The amount of promoter sequence precipitated by BAF57, BAF155, or BAF170 was also increased after CDCA treatment. However, little promoter DNA was present in anti-Brg-1 precipitates. These results indicate that the Swi/Snf complex is recruited to the promoter after bile acid treatment and that this complex appears to contain Brm as a central ATPase. Since Swi/Snf has been implicated in transcriptional activation in most studies, our finding that the complex was recruited to the CYP7A1 promoter during transcriptional repression was somewhat surprising.
ATPase activity of Swi/Snf is required for bile acid-induced chromatin remodeling at the CYP7A1 promoter.
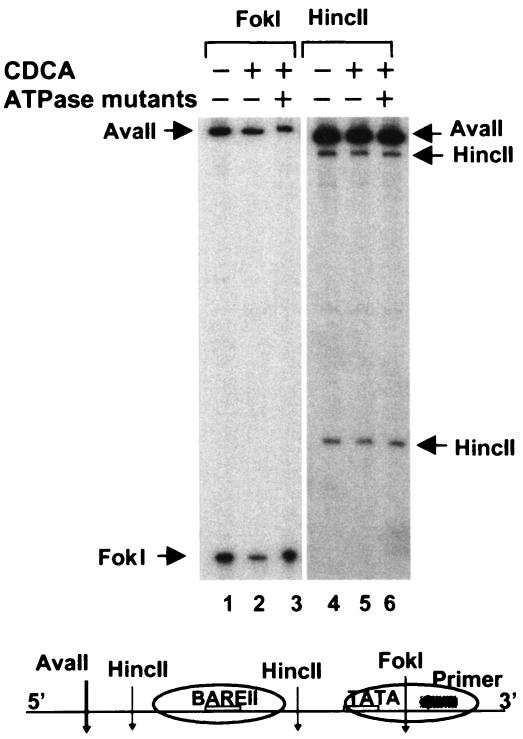
We wished to determine whether the decreased promoter accessibility at the endogenous CYP7A1 promoter after bile acid treatment required an active Swi/Snf complex. Therefore, we utilized Brm/Brg-1 ATPase mutants in which ATP hydrolysis was impaired by point mutations in the ATPase domains. These ATPase mutants act as dominant-negative mutants by forming nonfunctional Swi/Snf complexes in cells (9, 10). When the Brm gene was ablated in mice, Brg-1 expression was increased to compensate for the absence of Brm (41). In addition, since HepG2 cells contain high levels of both Brm and Brg-1 proteins (data not shown), plasmids for both ATPase mutants were transfected into cells, and the abilities of these mutants to block the bile acid-induced chromatin remodeling was evaluated in vivo. Bile acid treatment resulted in a decrease in cleavage by FokI at the core of nucleosome 3 (Fig. 3, lanes 1 and 2), consistent with previous results. However, decreased accessibility at the nucleosome core induced by CDCA was reversed by overexpression of the ATPase mutants (Fig. 3, lanes 1 to 3). In contrast, cleavages at HincII sites in the linker region were not affected by either CDCA treatment or expression of these mutants (lanes 4 to 6). These results suggest that inhibition of endogenous Swi/Snf function by overexpression of the ATPase mutants blocks chromatin remodeling at the promoter, suggesting that the recruited Swi/Snf complex mediates in vivo chromatin remodeling.
FIG. 3.
Bile acid-induced chromatin remodeling is reversed by overexpression of Swi/Snf ATPase mutants. HepG2 cells were transfected with mammalian expression plasmids for ATPase mutants of Brm/Brg-1 or empty vector by electroporation, and 36 h later the transfected cells were treated with CDCA or DMSO for 12 h and further subjected to restriction enzyme accessibility assays as described in Materials and Methods. Briefly, DNA in intact nuclei was partially digested with either FokI or HincII (10 to 25 U/100 μl) by incubation at 37°C for 15 min, and genomic DNA was purified and then digested to completion with AvaII in vitro. One microgram of purified genomic DNA was subjected to LMPCR analysis using the primer set A as indicated in the schematic diagram at the bottom. +, present; −, absent.
The corepressor mSin3A is associated with the Swi/Snf-Brm complex in HepG2 cells.
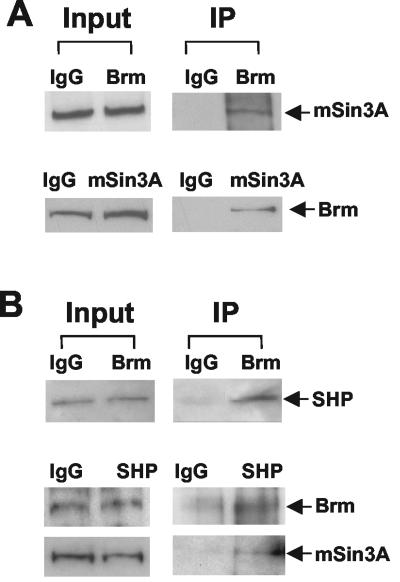
A recent biochemical study showed that the Swi/Snf-Brm and Swi/Snf-Brg-1 complexes in HeLa nuclear extracts copurified with components of the mSin3A corepressor complex (48). Thus, we examined whether mSin3A was associated with the Swi/Snf-Brm complex in HepG2 cells by using CoIP assays. Cell extracts were precipitated with either Brm or mSin3A antiserum, and the association of mSin3A or Brm, respectively, with the precipitates was detected by Western blotting. The mSin3A was detected in the anti-Brm immunoprecipitates, and Brm was present in the anti-mSin3A immunoprecipitates (Fig. 4A). We also detected BAF155 and BAF57 in mSin3A precipitates and HDAC-1 in the anti-Brm immunoprecipitates (data not shown). Our CoIP assays suggest that the mSin3A corepressor complex is associated with the endogenous Swi/Snf-Brm complex in HepG2 cells.
FIG. 4.
Association of mSin3A-Swi/Snf-Brm with SHP in HepG2 cells. (A) HepG2 cell extracts were subjected to immunoprecipitation (IP) using IgG or antibodies against Brm. Association of mSin3A with the anti-Brm precipitate was detected by Western blotting using mSin3A antiserum. Conversely, the cell extracts were incubated with either IgG or mSin3A antiserum, and association of Brm with the anti-mSin3A precipitate was detected by antiserum against Brm. Input, 5% of total cell lysates. (B) HepG2 cells were transfected with Gal4DBD-SHP by electroporation and subjected to CoIP. The association of SHP with Brm was detected by Western blotting using Gal4DBD antibody. Conversely, the cell extracts were immunoprecipitated with either IgG or Gal4DBD antibody. Association of Brm and mSin3A with SHP was detected by Western blotting.
SHP is associated with the endogenous mSin3A-Swi/Snf complex in HepG2 cells.
Next, we tested whether SHP could interact with the endogenous mSin3A-Swi/Snf-Brm complex in HepG2 cells. Due to the lack of a suitable SHP antibody, HepG2 cells were transfected with expression plasmids for Gal4DBD-SHP, so that SHP could be detected using Gal4DBD antiserum. The cell extracts were immunoprecipitated with either IgG or Brm antibody, and the association of SHP with the mSin3A-Swi/Snf complex was detected by Western blotting. SHP was present in anti-Brm precipitates (Fig. 4B). Conversely, the association of mSin3A and Brm with SHP was also confirmed (Fig. 4B). These results indicate that SHP is associated with the mSin3A-Swi/Snf complex in HepG2 cells.
SHP interacts with the mSin3A-Swi/Snf-Brm complex primarily through its C-terminal repression domain in vitro.
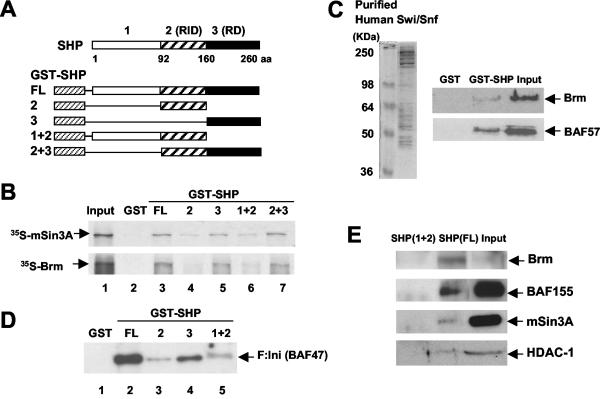
To analyze which domain of SHP directly interacts with the mSin3A-Swi/Snf complex, we performed in vitro GST pull-down assays (3). Both Brm and mSin3A interacted with full-length SHP or with SHP mutants (Fig. 5A) containing the C-terminal repression domain (Fig. 5B, lanes 3, 5, and 7). In contrast, Brm or mSin3A interacted weakly with the SHP mutants that lack this domain (Fig. 5B, lanes 4 and 6). These results indicate that SHP directly interacts with Brm or mSin3A primarily through its C-terminal intrinsic repression domain.
FIG. 5.
SHP interacts with mSin3A-Swi/Snf-Brm through its C-terminal repression domain in vitro. (A) Schematic diagrams of GST, full-length (FL) GST-SHP, and deletion mutants. RID and RD, receptor interacting and intrinsic repression domains, respectively; aa, amino acids. (B) 35S-labeled Brm and 35S-labeled mSin3A were synthesized in vitro, and GST pull-down assays were performed as described in Materials and Methods. Input, 20% of the total 35S-labeled proteins. (C) Swi/Snf complexes were purified using anti-Flag M2 agarose from nuclear extracts of HeLa cells stably expressing Flag-Ini 1 (F:Ini) (BAF47). GST or GST-SHP was incubated with purified Swi/Snf complex, and association of the complex with SHP was detected by Western blotting. Input, 20% of the amounts used in the binding reaction. (D and E) GST-SHP or the GST-SHP mutants were incubated with purified Swi/Snf complex (D) or HepG2 nuclear extracts (E), and association of the complex with SHP was analyzed by Western blotting using anti-Flag M2 antibody (D) or antisera against Brm, BAF155, mSin3A, and HDAC-1 (E).
Next, we examined whether SHP could interact with the entire Swi/Snf complex in vitro. Human Swi/Snf complexes were purified from nuclear extracts of HeLa cells that stably expressed Flag-tagged Ini 1 (BAF47), a component of the Swi/Snf complexes (48). Proteins of the expected molecular weights (20, 48) were observed (Fig. 5C), and some components of the Swi/Snf complex, Brm/Brg-1, BAF57, BAF155, BAF170, and BAF47, were subsequently identified by Western blotting (data not shown). Specific interaction between SHP and the purified Swi/Snf complex was detected by Western blotting using antibodies against Brm or BAF57 (Fig. 5C). SHP constructs containing the C-terminal repression domain efficiently interacted with Flag-Ini (BAF47), which was detected by Western blotting using the anti-Flag M2 antibody (Fig. 5D, lanes 2 and 4). Endogenous Brm and BAF155, as well as mSin3A and HDAC-1, in HepG2 nuclear extracts were preferentially associated with full-length SHP (Fig. 5E). A GST-SHP mutant that lacks the C-terminal repression domain did not interact with these proteins in HepG2 nuclear extracts. Together, these in vitro pull-down assays showed that SHP can interact with the purified Swi/Snf complex and directly interact with Brm and mSin3A through its intrinsic C-terminal repression domain.
Expression of a central ATPase of Swi/Snf, Brm, represses CYP7A1 promoter activity.
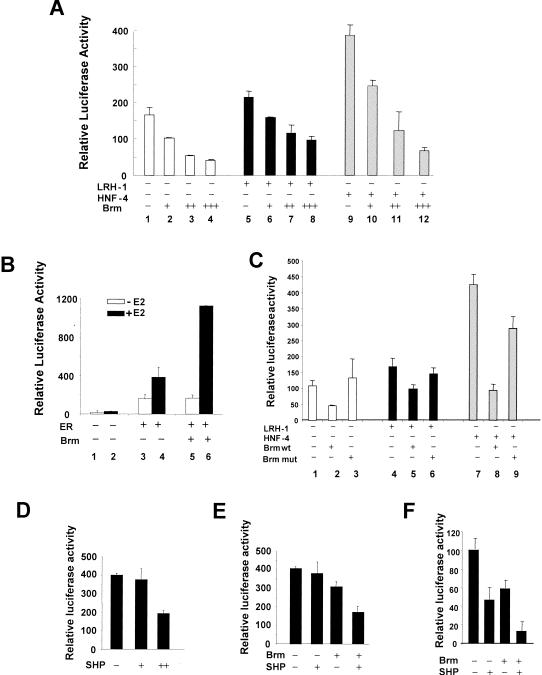
To assess the functional role of Swi/Snf in CYP7A1 regulation, we tested whether the expression of Brm suppressed promoter activity in SW13 cells. This human adrenal carcinoma cell line, in which Brm and Brg-1 are not detectable, has been utilized for functional analysis of Swi/Snf (13). SHP and the closely related DAX-1 have been reported to be abundantly expressed in adrenal cells (18, 19, 28, 46). Expression of Brm in SW13 cells suppressed basal human CYP7A1 promoter activity in a dose-dependent manner (Fig. 6A, lanes 1 to 4). Furthermore, Brm repressed the CYP7A1 promoter activity regulated by the hepatic transcriptional factor HNF-4 or LRH-1 (Fig. 6A, lanes 5 to 12).
FIG. 6.
Brm represses CYP7A1 promoter activity and further enhances SHP-mediated CYP7A1 repression. (A) SW13 cells were cotransfected with (+) 100 ng of 371CYP7A1-luc and either empty vector or LRH-1 or HNF-4 expression vector, along with 0 (−), 50 (+), 200 (++), or 700 (+++) ng of pcDNA3-Brm. (B) SW13 cells were transfected with 100 ng of 4ERE-tk-luc, 20 ng of CMV-hERα (ER), and 700 ng of pcDNA3-Brm (Brm). Twenty hours after transfection, ethanol or estradiol (E2; 10 nM) was added. (C) SW13 cells were transfected with 100 ng of CYP7A1-luc and either 100 ng of empty vector or expression vectors for LRH-1 or HNF-4, along with 200 ng of pcDNA3-Brm (Brm wt) or pcDNA3-Brm ATPase mutant (Brm mut). (D) HepG2 cells were transfected with 100 ng of CYP7A1-luc and CMV β-Gal internal control plasmid, along with 0 (−), 200 (+), or 800 (++) ng of pcDNA3-human SHP. (E) HepG2 cells were transfected with 100 ng of CYP7A1-luc, along with either 200 ng of pcDNA3-SHP, 200 ng of pcDNA3-Brm, or both plasmids. (F) SW13 cells were transfected with 100 ng of CYP7A1-luc, along with 250 ng of pcDNA3-Brm, 250 ng of pcDNA3-human SHP, or both plasmids. (A to F) Cytomegalovirus β-Gal was included as an internal control, and the values plotted are the ratios of luciferase activity to β-Gal. Standard errors of the mean are indicated by the error bars (n = 3). The empty vector pcDNA3 was added as needed so that the same amount of expression vector was present in each transfection. Reproducible results were obtained from three independent transfections of triplicate assays.
To ensure that Brm repression of the CYP7A1 gene in SW13 cells was promoter specific and not the result of cell toxicity related to Brm expression, we examined the effect of Brm on ER-mediated transactivation, which has been shown to be enhanced by Brg-1 (13). A luciferase reporter plasmid, 4ERE-tk-luc, was activated by ER and further activated by estradiol (Fig. 6B, lanes 1 to 4). When Brm was cotransfected, ligand-induced ER transactivation was increased two- to threefold (Fig. 6B, lanes 4 and 6), whereas ligand-independent ER transactivation was not affected (lanes 3 and 5). The enhancement of ER-mediated transactivation by Brm in the same cell line used for CYP7A1 studies indicates that CYP7A1 repression by Brm is promoter specific and is not related to general cell toxicity.
Functional ATPase activity by Brm is required for CYP7A1 repression.
To test whether the ATPase activity of Brm is required for Brm-mediated repression in transfected cells, we utilized a Brm mutant that contains a mutation in the ATPase domain. We analyzed the role of functional Brm in CYP7A1 regulation by using transient-transfection assays. SW13 cells were cotransfected with the CYP7A1 luciferase reporter and either Brm or the Brm ATPase mutant. The ability to repress CYP7A1 promoter activity was blunted in the ATPase mutant (Fig. 6C, lanes 1 to 3). Similar results were obtained from the reporter assays in which transcription of the CYP7A1 promoter was regulated by cotransfection of either HNF-4 or LRH-1 (lanes 4 to 9). These results suggest that ATP-dependent chromatin remodeling by Swi/Snf-Brm is associated with CYP7A1 repression in cells.
We also examined the effects of overexpression of Brm/Brg-1 mutants on endogenous CYP7A1 expression in HepG2 cells. However, we could not detect marked effects of inhibition of Swi/Snf function on endogenous CYP7A1 expression (data not shown). High expression levels of Brm and Brg-1 in cells could make it difficult to impair the function of Swi/Snf by dominant-negative mutants in this cell line. Furthermore, other mechanisms may contribute independently to bile acid-mediated repression of endogenous CYP7A1 so that the effect of inhibition of Swi/Snf and remodeling alone may not be sufficient for reversal of repression to be detected. Other mechanisms could include action of the corepressors mSin3A/HDAC-1 and SHP-independent JNK or PXR pathways (8, 16, 22, 50).
Brm potentiates SHP-mediated CYP7A1 repression.
We further tested whether Brm could enhance SHP-mediated repression, and conversely, if SHP could potentiate Brm-mediated repression. Ectopic expression of SHP in HepG2 cells suppressed CYP7A1 promoter activity in a dose-dependent manner (Fig. 6D). Overexpression of SHP or Brm alone resulted in modest reduction in CYP7A1 promoter activities in HepG2 cells (Fig. 6E). The small reduction in promoter activity is likely due to high expression levels of the endogenous Brm in these hepatic cells (data not shown). However, overexpression of both Brm and SHP proteins resulted in a substantial reduction in CYP7A1 promoter activity. Consistent with this result in HepG2 cells, in SW13 cells, expression of Brm or SHP repressed CYP7A1 promoter activity and expression of both proteins almost abolished the CYP7A1 promoter activity (Fig. 6F). Taken together, these functional studies suggest that the observed interaction in cells between SHP and the Swi/Snf complex is functionally significant.
Bile acid-induced recruitment of mSin3A-Swi/Snf and chromatin remodeling are dependent on SHP expression.
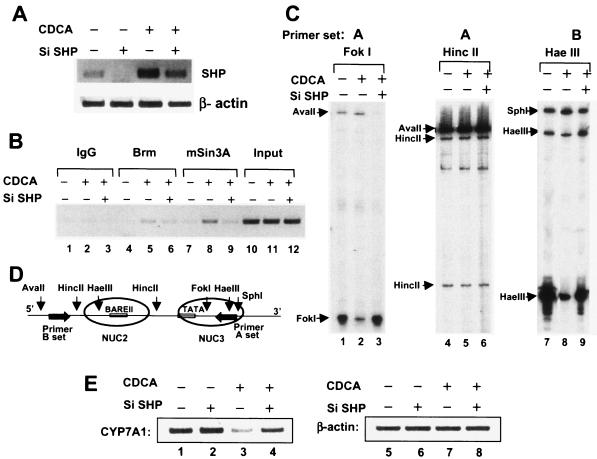
If bile acid-induced SHP directly recruits the Swi/Snf complex to the CYP7A1 promoter, then inhibition of endogenous SHP expression should impair the recruitment and further impair chromatin remodeling. We utilized the pSUPER siRNA system to inhibit SHP expression. CDCA treatment increased SHP mRNA levels as reported, and overexpression of the pSUPER vector producing SHP siRNA efficiently down-regulated endogenous SHP mRNA levels in HepG2 cells treated with either DMSO or CDCA (Fig. 7A). Bile acid-induced recruitment of Brm and mSin3A to the endogenous CYP7A1 promoter detected by ChIP analyses was also inhibited by overexpression of SHP siRNA (Fig. 7B, lanes 4 to 6 and 7 to 9). These results strongly suggest that SHP recruits mSin3A-Swi/Snf to the promoter.
FIG. 7.
Bile acid-induced recruitment of mSin3A-Swi/Snf-Brm, chromatin remodeling, and concomitant repression of endogenous CYP7A1 are dependent on SHP expression. (A) HepG2 cells were transfected with pSUPER vector carrying SHP siRNA (Si SHP) (+) or empty pSUPER vector (−) and treated with CDCA (+) or vehicle (−). Total RNA was isolated, and endogenous mRNA levels for SHP and β-actin were analyzed by RT-PCR. (B, C, and D) HepG2 cells were transfected with pSUPER vector carrying SHP siRNA (+) or empty pSUPER vector (−) by electroporation, and 72 h later, the cells were treated with CDCA or vehicle for 12 h and subjected to either ChIP assay (B) or restriction endonuclease accessibility assays (C). (B) The CYP7A1 promoter DNA precipitated by antibodies against mSin3A or Brm was subjected to PCR using CYP7A1 promoter-specific primers. Input, 1% of the total chromatin that was used for ChIP. (C) DNA in intact nuclei was partially digested with FokI, HincII, or HaeIII. After purification of the DNA, it was completely digested with AvaII or SphI. (D) Cleavages were detected by LMPCR using primer set A or B as shown in the schematic diagram. The weak bands for the AvaII fragments in the FokI experiment (C, lanes 1 to 3) are due to the high level of cleavage at the FokI site. (E) HepG2 cells were transfected with pSUPER SHP siRNA (+) or pSUPER (−), and after 72 h, the cells were treated with CDCA or vehicle in serum-free medium. Total RNA was isolated from the cells and subjected to RT-PCR. Semiquantitative PCR was performed using primer sets specific for human CYP7A1 and β-actin. Reproducible results were obtained from three ChIP assays, four remodeling assays, and three RT-PCR analyses.
Since bile acid-induced recruitment of Brm and mSin3A was impaired by SHP siRNA, we tested whether chromatin remodeling was also affected. As before, DNA in intact nuclei was partially digested with FokI, HincII, and HaeIII, and DNA was isolated and digested to completion with either AvaII or SphI, as appropriate. CDCA treatment decreased cleavage by FokI or HaeIII at their sites within the cores of the nucleosomes (Fig. 7C, lanes 1 versus 2 and 7 versus 8), but impressively, in cells expressing SHP siRNA, there was little change in cleavage at these sites after CDCA treatment (Fig. 7C, lanes 1 versus 3 and 7 versus 9). The variation in the intensities of the bands for AvaII digestion (Fig. 7C, lanes 1 to 3) is probably due to almost complete in vivo digestion of genomic DNA with FokI, so that increased digestion at FokI results in decreased extension of fragments to the AvaII site during LMPCR. As expected, cleavages at HincII sites in the linkers were not affected (lanes 4 to 6). These results indicate that bile acid-induced chromatin remodeling was substantially impaired in the cells expressing SHP siRNA. Our results provide compelling evidence that bile acid-induced SHP directly recruits the mSin3A-Swi/Snf complex to the endogenous CYP7A1 promoter, which results in concomitant chromatin remodeling.
Inhibition of SHP expression by SHP siRNA substantially reversed bile acid-mediated repression of endogenous human CYP7A1 expression.
Finally, we analyzed whether SHP siRNA completely or partially reverses bile acid-mediated CYP7A1 repression in HepG2 cells. CDCA treatment repressed CYP7A1 expression as expected (Fig. 7E, lanes 1 and 3). Reduction of the endogenous SHP expression by siRNA substantially reversed bile acid-mediated suppression of endogenous CYP7A1 expression (lane 4). β-Actin mRNA levels were not altered regardless of CDCA treatment or overexpression of SHP siRNA (lanes 5 to 8). These results firmly establish that SHP is a key mediator for bile acid-mediated repression of endogenous human CYP7A1 expression in hepatic cells.
DISCUSSION
Recent findings have demonstrated the key role of the orphan nuclear receptors FXR, LRH-1, and SHP in the negative feedback regulation of CYP7A1 gene expression by bile acids (15, 22, 32, 50). However, the changes in chromatin structure and the regulatory protein complex at the CYP7A1 promoter that lead to its repression by bile acids and bile acid-induced SHP are not understood. In the present study, we have shown that bile acid treatment results in SHP-mediated recruitment of mSin3A and Swi/Snf complex to the promoter, which is associated with chromatin remodeling and gene repression. Furthermore, coactivator HATs are dissociated from the promoter after bile acid treatment. Thus, both covalent modifications of histones and ATP-dependent remodeling of chromatin contribute to the bile acid-mediated repression of CYP7A1.
Potential role of ATP-dependent chromatin remodeling by the Swi/Snf complex in CYP7A1 repression.
Our analysis of chromatin at the CYP7A1 promoter indicates that nucleosome formation or nucleosome sliding did not occur at the promoter in vivo. Instead, chromatin remodeling, as assessed by accessibility to restriction endonucleases, occurs in the nucleosome cores containing the TATA and BARE II regions in the human CYP7A1 promoter. As suggested recently (36, 39), such remodeling may be catalyzed by the Swi/Snf-Brm complex by using the energy from ATP hydrolysis to twist the path of DNA over the histone core and increase the interaction between DNA and histones. The increased interaction may result in gene repression by decreasing the accessibility of the DNA to transcription activators or basal transcriptional machinery, which would be consistent with remodeling of the CYP7A1 promoter chromatin during bile acid-mediated repression of the gene.
Role of bile acid-induced SHP in CYP7A1 repression in a native chromatin context.
As proposed in a dual-repression mechanism for SHP-mediated gene repression (19, 29, 30), SHP competes for the binding of p160 coactivators to HNF-4 and/or LRH-1 and also directly recruits corepressors through a repression domain. This mechanism is relevant to the CYP7A1 system because LRH-1 and HNF-4 are key regulators of this gene (6, 11, 17). In this study, we found that the corepressors, mSin3A and HDAC-1, were recruited to the CYP7A1 promoter in vivo and the HATs, p300, CBP, and SRC-1, were dissociated, which is consistent with the earlier transfection studies (19, 29, 30). Important additions to the dual-repression mechanism resulting from the present work are the fact that chromatin remodeling by Swi/Snf comprises a crucial component of SHP-mediated repression of the endogenous CYP7A1 and that the mSin3A corepressor complex appears to be recruited to the promoter in a SHP-mSin3A-Swi/Snf complex.
Several lines of evidence suggest that the mSin3A-Swi/Snf complex is directly recruited to the CYP7A1 promoter by SHP. First, in GST pull-down experiments, SHP directly interacted with both mSin3A and Brm and with endogenous, as well as a purified human, Swi/Snf complex. The interaction was primarily through the intrinsic repression domain of SHP. Second, CoIP assays revealed that SHP was tightly associated with the mSin3A-Swi/Snf complex in cells. Finally, the most definitive evidence is that inhibition of SHP expression by SHP siRNA attenuated the recruitment of Brm and mSin3A to the endogenous promoter, impaired chromatin remodeling, and reversed the repression of the endogenous CYP7A1 expression. These results strongly suggest that bile acid-induced SHP directly recruits the mSin3A-Swi/Snf-Brm complex to the promoter.
Model of the action of SHP in negative feedback inhibition of CYP7A1 by bile acids.
Based on previous studies and our present studies, a model is proposed for the mechanism for bile acid-mediated repression of CYP7A1, as shown in Fig. 8. FXR is activated by bile acids, binds as a heterodimer with RXR to the SHP promoter, and induces the expression of SHP (15, 32). Bile acid-induced SHP interacts with the mSin3A-Swi/Snf complex in hepatic cells and brings this complex to the native CYP7A1 promoter. Bile acid treatment also results in dissociation of coactivator HATs, p300, CBP, and SRC-1, from the native promoter. The Swi/Snf complex catalyzes the ATP-dependent remodeling of the chromatin, which contributes to the repression. In addition, the mSin3A/HDAC-1 corepressor complex can further inhibit transcription by histone deacetylation by interacting with the basal transcription factors and/or by recruiting additional corepressors or histone-modifying enzymes. Furthermore, the dissociation of HATs would also favor deacetylation of the histones. Whether the ATP-dependent remodeling and the histone deacetylation are independent or mutually dependent events that contribute to repression is not known. It is possible that histone methylation might also play a role in SHP-mediated human CYP7A1 repression in bile acid signaling. Pal et al. recently reported that the arginine methyltransferase PRMT5 was recruited in an mSin3A-Swi/Snf-Brg-1 remodeling complex and was involved in transcriptional repression of a Myc target gene (38). Other mechanisms may also contribute to the bile acid-mediated repression of CYP7A1 independently of SHP. Both the bile acid-activated JNK pathway and the ligand-activated PXR/SXR have been implicated (8, 16, 22, 50), but the details of the mechanisms are not known (Fig. 8A).
FIG. 8.
Action of SHP on negative feedback regulation of CYP7A1 in bile acid signaling. Bile acid-induced SHP interacts with the mSin3A/Swi/Snf-Brm complex in cells and recruits the complex to the endogenous human CYP7A1 promoter (A), which results in chromatin remodeling at the promoter chromatin (B). (A) Bile acid treatment also causes dissociation of the p300/CBP/SRC-1 coactivator HAT complex from the promoter. (B) Recruitment of mSin3A/HDAC-1 and dissociation of HATs lead to histone deacetylation at the promoter. Bile acid-induced chromatin remodeling and histone deacetylation both contribute to transcriptional repression after bile acid treatment. (A) The bile acid-activated JNK or ligand-activated PXR (SXR) pathway may contribute to bile acid-mediated CYP7A1 repression independent of the SHP pathway, but the details of the mechanisms are not known, so they are indicated with dashed lines.
General relevance of the mSin3A-Swi/Snf complex in repressor SHP action in nuclear receptor activities.
Negative feedback regulation of bile acid production, as studied here, is a physiologically relevant and well-characterized biological system in which SHP plays a key regulatory role by interacting with an orphan nuclear receptor, LRH-1, at the CYP7A1 promoter (15, 32). SHP, however, interacts with a number of other nuclear receptors, including HNF-4, ERR, LXR, GR, ER, PPAR, TR, RXR, RAR, and CAR, and represses the transcriptional activities of these nuclear receptors (1-3, 19, 29, 30, 43, 45). SHP-mediated modulation of these nuclear receptor activities may affect diverse biological processes, including cell proliferation, reproduction, and metabolism of xenobiotics, cholesterol, bile acids, and glucose. In the regulation of cholesterol and bile acid metabolism, genes other than CYP7A1 that are negatively regulated by SHP include those for sterol 12α-hydroxylase (CYP8B1), apical sodium-dependent bile acid transporter, sodium taurocholate cotransporting polypeptide, and the scavenger receptor class B type I (5, 12, 44, 56). A role for SHP in the regulation of glucose and energy metabolism is suggested by the observation that a mutation in the SHP gene is associated with mild obesity and a metabolic disorder in Japanese subjects with early onset of diabetes (37). Consistent with this study, SHP has been reported to suppress expression of the PGC-1 and PEPCK genes, which play significant roles in glucose and energy metabolism (2, 54). The coordinated recruitment of the mSin3A corepressor complex and the Swi/Snf-Brm complex by SHP, as shown in this study for CYP7A1, may be applicable to these other genes. Our studies, therefore, may be relevant, not only for the regulation of cholesterol and bile acid metabolism, but also for glucose homeostasis and other important biological processes in which SHP plays key regulatory roles.
Acknowledgments
We thank W. Wang, R. Kingston, A. Imbalzano, S. Sif, J. Chiang, J. Auwerx, and D. Moore for providing valuable materials, including antibodies, a stable cell line, and plasmids for the study. We are grateful to S. Wu and C. Chiang from Case Western Reserve University for their advice on protein purification. We thank S. Okino and D. Burakov for technical advice on the LMPCR and ChIP assays, respectively. We also thank A. Nardulli and B. Kemper for helpful comments on the manuscript.
This study was supported by AHA grant 0130538Z and NIH DK062777 to J.K.K.
REFERENCES
- 1.Bae, Y., J. K. Kemper, and B. Kemper. 2004. Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP). DNA Cell Biol. 23:81-91. [DOI] [PubMed] [Google Scholar]
- 2.Borgius, L. J., K. R. Steffensen, J. A. Gustafsson, and E. Treuter. 2002. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J. Biol. Chem. 277:49761-49766. [DOI] [PubMed] [Google Scholar]
- 3.Brendel, C., K. Schoonjans, O. A. Botrugno, E. Treuter, and J. Auwerx. 2002. The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol. Endocrinol. 16:2065-2076. [DOI] [PubMed] [Google Scholar]
- 4.Burakov, D., L. Crofts, C. Chang, and L. Freedman. 2002. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J. Biol. Chem. 277:14359-14362. [DOI] [PubMed] [Google Scholar]
- 5.Chen, F., L. Ma, P. A. Dawson, C. J. Sinal, E. Sehayek, F. J. Gonzalez, J. Breslow, M. Ananthanarayanan, and B. L. Shneider. 2003. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J. Biol. Chem. 278:19909-19916. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, J. Y. L., R. Kimmel, C. Weinberger, and D. Stroup. 2000. Farnesoid X receptor responds to bile acids and represses cholesterol 7a-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275:10918-10924. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, J. Y. L., and D. Stroup. 1994. Identification and characterization of a putative bile acid-responsive element in cholesterol 7a-hydroxylase gene promoter. J. Biol. Chem. 269:17502-17507. [PubMed] [Google Scholar]
- 8.De Fabiani, E., N. Mitro, A. Anzulovich, A. Pinelli, G. Galli, and M. Crestani. 2001. The negative effects of bile acids and tumor necrosis factor-α on the transcription of cholesterol 7-α hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4. J. Biol. Chem. 276:30708-30716. [DOI] [PubMed] [Google Scholar]
- 9.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 10.de la Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 276:41486-41491. [DOI] [PubMed] [Google Scholar]
- 11.del Castillo-Olivares, A., and G. Gil. 2000. Role of FXR and FTF in bile acid-mediated suppression of cholesterol 7a-hydroxylase transcription. Nucleic Acids Res. 28:3587-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denson, L. A., E. Sturm, W. Echevarria, T. L. Zimmerman, M. Makishima, D. J. Mangelsdorf, and S. J. Karpen. 2001. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology 121:140-147. [DOI] [PubMed] [Google Scholar]
- 13.DiRenzo, J., Y. Shang, M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20:7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Wilson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, S., T. Stravitz, P. Dent, and P. Hylemon. 2001. Down-regulation of cholesterol 7 a-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-jun N-terminal kinase pathway. J. Biol. Chem. 276:15816-15822. [DOI] [PubMed] [Google Scholar]
- 17.Hayhurst, G., Y. Lee, G. Lambert, J. Ward, and F. J. Gonzalez. 2001. Hepatocyte nuclear factor 4a (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21:1393-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, M., R. Yu, and J. L. Jameson. 1997. DAX-1 inhibits SF-1 mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 17:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson, L., J. S. Thomsen, A. E. Damdimopoulos, G. Spyrou, J.-A. Gustafsson, and E. Treuter. 1999. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERα and ERβ. J. Biol. Chem. 274:345-353. [DOI] [PubMed] [Google Scholar]
- 20.Kadam, S., and B. M. Emerson. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377-389. [DOI] [PubMed] [Google Scholar]
- 21.Kadonaga, J. 1998. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92:307-315. [DOI] [PubMed] [Google Scholar]
- 22.Kerr, T., S. Saeki, M. Schneider, K. Schaefer, S. Burdy, T. Redder, B. Shan, D. Russell, and M. Schwaez. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J., and B. Kemper. 1997. Phenobarbital alters protein binding to the CYP2B1/2 phenobarbital-responsive unit in native chromatin. J. Biol. Chem. 272:29423-29426. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J., L. N. Petz, Y. S. Ziegler, J. R. Wood, S. J. Potthoff, and A. M. Nardulli. 2000. Regulation of the estrogen-responsive pS2 gene in MCF-7 human breast cancer cells. J. Steroid Biochem. Mol. Biol. 74:157-168. [DOI] [PubMed] [Google Scholar]
- 25.Kim, J., I. Rivera-Rivera, and B. Kemper. 2000. Tissue-specific chromatin structure of the CYP2B1 phenobarbital-responsive unit and proximal promoter and modulation by phenobarbital. Nucleic Acids Res. 28:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornberg, R. D., and Y. Lorch. 1999. Twenty five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 27.Kraus, W. L., and J. Wong. 2002. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur. J. Biochem. 269:2275-2283. [DOI] [PubMed] [Google Scholar]
- 28.Lee, H. K., Y. K. Lee, S. H. Park, Y. S. Kim, J. W. Lee, H. B. Kwon, J. Soh, D. D. Moore, and H. S. Choi. 1998. Structure and expression of the orphan nuclear receptor SHP gene. J. Biol. Chem. 273:14398-14402. [DOI] [PubMed] [Google Scholar]
- 29.Lee, Y., H. Dell, D. H. Dowhan, M. Hadzopoulou-Cladaras, and D. D. Moore. 2000. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell. Biol. 20:187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y., and D. D. Moore. 2002. Dual mechanism for repression of the monomeric orphan receptor liver receptor homologous protein-1 (LRH-1) by the orphan small heterodimer partner (SHP). J. Biol. Chem. 277:2463-2467. [DOI] [PubMed] [Google Scholar]
- 31.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 32.Lu, T., M. Makishima, J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 33.Makishima, M., A. Y. Okamoto, J. J. Repa, H. Tu, M. Learned, A. Luk, M. V. Hull, K. D. Lustig, D. J. Mangelsdorf, and B. Shan. 1999. Identification of a nuclear receptor for bile acids. Science 284:1362-1365. [DOI] [PubMed] [Google Scholar]
- 34.McPherson, C., E. Shim, D. Friedman, and K. Zaret. 1993. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75:387-397. [DOI] [PubMed] [Google Scholar]
- 35.Min, G., H. Kim, Y. Bae, L. Petz, and J. K. Kemper. 2002. Inhibitory crosstalk between estrogen receptor (ER) and constitutive activated androstane receptor (CAR). J. Biol. Chem. 277:34626-34633. [DOI] [PubMed] [Google Scholar]
- 36.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 37.Nishigori, H., H. Tomura, N. Tonooka, M. Kanamori, S. Yamada, K. Sho, I. Inoue, N. Kikuchi, K. Onigata, I. Kojima, T. Kohama, K. Yamagata, Q. Yang, Y. Matsuzawa, T. Miki, S. Seino, M. Y. Kim, H. S. Choi, Y. K. Lee, D. D. Moore, and J. Takeda. 2001. Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc. Natl. Acad. Sci. USA 98:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 40.Pina, B., U. Bruggemeier, and M. Beato. 1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60:719-731. [DOI] [PubMed] [Google Scholar]
- 41.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, D. W. 1999. Nuclear orphan receptors control cholesterol catabolism. Cell 97:539-542. [DOI] [PubMed] [Google Scholar]
- 43.Sanyal, S., J. Y. Kim, H. J. Kim, J. Takeda, Y. K. Lee, D. D. Moore, and H. S. Choi. 2002. Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J. Biol. Chem. 277:1739-1748. [DOI] [PubMed] [Google Scholar]
- 44.Schoonjans, K., J. S. Annicotte, T. Huby, O. A. Botrugno, E. Fayard, Y. Ueda, J. Chapman, and J. Auwerx. 2002. Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 3:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seol, W., H. Choi, and D. D. Moore. 1996. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science 272:1336-1339. [DOI] [PubMed] [Google Scholar]
- 46.Seol, W., M. Chung, and D. D. Moore. 1997. Novel receptor interaction and repression domains in the orphan receptor SHP. Mol. Cell. Biol. 17:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang, Y., X. Hu, J. DiRenzo, M. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 48.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soutoglou, E., and I. Talianidis. 2002. Cordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 50.Wang, L., Y. Lee, D. Bundman, Y. Han, S. Thevananther, C. Kim, S. Chua, P. Wei, R. Heyman, M. Karin, and D. Moore. 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2:721-731. [DOI] [PubMed] [Google Scholar]
- 51.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 52.Wu, S., and C. M. Chiang. 2001. Expression and purification of epitope tagged multisubunit protein complexes from mammalian cells, p. 16.22.1-16.22.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 53.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 54.Yoon, J. C., P. Puigserver, G. Chen, J. Donovan, Z. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, C. B. Newgard, and B. M. Spiegelman. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131-138. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, M., and J. Y. Chiang. 2001. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): roles of hepatocyte nuclear factor 4α in mediating bile acid repression. J. Biol. Chem. 276:41690-41699. [DOI] [PubMed] [Google Scholar]