Abstract
Background
Cholera, one of the world’s deadliest infectious diseases, remains rampant and frequent in Tanzania and thus hinders existing control measures. The present study was undertaken to evaluate the occurrence of toxigenic Vibrio cholerae O1 in wastewater, fish and vegetables during a non-outbreak period in Morogoro, Tanzania.
Methods
From October 2014 to February 2015, 60 wastewater samples, 60 fish samples from sewage stabilization ponds and 60 wastewater irrigated vegetable samples were collected. Samples were cultured for identification of V. cholerae using conventional bacteriological methods. Isolates were confirmed as V. cholerae by detection of the outer membrane protein gene (ompW) using polymerase chain reaction (PCR). Isolates were further tested for antibiotic susceptibility and presence of virulence genes including, cholera enterotoxin gene (ctx), the toxin co-regulated pilus gene (tcpA) and the haemolysin gene (hlyA).
Results
The prevalence of V. cholerae in wastewater, vegetables and fish was 36.7, 21.7 and 23.3 %, respectively. Two isolates from fish gills were V. cholerae O1 and tested positive for ctx and tcpA. One of these contained in addition the hlyA gene while five isolates from fish intestines tested positive for tcpA. All V. cholerae isolates were resistant to ampicillin, amoxicillin and some to tetracycline, but sensitive to gentamicin, chloramphenicol, and ciprofloxacin.
Conclusions
Our results show that toxigenic and drug-resistant V. cholerae O1 species are present and persist in aquatic environments during a non-cholera outbreak period. This is of public health importance and shows that such environments may be important as reservoirs and in the transmission of V. cholerae O1.
Keywords: Vibrio cholerae O1, Wastewater, Fish, Vegetables, Antibiotic susceptibility
Background
Climate change is one of the leading causes of water scarcity which is a serious problem in the world and in developing countries in particular. For populations experiencing water scarcity, a main source of water is reclaimed wastewater which is used for a large range of activities including fish culture [1]. Fish culture and vegetables irrigation using wastewater is practiced by populations living in water scarce areas in Tanzania [1]. This situation is due to the depletion of the natural fish stock and harvests from freshwater and ocean fisheries [2] and the increased demand of vegetables alongside high water scarcity. However, fish and vegetables grown in such unhygienic conditions may represent food safety hazards to consumers, i.e. as documented by the isolation of a number of pathogens including Vibrio cholerae, the causative agent of cholera [3, 4]. In cholera endemic areas, the disease often has a seasonal pattern [5, 6] and cholera remains highly frequent in Tanzania due to multiple reasons that need to be addressed [7, 8]. Since the seventh cholera pandemic reached the country in 1974, the disease has been reported almost every year in Tanzania regardless of seasons [7, 8]. It is therefore likely that neglected reservoirs of V. cholerae exist in Tanzania. Inadequate information on possible sources of toxigenic strains of V. cholerae complicates cholera prediction, prevention and control and contributes to frequent and unexpected outbreaks of cholera in Tanzania. Therefore, there is a need to assess to what extent toxigenic strains of V. cholerae occur in the aquatic environment during non-cholera outbreak periods. Widespread use of antibiotics as prophylaxis during cholera outbreaks worldwide has contributed to the development of multidrug resistant strains of V. cholerae [9]. Information and surveillance of the antibiotic susceptibility of the organism not only in clinical, but also in environmental isolates in endemic regions is thus needed. The present study aimed to determine the occurrence of toxigenic V cholerae and their antibiotic susceptibility in isolates obtained from fish and vegetables grown in wastewater during a non-cholera outbreak period in Morogoro, Tanzania.
Methods
Study sites
The study was carried out at Mafisa and Mzumbe wastewater treatment units and at a vegetable production site called Funga–Funga near the Morogoro River in urban and peri-urban Morogoro, Tanzania. The Mafisa wastewater treatment unit receives municipal wastewater mainly from households, commercial areas and hospitals around Morogoro. Treated wastewater from Mafisa is used to irrigate a large area of rice fields. Funga–Funga is the main and largest vegetable cultivation site located alongside Morogoro River with fields being irrigated with water from the river. Mzumbe wastewater treatment unit is located at the University of Mzumbe and receives wastewater from housing facilities, cafeterias and the hospital. The treated wastewater is discharged through an informal network of earth canals and used to irrigate a variety of vegetables.
Collection of samples
A cross-sectional study was conducted from October 2014 to February 2015 including samples of wastewater, Chinese cabbage (Brassica rapa), Nile tilapia (Oreochromis niloticus), and African sharp tooth catfish (Clarias gariepinus). Water samples were collected during five samplings at two weeks interval. Fifteen water samples were collected from different points of Mafisa and 20 water samples were collected along Morogoro River and 25 samples from Mzumbe. From the ponds, water samples were collected from the inlets and the outlets (Fig. 1). Volumes of 100 ml were collected in sterile bottles according to WHO guidelines for microbiological drinking water analysis [10]. A total of ten Chinese cabbage heads were collected from each field irrigating with treated effluent from the Mzumbe and Funga–Funga production sites and placed in sterile plastic bags during each of three monthly samplings with cabbage heads obtained from different furrows during each sampling. Thirty tilapia and 30 African sharp tooth catfish of at least 300-grs size were collected by cast net from Mzumbe stabilisation ponds early in the morning and individually placed in sterile plastic bags. Samples were placed in an insulated box with cooling elements and transported to the laboratory at the Sokoine University of Agriculture in Morogoro where the microbiological analysis was initiated within 2 h of sampling.
Fig. 1.
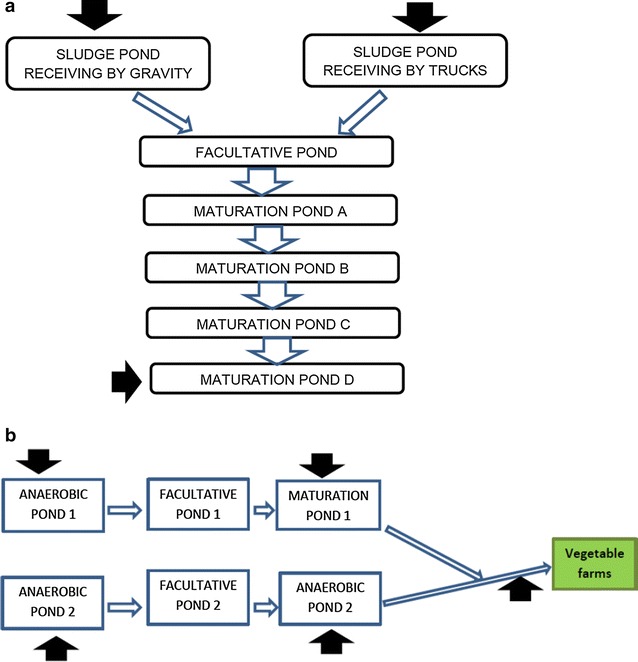
Water sampling points at Mafisa (a) and Mzumbe (b) wastewater treatment units. Black arrows indicate sampling points
Isolation of Vibrio cholerae
In the laboratory, 25 ml of each well mixed water sample was added to 225 ml of Alkaline Peptone Water (APW; (Sigma, Steinheim, Germany)) for enrichment. Approximately, 25 g of cabbage leaves were cut off and added to 225 ml of APW for enrichment. For fish samples, subsamples were individually taken from gills and intestines (10 g of each) and added to 90 ml of APW for enrichment before culture. APW samples were enriched for 18 h at 37 °C then subcultured onto thiosulfate citrate bile sucrose (TCBS) agar (ACCUMIX, Verna, India). Following 24 h incubation at 37 °C, yellow colonies (sucrose fermenting, 2–3 mm diameter) suspected as Vibrio cholerae were purified on trypticase soy agar (Merck, Darmstadt, Germany) for 24 h at 37 °C. Purified colonies were characterized by Gram staining (Gram-negative comma shaped rods), positive oxidase reaction (colour change to blue dark within 5 s) and reaction in triple sugar iron agar (yellow slants with no gas formation) [11]. Presumptive V. cholerae isolates were confirmed by sero-agglutination test with the polyvalent V. cholerae O1 serum (Bio-Rad, Marnes-la-conquette, France).
Molecular characterization of Vibrio cholerae
DNA was extracted from colonies picked from TSA by the boiling lysis method. Following removal of cell debris by centrifugation at 12,000 rpm for 3 min, the supernatant containing the template DNA was stored at −20 °C for PCR [12]. Isolates were confirmed as V. cholerae by PCR for the outer membrane protein encoding gene (OmpW) [13, 14]. PCR was also done to detect the ctx, tcpA and hlyA genes which are all important virulence factors in V. cholerae using primers as listed in Table 1.The ctx gene is the main virulence factor of V. cholerae and encodes the production of cholera toxin which causes the severe diarrhoea seen in cholera patients. tcpA intervenes in fimbriae synthesis enabling V. cholerae to adhere to the host’s intestinal epithelium whereas the hlyA gene is associated with blood cell lysis in the infected host. Electrophoresis of amplicons was done in a 1.5 % agarose gel (Advance, Tokyo, Japan) at 100 V for 1 h. The gel was visualized under UV light using Gel Doc EZ Imager apparatus (Bio Rad, California, USA). Double distilled DNase free water was used as negative control and V. cholerae O139 NCTC 12945 (ATCC 51394) as a positive control.
Table 1.
Primers sequences used for the PCR
| Targeted genes | Primer Sequences (5′–3′) | Size (bp) | Source |
|---|---|---|---|
|
Ctx
Cholera toxin gene |
F-CAGTCAGGTGGTCTTATGCCAAGAGG R-CCCACTAAGTGGGCACTTCTCAAACT |
167 | [27] |
|
OmpW
Outer membrane protein |
F-CACCAAGAAGGTGACTTTATTGTG R-GAACTTATAACCACCCGCG |
588 | [3] |
|
TcpA
Toxin coregulated pilus |
F-CAC GAT AAG AAA ACC GGT CAA GAG R-CGA AAG CAC CTT CTT TCA CGT TG |
453 | [26] |
|
hlyA
Haemolysin |
F-GGC AAA CAG CGA AAC AAA TAC C R-CTC AGC GGG CTA ATA CGG TTT A |
727 | [6] |
Antimicrobial susceptibility testing
All confirmed V. cholerae isolates were subjected to antimicrobial susceptibility testing on Muller Hinton agar plates (OXOID, Basingstoke, Hampshire, England) using the Kirby-Bauer disc diffusion method to the following antibiotics: tetracycline (20 µg), gentamicin (10 µg), ciprofloxacin (5 µg), chloramphenicol (30 µg), ampicillin (10 µg) and amoxicillin (10 µg) (OXOID). Measured inhibition zone diameters were interpreted according to CLSI guidelines [15].
Data analysis
Proportions of positive Vibrio cholerae samples at different sites and different sample types were calculated then compared by Chi square and Fisher exact tests based on the total sizes using EPI-INFO 7 statistical software. Statistical significance was defined at a probability of p = 0.05.
Results
Detection of Vibrio cholerae in water, vegetables and fish
Out of the 60 water samples, 22 (36.7 %) were positive for V. cholerae as shown by a 588 bp sized PCR amplicon of the OmpW gene; however none of the isolates did agglutinate the V. cholerae O1 antiserum. Water samples collected from the Mafisa unit contained the highest prevalence of V. cholerae non-O1 (46.7 %, n = 15), followed by Morogoro River (35.0 %, n = 20) and the Mzumbe unit (32.0 %, n = 25). V. cholerae non-O1 was significantly (p < 0.05) more often found in outlet as compared to inlet water at both treatment units.
Vibrio cholerae was isolated from 13/60 (21.7 %) Chinese cabbage samples collected from the Mzumbe and Funga–Funga vegetables production sites; however, none of these isolates were V. cholerae O1. Cabbage from Funga–Funga site contained a higher V. cholerae non-O1 prevalence (36.7 %, n = 30) than did cabbage from Mzumbe production site (6.7 %, n = 30) (p < 0.05).
A total of 15 (25.0 %, n = 60) intestinal samples from all the sampled fish contained V. cholerae non-O1 and 13 (21.7, n = 60 %) gill samples contained V. cholerae non-O1. Two (7.1 %) of the gill isolates were confirmed as V. cholerae O1 in the slide agglutination test. Actually, V. cholerae was not found in any catfish samples but only from tilapia.
Characterization of Vibrio cholerae O1 and non-O1
PCR confirmed that the cholera toxin gene (ctx) was present in the two V. cholerae O1 isolates and the positive control strain as shown by a 167 bp sized amplicon (Fig. 2). The toxin co-regulated pilus subunit A gene (tcp-A) was amplified (453-bp, Fig. 3) in seven V. cholerae isolates from tilapia including the two V. cholerae O1 isolates. Only one of the V. cholerae O1 isolates contained the hlyA gene (727 bp-sized amplicon, Fig. 4). The two V. cholerae O1 isolates, as well as the non-O1 isolates were resistant to ampicillin, amoxicillin and tetracycline, but sensitive to gentamicin, chloramphenicol, and ciprofloxacin.
Fig. 2.
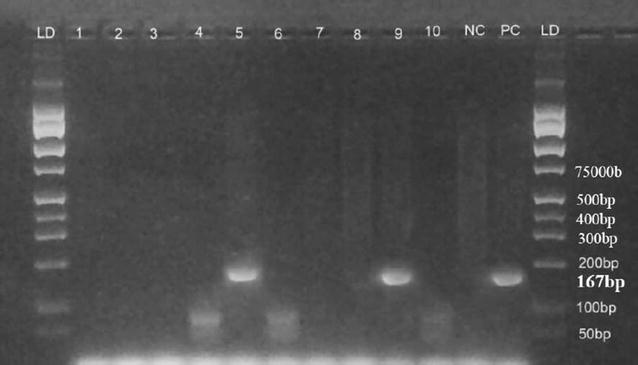
Cholera toxin gene (ctx) detected in fish isolates using PCR at 167 bp. LD DNA ladder; lane 1 to 10 are V. cholerae DNA samples; NC negative control (DNA free water); PC positive control (VC O139, ATCC 51394)
Fig. 3.
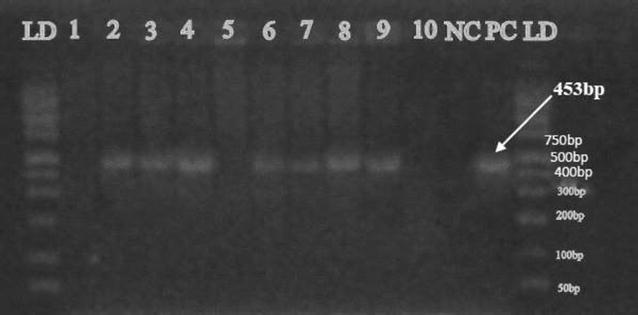
Cholera toxin co-regulated pilus gene (tcpA) detected in fish isolates using PCR at 453 bp. LD DNA ladder; Lane 1 to 10 are V. cholerae DNA samples; NC negative control; PC positive control
Fig. 4.
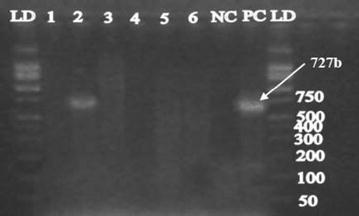
Cholera haemolysin gene (hlyA) detected in fish isolates using PCR at 727 bp. LD DNA ladder, lane 1 to 6 are V. cholerae DNA samples, NC negative control, PC positive control
Discussion
The isolation of ctx-positive V. cholerae O1 in a non-cholera outbreak period from fish samples in Tanzania is of food safety and public health concern [8–16].The presence of the toxin co-regulated pilus (tcp-A) is important as this gene encodes for fimbriae synthesis which allows the bacteria to adhere to the host’s intestinal epithelium [17]. The haemolysin gene (hlyA) present in these isolates enables blood cell lysis leading to anaemia in the infected individuals [6]. As the first to ascertain the occurrence of V. cholerae O1 in fish from a non-outbreak period in Tanzania, this study demonstrates the wide diversity of reservoirs of toxigenic V. cholerae O1. This observation is supported by the results of Nkoko et al. [18], who reported various environmental and aquatic reservoirs of V. cholerae including blue-green algae and chironomid eggs. Several other studies have illustrated the ability of V. cholerae to associate with a number of zooplankton, phytoplankton, blue-green algae (cyanobacteria) that prolong its survival [19, 20]. The isolation of V. cholerae O1 from fish samples is of food safety and health concern mainly due to risks for cross- contamination and potential for multiplication in different types of foods. Senderovich et al. [21] confirmed different fish species including tilapia (Sarotherodon galilaeus) from aquatic environments as reservoirs of V. cholerae. Furthermore, Onyuka et al. [22] isolated V. cholerae O1 from fish samples in Kenya during a non-outbreak period and concluded that in cholera endemic areas these microorganisms exist in biofilm-like aggregates in which the cells are in conditional viable state. Similar results were also reported by Sathiyamurthy et al. [3] and Mrityunjoy et al. [4]. Physico-chemical conditions mainly pH and salinity influences the survival of V. cholerae O1 [23]. Therefore, the physico-chemical characteristics and conditions in fish gills (the identified reservoir of this study) may favour persistence of such toxigenic organisms in fish. Moreover, Evans et al. [24] reported that in osmoregulation and iron balance in fish, high concentrated chloride solution is secreted by fish gills whereby the mechanisms of NaCl secretion by the gill epithelium are greater than the NaCl uptake from the environment. This suggests that the persistence of toxigenic V. cholerae O1 isolates in gills may be associated with NaCl content in this type of tissue. Also, Faruque et al. [19] reported that in natural ecological settings, unidentified environmental factors induce lysogenic phage CTXФ in toxigenic V. cholerae, resulting in the release of extracellular CTXФ particles into the aquatic environment. Therefore, the cell-free phage particles may be associated with the emergence of novel toxigenic strains of V. cholerae through interactions with non-toxigenic strains. It is uncertain to what extend lysogenic phages like CTXФ play a role in the emergence of toxigenic V. cholerae O1 and associated cholera outbreaks in Tanzania where they occur every year in many regions of the country [16]. The frequent cholera outbreaks experienced in Tanzania in recent years may be associated with the ecology and potential aquatic reservoirs of V. cholerae, i.e. fish and aquatic organisms. Also, the neighbouring countries of Tanzania are often experiencing cholera outbreaks, e.g. associated with large displacements of people due to social unrest, resulting in discharge and introduction of V. cholerae O1 into aquatic environments like the African Great Lakes to which access is shared by Tanzania. Since cholera is a hygiene related disease, most cholera control policies are directed towards household hygiene and sanitation with less attention paid to potential reservoirs and transmission routes of V. cholerae O1. An analysis of cholera outbreaks in countries around the African Great Lakes revealed a strong association of cholera with access to and location close to the Lakes [18].
The frequent finding of non-O1 V. cholerae in the sewage system may be ecologically normal but the presence of these organisms on irrigated vegetables could represent food safety and human health hazards. Sathiyamurthy et al. [3] demonstrated the clinical and epidemiological importance of non-O1 V. cholerae as causes of diarrhoea and gastroenteritis in humans. For instance, non-O1 and non-O139 strains of V. cholerae have been associated with occasional diarrhoea outbreaks resembling cholera [25]. Therefore, appropriate measures should be taken to curb the public health threats of diarrhoea associated with consumption of wastewater irrigated vegetable in Tanzania [1]. The prevalence of V. cholerae non-O1 in the effluents water of both studied sewage treatment units was higher than those of the inlets. These findings suggest that there is growth and proliferation of the bacteria throughout the system. According to Faruque et al. [19], V. cholerae are autochthonous of aquatic environments and can therefore be recovered at any point in a sewage plant independently on those isolated at the inlet. Also, V. cholerae may survive better in the latter parts of the treatment system, i.e. in the facultative and the maturation ponds where water is cleaner and contains less competitive microorganisms. Besides, studies have illustrated the ability of V. cholerae to associate with a variety of zooplankton, phytoplankton, blue-green algae (cyanobacteria) that prolong its survival and facilitate multiplication [19, 20]. Such organisms are often seen in high concentrations in the maturation ponds. The recovery of high concentrations of V. cholerae in effluent water indicates that wastewater stabilization ponds are unable to effectively remove these pathogens and could be considered as reservoirs of toxigenic V. cholerae O1 during non-cholera outbreak period.
Although rehydration plays a pivotal role in reducing mortality during cholera epidemics, antibiotics have been used to reduce the shedding of the organism (thereby reducing spread of the disease), treating severe illness (by reducing volume of diarrhoea), and also to reduce duration of disease and hospitalisation [9]. However, the resistance of microorganisms including V. cholerae to antibiotics has become a serious health challenge worldwide. Our V. cholerae O1 isolates were resistant to ampicillin, amoxicillin and tetracycline; a resistance pattern that has also been reported elsewhere for V. cholerae O1 [23–28]. Such similarity could be due to the misuse of the same antibiotics all over the world because in this era of antibiotics resistance, effective antibiotics are communicated and prescribed worldwide leading to simultaneous occurrence of resistance around the world. The resistance to tetracycline is particularly worrisome as tetracycline has been shown to be effective treatment for cholera and quite superior to other antimicrobials in reducing cholera morbidity. It should be noted that our V. cholerae O1 strains were sensitive to gentamicin, chloramphenicol and ciprofloxacin with the latter being a recommended alternative drug for treatment of cholera cases. The misuse and release of antibiotics in the sewage could be one of the reasons of occurrence of antibiotics resistant organisms in these environmental isolates.
Conclusion
Different serotypes of V. cholerae were isolated from wastewater and its irrigated vegetables and fish in Morogoro, Tanzania from October 2014 to February 2015 during a non-outbreak period. Two isolates were confirmed V. cholerae O1. Three main virulence genes were detected in these isolates notably the cholera toxin gene, the leading cause of the disease and others virulent genes mainly the toxin co-regulated pilus and the haemolysin gene which play important roles in the pathogenesis of the bacteria. The V. cholerae O1 isolates demonstrated resistance to ampicillin, amoxicillin and tetracycline, but sensitivity to gentamicin, chloramphenicol, and ciprofloxacin. Thus, toxigenic V. cholerae O1 were present and seem to persist in environmental samples during a non-cholera outbreak period. Such an environmental reservoir is of food safety and public health concern and shows that environmental reservoirs of V. cholerae O1 should be studied and monitored as part of cholera prevention and control programmes.
Authors’ contributions
All authors contributed equally in the achievement of this study since the designing up to final submission of the manuscript. Specifically, YMGH was involved in study design, samples collection and laboratory analyses, statistics and the final manuscript writing. Laboratory analyses were assisted by JM, GM, OJM and ESM. Data analyses and interpretation and manuscript editing were assisted by TVD and OJM. RHM and AD were the supervisors of the study and involved in study design, resources mobilisation, accuracy of methodology, manuscript editing and proof-reading. All authors read and approved the final manuscript.
Acknowledgements
Authors thank the Sokoine University of Agriculture for providing the technical setting for this study. They also present their sincere gratitude to the below mentioned funders. Particular gratitude is also expressed to Professor Dominic Kambarage, Current Vice-Chancellor of Mwalimu Julius Nyerere University of Agriculture and Technology in Tanzania, to Professor Phyllis Addo from Noguchi Memorial Institute for Medical Research, University of Ghana and to Professor Ezron Karimuribo from Sokoine University of Agriculture for their respective supports.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
The current manuscript contains no individual person’s data. Therefore consent to publish is not applicable.
Ethics
The present study required no ethical approval since the samples were from the environment. Although fish samples were collected in this study, approval was not required for such studies according to the Animal welfare committee of Sokoine University of Agriculture that provides ethical clearance for animal studies in Tanzania. However, Research permits where obtained from Morogoro Municipal Council for water and vegetables sampling from Morogoro River and Funga–Funga vegetable production site; Mzumbe University Health Centre for fish, water and vegetables sampling from Mzumbe sewage ponds and MORUWASA (Morogoro Urban Water Supply and Sanitation Authority) for water sampling from Mafisa sampling site. All these permits are uploaded in the supplementary materials of this paper.
Funding
The study was co-funded by the “Enhancing the Community of Practice in One Health for Infectious Diseases Project” of the INTRA-ACP Academic Mobility scheme and “Safe Water for Food (SAWAFO) project” of Danish International Development Agency (DANIDA).
Contributor Information
Yaovi M. G. Hounmanou, Email: gilmahu@yahoo.fr
Robinson H. Mdegela, Email: rmdegela2012@gmail.com
Tamègnon V. Dougnon, Email: victorien88@hotmail.com
Ofred J. Mhongole, Email: ojmmhongole@yahoo.co.uk
Edward S. Mayila, Email: edwardbabasam@yahoo.com
Joseph Malakalinga, Email: malakalingajoseph@yahoo.com.
George Makingi, Email: makingi19@yahoo.co.uk.
Anders Dalsgaard, Email: adal@sund.ku.dk.
References
- 1.Foeken D, Sofer M, Mlozi M. Urban agriculture in Tanzania, Issues of sustainability African Studies Centre Research Report 75. Leiden: African Studies Centre; 2004. [Google Scholar]
- 2.Murnyak D. Fish Farming: Basics of Raising Tilapia & Implementing Aquaculture Projects. North Fort Myers: Echo Technical Note; 2010. [Google Scholar]
- 3.Sathiyamurthy K, Athmanathan B, Subbaraj DK. Prevalence of Vibrio cholerae and other Vibrios from environmental and seafood sources, Tamil Nadu, India. Br Microbiol Res J. 2013;3(4):538–549. doi: 10.9734/BMRJ/2013/4805. [DOI] [Google Scholar]
- 4.Mrityunjoy A, Kaniz F, Fahmida J, Shanzida JS, Md-Aftab U, Rashed N. Prevalence of Vibrio cholerae in different food samples in the city of Dhaka, Bangladesh. Int Food Res J. 2013;20(2):1017–1022. [Google Scholar]
- 5.Mishra A, Taneja N, Sharma M. Environmental and epidemiological surveillance of Vibrio cholerae in a cholera-endemic region in India with freshwater environs. J Appl Microbiol. 2012;112(1):225–237. doi: 10.1111/j.1365-2672.2011.05191.x. [DOI] [PubMed] [Google Scholar]
- 6.Fooladi IAA, Islamieh ID, Doust HR, Karami A, Marashi SM. Design of a multiplex PCR method for detection of toxigenic pathogenic in Vibrio cholerae. Asian Pac J Trop Med. 2013;6(2):115–118. doi: 10.1016/S1995-7645(13)60005-X. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Cholera country profile: United Republic of Tanzania. 2008.http://www.who.int/cholera/countries/TanzaniaCountryProfile2008.pdf. Accessed 12 February 2015.
- 8.World Health Organization. Cholera: situation in WHO African region as of 03 June 2013. 2013. http://www.afro.who.int/en/clusters-a-programmes/dpc/epidemic-a-pandemic-alert-and-response/3852-cholera-situation-in-the-who-african-region-as-of-03-june-2013.html. Accessed 20 Feb 2015.
- 9.Mandal J, Dinoop KP, Parija SC. Increasing antimicrobial resistance of Vibrio cholerae O1 biotype El Tor strains isolated in a tertiary-care centre in India. J Health Population Nutr. 2012;30(1):12–16. doi: 10.3329/jhpn.v30i1.11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . Guidelines for drinking-water quality. 2. Geneva: World Health Organization; 1997. [Google Scholar]
- 11.Elliot EL. Vibrio cholerae, V parahaemolyticus, V vulnificus, and other Vibrio spp. In: United States Food and Drug Administration, editor. Bacteriological analytical manual online. Washington: US Government; 2001. pp. 171–187. [Google Scholar]
- 12.Park JY, Jeon S, Kim JY, Park M, Kim S. Multiplex Real-time polymerase chain reaction assays for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Osong Public Health Res Perspect. 2013;4(3):133–139. doi: 10.1016/j.phrp.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido-Maestu A, Chapela M, Peñaranda E, Vieites JM, Cabado AG. In-house validation of novel multiplex real-time PCR gene combination for the simultaneous detection of the main human pathogenic vibrios (Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus) Food Control. 2014;37:371–379. doi: 10.1016/j.foodcont.2013.09.026. [DOI] [Google Scholar]
- 14.Wei S, Zhao H, Xian Y, Hussain MA, Wu X. Multiplex PCR assays for the detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an internal amplification control. Diagn Microbiol Infect Dis. 2014;79:115–118. doi: 10.1016/j.diagmicrobio.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute, CLSI. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. M100. 2007;27(1):182.
- 16.World Health Organization (2015). Cholera—United Republic of Tanzania. Disease outbreak news. 2015. http://www.who.int/csr/don/26-november-2015-cholera-tanzania/en/. Accessed 27 November 2015.
- 17.Sánchez J, Jan H. Cholera toxin-A foe & a friend. Ind J Med Res. 2011;133:153–163. [PMC free article] [PubMed] [Google Scholar]
- 18.Nkoko DB, Giraudoux P, Plisnier PD, Tinda AM, Piarroux M, Sudre B, Horion S, et al. Dynamics of Cholera outbreaks in great lakes region of Africa, 1978–2008. Emerg Infect Dis. 2011;17:2026–2034. doi: 10.3201/eid1711.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque SM, Asadulghani Kamruzzaman M, Nandi RK, Ghosh AN, Nair GB, Mekalanos JJ, Sack DA. RS1 Element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTX phage. Infect Immun. 2002;70(1):163–170. doi: 10.1128/IAI.70.1.163-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faruque SM, Mekalanos JJ. Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence. 2012;3(7):1–10. doi: 10.4161/viru.22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senderovich Y, Izhaki I, Halpern M. Fish as reservoirs and vectors of Vibrio cholerae. PLoS ONE. 2010;5(1):1–5. doi: 10.1371/journal.pone.0008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onyuka JHO, Kakai R, Onyango DM, Arama PF, Gichuki J, Ofulla AVO. Prevalence and antimicrobial susceptibility patterns of enteric bacteria isolated from water and fish in lake Victoria Basin of Western Kenya. World Acad Sci Eng Technol. 2011;5:3–22. [Google Scholar]
- 23.Akoachere JTK, Masalla TN, Njom HA. Multi-drug resistant toxigenic Vibrio cholerae O1 is persistent in water sources in New Bell-Douala, Cameroon. BMC Infect Dis. 2013;13:366. doi: 10.1186/1471-2334-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev. 2005;85:97–177. doi: 10.1152/physrev.00050.2003. [DOI] [PubMed] [Google Scholar]
- 25.Singh DV, Sree RI, Colwell RR. Development of a hexaplex PCR assay for rapid detection of virulence and regulatory genes in Vibrio cholera and Vibrio mimicus. J Clin Microbiol. 2002;40(11):4321–4324. doi: 10.1128/JCM.40.11.4321-4324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenpeng G, Yin J, Yang J, Li C, Chen Y, Yin J, Xu W, Zhao S, Liang J, Jing H, Fu X. Characterization of Vibrio cholerae from 1986 to 2012 in Yunnan Province, southwest China bordering Myanmar. Infect Genet Evol. 2014;21:1–7. doi: 10.1016/j.meegid.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Wong H, You W, Chen S. Detection of toxigenic Vibrio cholerae, V. parahaemolyticusand V. vulnificus in oyster by multiplex-pcr with internal amplification control. J Food Drug Anal. 2012;20(1):48–58. [Google Scholar]
- 28.Chikwendu CI, Ibe SN, Okpokwasili GC. Multiple antimicrobial resistance in vibrio spp isolated from river and aquaculture water sources in Imo state, Nigeria. Br Microbiol Res J. 2014;4(5):560–569. doi: 10.9734/BMRJ/2014/4896. [DOI] [Google Scholar]


