Abstract
The B-subunit (p70/Pol12p) of the DNA polymerase α-primase (Polα-primase) complex is thought to have a regulatory role in an early stage of S phase. We generated a panel of fission yeast thermosensitive mutants of the B-subunit (termed Spb70) to investigate its role in initiation of DNA replication by genetic and biochemical approaches. Here, we show that the fission yeast Spb70 genetically interacts and coprecipitates with origin recognition complex proteins Orp1/Orc1 and Orp2/Orc2 and primase coupling subunit Spp2/p58. A fraction of Spb70 associates with Orp2 on chromatin throughout the cell cycle independent of the other subunits of Polα-primase. Furthermore, primase Spp2/p58 subunit preferentially associates with the unphosphorylated Orp2, and the association requires Spb70. Mutations in orp2+ that abolish or mimic the Cdc2 phosphorylation of Orp2 suppress or exacerbate the thermosensitivity of the spb70 mutants, respectively, indicating that an unphosphorylated Orp2 promotes an Spb70-dependent replication event. Together, these results indicate that the chromatin-bound B-subunit in association with origin recognition complex mediates recruiting Polα-primase complex onto replication origins in G1 pre-Start through an interaction with primase Spp2/p58 subunit. Our results thus suggest a role for the recruited Polα-primase in the initiation of both leading and lagging strands at the replication origins.
Initiation of eukaryotic chromosome replication requires the assembly of multiprotein complexes onto the chromosomal replication origins. Upon activation of the multiprotein complexes by two types of kinases, cyclin-dependent kinases (CDK) and Cdc7/Dbf4 kinase (DDK), cells transit from prereplication stage in G1 to replication in S phase (7). A principal player in the replication complex for initiation of replication is DNA polymerase α-primase (Polα-primase). Polα-primase is a heterotetrameric enzyme complex, which is unique among the replicative polymerases for its ability to initiate de novo DNA synthesis of a short RNA primer on the leading and lagging strand templates by the primase activity and to extend RNA primer into a 35-nucleotide RNA-DNA primer, termed initiator DNA, by the DNA polymerase activity (6, 42, 44). The largest subunit of Polα-primase complex is of 180 kDa (p180), and it contains the DNA polymerase catalytic activity. The primase activity resides in two smaller subunits of 58 (p58) and 49 (p49) kDa. The p49 is the primase catalytic subunit that synthesizes the RNA primer. The p58 subunit is the coupling subunit of the p49 to p180 and is thought to play a role in regulating the length of RNA primer synthesis (3, 11, 37). The 70-kDa subunit, also named B-subunit, has no detectable enzymatic activity. B-subunit is essential for budding yeast (Saccharomyces cerevisiae) cell viability and has been proposed to execute a critical function in an initial stage of S phase prior to the hydroxyurea (HU) arrest point. Thus, B-subunit has been proposed to play a regulatory role in initiation of S phase (15). The B-subunits in human cells and budding yeast are both phosphorylated in a cell cycle-regulated manner. These findings further support its regulatory role (14, 29). An earlier study of the human B-subunit has shown that the B-subunit interacts with simian virus 40 (SV40) T antigen at the SV40 replication origin and suggested that the B-subunit tethers the Polα catalytic subunit (p180) to interact with T antigen in promoting initiation at the SV40 replication origin and facilitating the subsequent priming and synthesis of DNA chain (10). A recent study of the reconstituted SV40 replication assay has further demonstrated the requirement of B-subunit for initiation of in vitro SV40 replication and priming and elongation of replication protein A (RPA)-coated single-stranded DNA template (31).
Initiation of replication begins with assembly of the prereplication complex (pre-RC) at origins of replication during M phase to G1 phase; it is an event involving origin recognition complexes (ORCs), Cdc6, Cdt1, and minichromosome maintenance 2-7 (MCM2-7) (7). After CDK and DDK activation, additional replication factors that promote DNA unwinding and DNA synthesis including the replicative DNA polymerases assemble into the RC. Experiments in Xenopus egg extract, budding yeast, fission yeast, and human cells have shown that loading of Polα-primase onto chromatin and replication origin requires Cdc45 (1, 22, 26, 40, 47). In addition to Polα-primase, Cdc45 also associates with ORC/Orp, RPA, Polɛ, and the MCM complex (21, 22, 34, 38, 39, 43, 47). Together, these studies suggest that Cdc45 plays a critical role in coordinating origin sequence unwinding by MCMs and initiation of DNA synthesis by Polα-primase. Studies of fission yeast initiation have suggested that Polα-primase associates with MCM proteins at G1/S through interaction with Cdc45 (40). These chains of protein-protein interactions are thought to promote transition from pre-RC to RC at the replication fork in coordination with the cell cycle transition from G1 to S phase.
An early study of budding yeast B-subunit of Polα-primase has shown that the B-subunit executes at a step in DNA replication prior to the HU arrest point. It has been proposed in this budding yeast study that B-subunit (POL12 gene product) interacts with Orcs to mediate the interaction between Polα-primase complex and Orcs (15). Thus far, there is no experimental evidence to support this hypothesis. A later budding yeast study has also shown that Polα-primase associates with chromatin in G1 before Start in a cell cycle-regulated manner. The Polα-primase chromatin association is independent of Cdc6 and enhanced by inhibition of Cdc28/Clb kinase, and the association requires the primase subunits (12). The chromatin association of Polα-primase in G1 has been proposed in this study as a Cdc6-independent mitotic resetting event (12). A study of fission yeast has suggested that loading of Polα onto replication origins occurs by two steps: Polα is loaded onto chromatin at pre-Start and also associates with pre-RC on chromatin at G1/S junction through Cdc45, which interacts with MCM (40). Despite all of these studies of budding and fission yeast, the role of Polα-primase being loaded onto chromatin in early G1 is still unclear.
In fission yeast, after assembly of the pre-RC, Cdc2 kinase is activated, and along with DDK (termed Hsk1 kinase in fission yeast), these trigger the transition from G1 to S phase (13, 19, 20, 23, 27). Cdc2 kinase also regulates the fission yeast ORC (termed Orps) at the replication origin in a cell cycle-regulated manner (24, 41). Cdc2 kinase phosphorylates a subunit of the replication ORC Orp2 (homolog of budding yeast Orc2p). The phosphorylation of Orp2 by Cdc2 kinase is a redundant process to prevent rereplication during S and G2, it is not for triggering the transition from G1 to S phase (41, 45). In G1 pre-Start stage, Orp2 is not phosphorylated, consistent with the finding that a low level of CDK kinase activity is a prerequisite for pre-RC assembly (33).
Here, we generate a panel of fission yeast B-subunit thermosensitive mutants. We show that the B-subunit in fission yeast (Spb70) physically and genetically interacts with origin recognition proteins Orp1 (Orc1) and Orp2 (Orc2). A fraction of the Spb70 is tightly chromatin bound independent of the other Polα-primase subunits and associates with Orps throughout the cell cycle. The phosphorylation status of Orp2 profoundly influences the thermosensitive growth of sbp70 mutants. In G1 pre-Start, the primase coupling subunit Spp2/p58 in the Polα-primase complex physically associates with the unphosphorylated Orp2/Orc2 on the chromatin prior to MCMs loading onto chromatin. Furthermore, the association between Spp2/p58 and Orp2 requires the B-subunit. Our data indicate that the chromatin-bound B-subunit in association with the Orp complex recruits the Polα-primase complex onto chromatin during G1 pre-Start. The biological role of the Polα-primase recruited by the chromatin-bound B-subunit Orps to the replication origin during G1 pre-Start is proposed.
MATERIALS AND METHODS
Strains.
Unless otherwise stated, the strains used were all derived from either KG2 h− ade6-M216 ura4-D18 leu1-32 his3-D1 or KG3 h+ ade6-M210 ura4-D18 leu1-32 his3-D1 (9). All of the mutant strains and tagged strains were integrants at their respective endogenous gene loci under the control of their own promoters. The presence of epitope tags in all strains used in this study did not cause any growth defects under all experimental conditions. Strains used in this study are listed in Table 1.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| KG2 | h−ade6-M216 leu1-32 ura4-D18 his3-D1 | 9 |
| KG3 | h+ade6-M210 leu1-32 ura4-D18 his3-D1 | 9 |
| spb70-U01 | h−spb70::spb70-U01 ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U02 | h−spb70-U02 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U03 | h−spb70-U03 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U04 | h−spb70-U04 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U05 | h−spb70-U05 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U12 | h−spb70-U12 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U13 | h−spb70-U13 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U14 | h−spb70-U14 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spb70-U15 | h−spb70-U15 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| mhp70 | h− two-myc-six-His-spb70 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| mhu02 | h− two-myc-six-His-spb70-U02 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| mhu03 | h− two-myc-six-His-spb70-U02 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| mhu04 | h− two-myc-six-His-spb70-U02 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| mhu12 | h− two-myc-six-His-spb70-U02 ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| DBts131 | h+polαt13 ade6-M210 leu1-32 ura4-D18 | 8 |
| goa1-U53 | h−spcdc45goal-U53 ade6-M216 leu1-32 ura4-D18 | 39 |
| ST108 | h−spp2::spp2-8 ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | 37 |
| JLP445A | h+orp2::orp2-T4A leu1+ ade6-M210 leu1-32 ura4-D18 his7-366 | 41 |
| JLP605 | h+orp2::orp2-T3D leu1+ ade6-M210 leu1-32 ura4-D18 his7-366 | 41 |
| Spp2-HA | h−spp2::spp2-three-HA ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| Orp2-FLAG | h−orp2::orp2-three-FLAG his3+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| orp1-4 | h−orp1-4 ade6-M216 leu1-32 ura4-D18 | 16 |
| HA-orp1 | h−HA-orp1 | 16 |
Media.
For all genetic analyses, including fluorescence-activated cell sorter (FACS) analysis, YES media (yeast extract, 2% glucose, and appropriate supplements) were used. Solid media are 2% agar plates of YES media containing 3 μg of phloxine B/ml. Edinburgh minimal medium (EMM) and appropriate supplements were used for all biochemical analyses.
Genetic and cytological analysis.
All standard genetic analyses were performed as described previously in reference 28. To characterize the temperature sensitivity and genetic interaction, we utilized the serial dilution growth assay as described in reference 37. For cytological analysis, cells were fixed in 70% ethanol and stained by either 5 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml only or 5 μg of DAPI/ml and 1 μg of calcofluor/ml.
Molecular biology techniques.
Unless otherwise stated, all molecular biology techniques were as described previously (4). Transformation of yeast strains was performed by the lithium acetate procedure. DNA content measured by FACS analysis was performed as described in reference 35. PCR was conducted by ExTaq polymerase (TAKARA) to eliminate the possible amplification mistakes for cloning. For mutagenesis, PCR was conducted with Taq polymerase.
Strain constructions. (i) Temperature-sensitive mutants.
The genomic fragment from the KpnI site (307 bp upstream of the Start codon) to 337 bp downstream from the NurI site containing the entire coding region of spb70+ (SPCC553.09c) was amplified by standard PCR technique from KG2 genomic DNA and cloned onto KpnI-PstI of pUC18 vector. The HindIII fragment containing the ura4 gene from pREP4 vector was cloned in the NurI site (258 bp downstream of the Stop codon) for spb70-U01 to spb70-U12. To facilitate the pop-out of the ura4 marker by 5′-fluoro-orotic acid (FOA) counterselection (18), the EcoRV (298 bp upstream of the Stop codon) to NurI fragment was cloned in the BlpI site at the end of the ura4 gene fragment for spb70-U02 to spb70-U12. Another gene fragment from 151 bp upstream of KpnI to the SacI site (82 bp upstream of the Stop codon) was cloned onto PstI-SacI of pUC18 vector, and then the two insertions, the ura4 gene and the above 151-bp fragment to the KpnI site, were introduced at the KpnI site for spb70-U13 to spb70-U15. The KpnI-PstI fragment and PstI-SacI fragment were amplified by PCR, and the amplified fragments, carrying random mutations, were transformed into the KG2 strain. The transformed cells were plated onto EMM without uracil. The ura4+ cells were examined for the temperature sensitivity by replica plating. Potential clones were tested by PCR with primers, one residing in the ura4 gene and the other residing outside of the fragments, to confirm that the gene fragment was integrated into the correct locus and further tested by transforming pREP41X-spb70+ to test the ability of spb70+ to complement the temperature sensitivity. The mutants, spb70-U02 to spb70-U15, were plated onto YES plates with 5′-FOA to obtain the strain without the ura4 marker. Candidates were examined by PCR to confirm whether the ura4 gene was successfully removed from the locus adjacent to spb70. The mutant alleles were determined by sequence analysis of the PCR-amplified fragment from the genomic DNA of each mutant. The individual mutation allele was independently subcloned into the KG2 strain, followed by testing each single mutant for thermosensitivity as described above.
(ii) Heterozygotic diploid strain.
The KpnI-PstI fragment in pUC18 vector was used to construct the deletion fragment. The HindIII fragment containing the ura4+ gene from pREP4 vector was cloned in place of the StuI (151 bp downstream of the Start codon) to NurI region. The resulting KpnI-PstI fragment was amplified by PCR and transformed into a KG2 strain carrying pREP41X-spb70+. Candidates with deletions were tested by PCR. The haploid deletion strain was then mated with the KG3 strain and plated on EMM containing leucine but lucking adenine to obtain a heterogeneous diploid strain carrying spb70+/spb70Δ without the pREP41X-spb70+ plasmid.
(iii) Epitope-tagged spb70 strains.
The PstI-SacI fragment on pUC18 vector was utilized to construct two-myc- and six-His-tagged spb70+. In place of the Start codon of spb70+, XhoI and BglII sites were introduced by PCR. The two-myc-six-His fragment was amplified by PCR and introduced into the XhoI-BglII sites. For tagging thermosensitive mutants, mutated versions of PstI-SacI fragments were constructed, amplified by PCR, and transformed into KG2 and spb70 thermosensitive mutant strains. The candidates were tested by PCR and Western blotting with anti-myc antibody. The ura4 markers were then removed by the counterselection with 5′-FOA.
Epitope-tagged orp2 and spp2 strains.
The C-terminal epitope tagging was conducted as described in reference 36. The kanr fragment of pFA6a-3XHA-kanMX6 (5) was replaced by ura4 for tagging the orp2+ and by his3 for tagging the spp2+ gene fragment. To construct the FLAG tag, the three-hemagglutinin (HA) fragment of pFA6a-3XHA-kanMX6 was replaced by the three-FLAG (Sigma-Aldrich) fragment. The 1,082-bp C-terminal fragment of orp2+ (from NaeI to end) and 685-bp C-terminal fragment of spp2+ (from PshAI site to the termination) were amplified by PCR from KG2 genomic DNA and cloned into the PvuII-BamHI region of the tagging vectors. The resulting plasmids, named pFA64-orp2-3XFLAG and pFA63-spp2-3XHA, were linearized by XbaI and SalI, respectively, and transformed into the KG2 strain. The possible integrants were examined by PCR and Western blotting with appropriate antibodies.
Preparation of cell extracts and protein analysis.
Harvested cells were immediately frozen at −80°C prior to all of the extraction process. Cell fractionation is outlined in Fig. 1C.
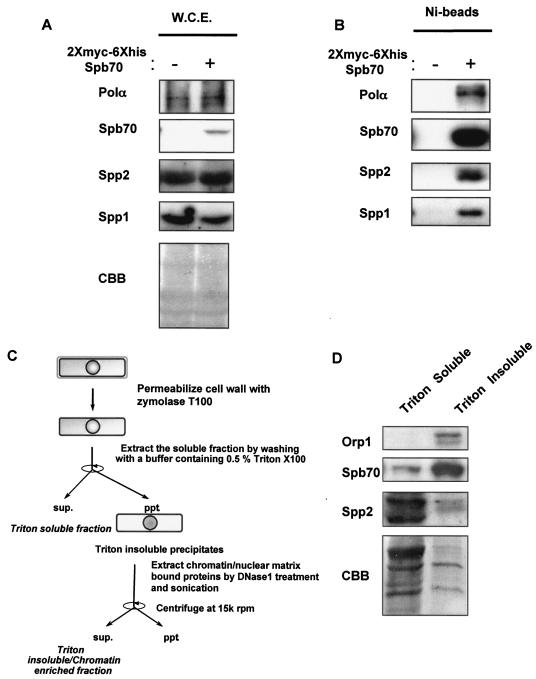
FIG. 1.
Identification of the fission yeast B-subunit of DNA Polα-primase complex. (A) Expression of two-myc-six-His-tagged Spb70 in cells. The two-myc- and six-His-tagged spb70+ and untagged spb70+ strain were grown at 25°C in EMM to mid-log phase. Cells were harvested and whole-cell extracts (W.C.E.) were prepared as described in Materials and Methods. The expression of Polα/p180, Spp2/p58, and Spp1/p49 and absence of two-myc-six-His-tagged Spb70 in whole-cell extracts from cells containing spb70+ with no epitope tag (−) and expression of two-myc-six-His-tagged Spb70 in whole-cell extracts from cells containing the two-myc-six-His-tagged spb70+ (+) are shown by Western blotting with their respective antibodies and anti-myc antibody. CBB, the Coomassie brilliant blue staining of the Western blot membrane. (B) Spb70 physically associates with Polα/p180 and primase Spp1/p49 and Spp2/p58. Ni bead pull-down fractions of whole-cell extracts from the untagged spb70+ strain (−) and from the two-myc-six-His-tagged spb70+ strain (+) were blotted with their respective antibodies. (C) Schematic diagram of cell fractionation. (D) Spb70 localizes in both Triton X-100-soluble and -insoluble fractions. The strain containing two-myc-six-His-spb70+ and HA-orp1+ was grown at 25°C in EMM to mid-log phase. Cells were harvested, and Triton X-100-soluble and -insoluble (chromatin-enriched) fractions were prepared as described in Materials and Methods and outlined in panel C. HA-tagged Orp1 was used as the Triton X-100-insoluble protein control in chromatin-enriched fraction; two-myc-six-His-Spb70 and Spp2/p58 in each fraction was shown by Western blotting with their respective antibodies. As described in Materials and Methods, the intensity of each protein shown in Western blotting does not represent their molar quantities in cell extracts and Ni bead pull-downs due to the difference in reactivity of each antibody. CBB, the Coomassie brilliant blue staining of the immunoblot membrane.
Whole-cell extracts.
Harvested cells were washed twice with 3 volumes of cold water and then washed twice by 3 volumes of EBT buffer (25 mM HEPES, pH 7.6, 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM NaF, 0.1 mM vanadate, 1 mM dithiothreitol [DTT], 20 mM β-glycerophosphate, 0.25% Triton X-100, and proteinase inhibitors). Cells were then resuspended in 1 volume of EBT. An equal volume of acid-washed glass beads was then added. Cells were broken by FastPrep (Qbiogene), and glass beads were removed by filtration. DNase I was added into crude cell lysates including cell debris to a final concentration of 100 to 200 U/ml and incubated at 32°C for 5 min, followed by further treatment by sonication. The sonicated cell lysates were then centrifuged in a TOMY MRX150 microcentrifuge at 15,000 rpm for 25 min. The supernatants were used as whole-cell extracts. For the Ni bead pull-down experiments, EBN buffer (25 mM HEPES, pH 7.6, 150 mM NaCl, 2 mM MgCl2, 1 mM NaF, 0.1 mM vanadate, 6 mM imidazole, 20 mM β-glycerophosphate, 0.25% Triton X-100, and proteinase inhibitors) was used instead of EBT buffer.
Chromatin-enriched fractions.
Cells were washed twice with 3 volumes of cold water followed by 3 volumes of SB buffer (25 mM HEPES, pH 7.6, 1.2 M sorbitol, 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM NaF, 0.1 mM vanadate, 1 mM DTT, 20 mM β-glycerophosphate, and proteinase inhibitors). To generate spheroplasts, cells were resuspended in an adjusted volume of SB buffer containing Zymorase T100 based on the weight of each cell pellet to ensure equal amounts of total protein analyzed in different time points or fractions and incubated 15 min at 32°C. After washing three times with 3 volumes of SB buffer, the spheroplasts were extracted by incubation in 2 volumes of EBT buffer (25 mM HEPES, pH 7.6, 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM NaF, 0.1 mM Vanadate, 1 mM DTT, 20 mM β-glycerophosphate, 0.5% Triton X-100, and proteinase inhibitors) at 4°C for 30 min followed by low-speed centrifugation. The supernatant was designated as a Triton X-100-soluble fraction. The Triton X-100-insoluble pellets were washed four times with 3 volumes of EBT and resuspended in 1 volume of EBT. DNase I was added to a final concentration of 100 to 200 U/ml and further incubated at 32°C for 5 min followed by sonication and centrifugation at 15,000 rpm for 25 min. The supernatant fraction is the Triton X-100-insoluble chromatin-enriched fraction. For simplicity, we designated this fraction as chromatin fraction.
Immunoprecipitation and Ni bead pull-down.
Immunoprecipitations were performed as previously described in reference 32, with the following modification. EBT buffer containing the same concentration of Triton X-100 as in the whole-cell extract was used as washing buffer. An equal volume mixture of protein A and protein G Sepharose beads was used for immunoprecipitation. Extracts were first precleared by incubating with protein A/G Sepharose beads without antibody to remove nonspecific proteins absorbed to protein A/G Sepharose. For Ni bead pull-down, TALON metal affinity resin (BD Biosciences) was used to precipitate six-His-tagged proteins and washed with EBN. Proteins bound to either immunoaffinity beads or Ni affinity beads were washed four times by appropriate washing buffers and resuspended directly with sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample loading buffer. The proteins in the extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% acrylamide with 29:1 acrylamide to bisacrylamide) and then transferred onto polyvinylidene difluoride membranes by semidry blotting apparatus. The transferred membranes were then subjected to Western blotting with appropriate antibodies.
It is important to mention here that there are differences in each antibody's reactivity against the respective protein. The intensity of each protein band shown by Western blotting does not reflect the molar quantities of these proteins in cell extracts, Ni bead pull-downs, and immunoprecipitates. Notably, the antibody against Polα/p180 is an immunoglobulin Y (32), which is a rather weak antibody and often has difficulty detecting Polα/p180 protein in crude cell extracts by Western blotting.
RESULTS
Identification of the fission yeast B-subunit of DNA Polα-primase.
Analysis of the fission yeast genome database revealed that SPCC553.09c contains 30% identify to the B-subunit of budding yeast POL12 and human B-subunit (p70) of Polα-primase complex. To investigate whether this clone encodes the fission yeast ortholog of budding yeast POL12 and the human B-subunit of Polα-primase, we first tested if SPCC553.09c is essential for growth by constructing a heterozygotic diploid strain with one copy of this gene disrupted. Germinating spores from this diploid yielded two viable spores, indicating that SPCC553.09c, similar to the budding yeast POL12, is essential for cell growth. To ensure that this gene encodes the ortholog of budding yeast POL12 and the B-subunit of Polα-primase in mammalian cells, we constructed a strain containing a two-myc-six-His epitope tag at the start site of the gene at its endogenous chromosomal locus. Western blotting of the whole-cell extracts that contain the two-myc-six-His epitope-tagged SPCC553.09c with antibodies against Polα/p180 (32), Spp1/p49 (17), and Spp2/p58 (37) showed the presence of Polα/p180, the primase catalytic subunit Spp1/p49, and the primase coupling subunit Spp2/p58 (Fig. 1A, left panel). Western blotting with anti-myc antibody revealed a protein of molecular mass comparable to that of the budding yeast Pol12p and human B-subunit/p70 plus the molecular mass of the two-myc-six-His tag (Fig. 1A, right panel).
To further ensure that this gene encodes the ortholog of fission yeast B-subunit of Polα-primase complex, we used Ni beads to pull down the two-myc-six-His-tagged protein from the whole-cell extracts as described in Materials and Methods. Polα/p180 and the primase subunits, Spp1/p49 and Spp2/p58, were readily detected by their respective antibodies in the Ni bead pull-down fraction from whole-cell extracts containing the two-myc-six-His epitope-tagged SPCC553.09c, whereas none of these proteins were detected from extracts of cells without the two-myc-six-His epitope-tagged SPCC553.09c (Fig. 1B). These results confirm that the fission yeast SPCC553.09c gene indeed encodes the ortholog of budding yeast POL12 and the mammalian B-subunit/p70 of Polα-primase complex. We named the gene spb70+ (Schizosaccharomyces pombe B-subunit/p70) and the gene product Spb70.
Although the two-myc-six-His-tagged Spb70 protein was found in the soluble cellular fraction, a fraction of the two-myc-six-His-tagged Spb70 protein was also found in the chromatin-enriched fraction (Fig. 1C and D; also see Materials and Methods).
Characterization of cells with spb70+ mutations.
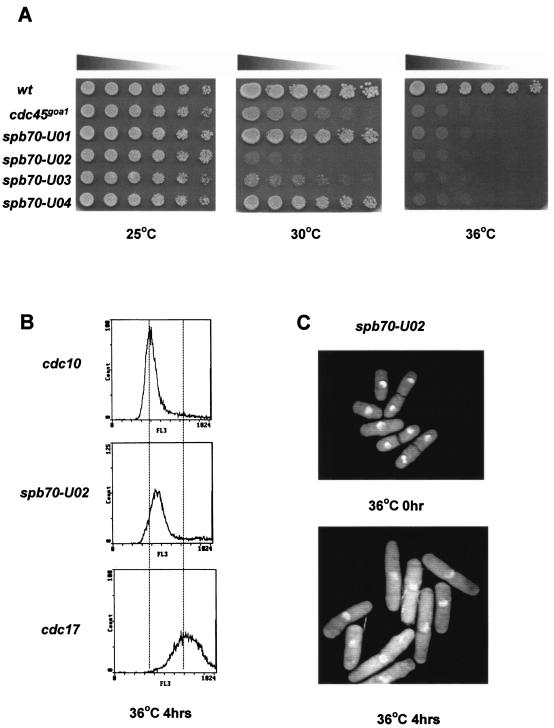
We generated a panel of thermosensitive mutants of spb70+ to investigate the role of Spb70 in DNA replication. The mutants were constructed by replacing the genomic copy of spb70+ with the mutated spb70+ fused with ura4+ by two different approaches. One had the ura4+ marker in the 3′ noncoding region, and the other approach had the ura4+ marker in the 5′ noncoding region, as described in Materials and Methods. Six mutants were isolated from the first approach and three mutants were isolated from the second approach. These mutants were confirmed by several rounds of testing for their growth at 25, 30, and 36°C, and the mutation alleles were identified by sequencing (Table 2). Two mutants, spb70-U02 and spb70-U14, exhibited severe thermosensitivity and failed to grow at 30°C, and spb70-U14 even exhibited a growth defect at 25°C (Table 2). Mutants that contain more than one mutation allele were further tested for mutation in each amino acid residue. Residues in each mutant responsible for thermosensitive growth are listed in Table 2. Mutant spb70-U05 contains mutations in two residues. Mutation in either one of the two residues in spb70-U05 did not exhibit temperature sensitivity of cell growth, indicating that the combination of both mutant alleles contributes to the thermosensitivity of growth (Table 2). At 30°C, these mutants show a different extent of thermosensitivity. Two mutants, spb70-U02 and spb70-U03, exhibited severe growth defects, whereas spb70-U01 and spb70-U04 exhibit a wild-type-like growth at 30°C (Fig. 2A). Flow cytometry analysis of these mutants (shown with spb70-U02 as the representative) after a 4-h shift to 36°C indicated that all of these mutants arrested in early to mid S phase, similar to that of the polαts mutant (8) (Fig. 2B). The terminal phenotype of these mutants (shown by using spb70-U02 as the representative) 4 h after a shift to 36°C showed that the spb70 mutants arrested with single nuclei and cdc phenotype, indicating that these mutants have an intact mitotic checkpoint (Fig. 2C).
TABLE 2.
spb70 mutants
| Mutant | Allelea | Growth at (°C)b:
|
|
|---|---|---|---|
| 25 | 30 | ||
| spb70-U01 | Q469R | ++ | + |
| spb70-U02 | L369S, H471Y | ++ | − |
| spb70-U03 | F450L, N527H | ++ | +/− |
| spb70-U04 | Q469K | ++ | + |
| spb70-U05 | H349L, V523A | ++ | ++ |
| spb70-U12 | L442P, N485S | ++ | + |
| spb70-U13 | S314P, 3 others | ++ | ND |
| spb70-U14 | G311R, S314P | +/− | − |
| spb70-U15 | S314P, 3 others | ++ | ND |
Bold allele is the mutation responsible for temperature sensitivity; italic allele causes constitutive growth defect.
++, no growth defect; +, mild growth defect; +/−, significant growth defect; −, no growth; ND, not determined. No growth was observed with any mutant at 36°C.
FIG. 2.
Thermosensitive mutants of spb70+. (A) Thermosensitivity of spb70 mutants. Wild type (wt), cdc45goa1, spb70-U01, spb70-U02, spb70-U03, and spb70-U04 were first cultured in liquid YES medium at 25°C. Equal numbers of log-phase cells were spotted on YES plates in threefold serial dilution and incubated at 25, 30, and 36°C for 3 days. (B) FACS profile of spb70-U02. cdc10-M17, spb70-U02, and cdc17-K42 were first grown at 25°C in YES medium followed by shifting to 36°C for 4 h. cdc10 and cdc17 were used as 1C and 2C DNA content standards, respectively. (C) Terminal phenotype of spb70-U02. Exponentially growing cells were shifted to 36°C for 4 h and stained with DAPI.
Effect of mutations in spb70+ on Polα and primase.
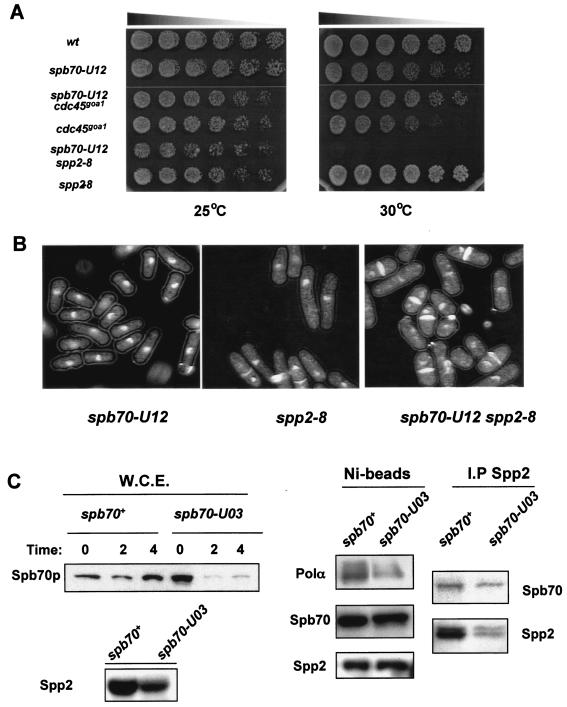
Previous studies have shown that polαts13 or spp2-8 mutants exhibit a mixed phenotype with a small fraction of the cells displaying mitotic catastrophic phenotype at the restrictive temperature of 36°C (8, 37). The finding that spb70 mutants exhibit cdc phenotype suggests that Spb70 might have a distinct role in initiation of replication that is different from that of the Polα catalytic subunit and primase coupling subunit Spp2/p58. To investigate this possibility, we first analyzed the genetic interactions of the spb70 mutants with polαts13 (8) and spp2-8 (37). Cdc45 was known to promote Polα-primase loading onto chromatin at G1/S transition (7). We thus also analyzed the genetic interaction of the spb70 mutant with cdc45goa1 (39, 40). Mutant spb70-U12, which exhibited a semipermissive growth at 30°C (Table 1 and Fig. 3A), was used as the representative for the analysis. Double mutant spb70-U12 polαts13 grew better than either the spb70-U12 or polαts13 single mutant at 30°C (data not shown). Similarly, the spb70-U12 cdc45goa1 double mutant at 30°C was also found to grow better than either of the single mutants (Fig. 3A). These results indicate that spb70-U12 has a suppressive genetic effect on polαts13 and cdc45goa1. In striking contrast, spb70-U12 exhibited an overt genetic interaction with spp2-8. Double mutant spb70-U12 spp2-8 showed apparent growth defects at 25°C. spp2-8 and spb70-U12 were synthetic lethal at 30°C (Fig. 3A). After 4 h at 36°C, spb70-U12 displayed a slight cdc phenotype with normal nuclear morphology, whereas spp2-8 displayed a mixed phenotype of cdc with a small population of cells exhibiting cut phenotype (37). In contrast, a majority of the double mutant spb70-U12 spp2-8 cells exhibited mitotic catastrophic cut phenotype (Fig. 3B).
FIG. 3.
Genetic and physical characterizations of spb70 mutants. (A) Genetic interactions. Wild type (wt), single mutants spb70-U12, cdc45goa1, and spp2-8, and double mutants spb70-U12 cdc45goa1 and spb70-U12 spp2-8 were cultured in liquid YES medium at 25°C. Equal numbers of log-phase cells were spotted on YES plates in threefold serial dilution and incubated at 25 and 30°C for 3 days. (B) Terminal phenotypes. Single mutants spb70-U12 and spp2-8 and double mutant spb70-U12 spp2-8 were first grown in liquid YES medium at 25°C then shifted up to 36°C for 4 h. Cells were harvested and stained with DAPI and calcofluor. (C) Stability of Spb70 protein and interaction of Spb70 with other Polα-primase subunits. Wild-type two-myc-six-His-spb70+ and mutant two-myc-six-His-spb70-U03 cells were cultured at 25°C in EMM to mid-log phase and then shifted to 36°C. At the indicated time, cells were harvested and whole-cell extracts (W.C.E.) were prepared. The Spb70 protein levels in whole-cell extracts from spb70+ and spb70-U03 cells were detected by anti-myc antibody (W.C.E. panel). Spp2/p58 protein levels in whole-cell extracts from spb70+ and spb70-U03 cells after the shift to 36°C for 4 h used for the Ni bead pull-down are shown in the bottom panel. Ni bead pull-down fractions from wild-type two-myc-six-His-spb70+ and mutant two-myc-six-His-spb70-U03 whole-cell extracts after shifting the cells to 36°C for 4 h were analyzed by Western blotting with anti-Polα antibody, anti-HA for Spp2/p58, and anti-myc antibody for Spb70 for association of Polα/p180 and Spp2/p58 with Spb70 protein, respectively (Ni-beads panel). HA-tagged Spp2 proteins were immunoprecipitated from whole-cell extracts of the wild type containing two-myc-six-His-spb70+ and spp2+-three-HA and the mutant containing two-myc-six-His-spb70-U03 and spp2+-three-HA with anti-HA antibody. The immunoprecipitates were analyzed by Western blotting with anti-myc for Spb70 and anti-HA antibody for Spp2 (I.P Spp2 panel).
Mutation in spp2-8 destabilizes the Polα-primase complex and cells with the spp2-8 mutation fail to activate HU-induced Cds1 kinase activity. These findings indicate that primase coupling subunit Spp2/p58 has a critical role in activation of the S-phase checkpoint and the mitotic checkpoint that depends on the synthesis of initiator DNA (8, 37). The finding that spp2-8 and spb70-U12 are synthetic lethal at 30°C prompted us to investigate whether mutations in spb70+ could also compromise the stability of the Polα-primase complex. Four mutant alleles spb70-U02, spb70-U03, spb70-U04, and spb70-U12 were individually epitope tagged with two-myc-six-His at the chromosomal locus. The Spb70 protein in each epitope-tagged spb70 mutant was tested for its ability to associate with Polα/p180 and Spp2/p58. Spb70 mutant protein from spb70-U03, which exhibits a moderate growth defect at 30°C (Table 2), was shown as the representative of the test; similar results were seen in other mutant strains. After 4 h at 36°C, Spb70 protein levels were significantly reduced in cell extracts from the spb70-U03 mutant compared to that from wild-type cells. However, Spp2/p58 was detectable in spb70-U03, albeit in a slightly reduced level after 4 h at 36°C, indicating that the other subunits are not as unstable as Spb70 (Fig. 3C, left bottom panel). Due to the weak reactivity of anti-Polα antibody (32) in detecting Polα/p180 protein in crude cell extract, Polα was detected in spb70-U03 crude cell extract in poor quality (data not shown). Despite having a substantially lower level of Spb70 protein in the spb70-U03 mutant, Ni beads were able to pull down the Spb70 mutant protein after 4 h at 36°C from whole-cell extracts of the mutant. With equal amounts of Spb70 proteins from the Ni bead pull-down analyzed by Western blotting, Polα/p180 and Spp2/p58 proteins were detected in the Ni bead pull-down fraction (Fig. 3C). Because of the difference in reactivity of anti-Polα and anti-Spp2, the intensities of these proteins do not represent their respective molar quantities associated with Spb70 in the Ni bead pull-down. To test whether the mutant Spb70 protein might compromise the association with Spp2/p58, wild-type two-myc-six-His-tagged spb70+ and the two-myc-six-His-tagged spb70-U03 mutant were independently constructed with three-HA-tagged spp2+. Immunoprecipitates of Spp2-three-HA were probed with anti-HA for the presence of Spp2/p58, and anti-myc antibody was used to detect the coprecipitation of Spb70. Spb70 protein was detected in the Spp2 anti-HA immunoprecipitates from the spb70+ cell extracts and the spb70 mutant cell extracts at a lower level (Fig. 3C). These experiments indicate that spb70 mutants contain intact Polα-primase complex; however, the protein level of Polα-primase complex could be lower in the spb70 mutants than the wild-type cells at 36°C due to the instability of the mutant Spb70.
Spb70 genetically and physically interacts with the ORC proteins Orp1/Orc1.
We then investigated whether the growth defect of the spb70 mutant at 30°C and the cell cycle arrest at 36°C are caused by defective interaction of the Spb70 mutant protein with another factor(s) essential for initiation of replication. We crossed the spb70-U03 mutant with various cdc mutants defective in initiation of replication. The double mutant of spb70-U03 orp1-4 (orp1-4 is a mutant of orp1 [ORC1]), a subunit of the origin recognition protein complex (16), was synthetic lethal at 30°C (Fig. 4A).
FIG. 4.
Spb70 genetically and physically interacts with ORC proteins. (A) Genetic interaction. Wild type (wt), orp1-4, spb70-U03, and double mutant spb70-U03 orp1-4 were cultured in liquid YES medium at 25°C. Equal numbers of log-phase cells were spotted on YES plates in threefold serial dilution and incubated at 25 and 30°C for 4 days. (B) Spb70 protein physically associates with Orp1/Orc1. HA-orp1+/orc1 and two-myc-six-His-spb70+ HA-orp1+ strains were grown at 25°C in EMM to mid-log phase and whole-cell extracts (W.C.E.) were prepared from each strain. Presence of Spb70 and Orp1 in whole-cell extracts was detected by anti-myc and anti-HA antibody, respectively (W.C.E. panel). Ni bead pull-down fractions were Western blotted with anti-myc for Spb70 and anti-HA for Orp1 (Ni-beads panel). (C) Spb70 protein physically associates with Orp2/Orc2, and a mutation in spb70+ compromises the interaction. Wild-type cells containing two-myc-six-His-spb70+ and orp2+-three-FLAG and the mutant containing two-myc-six-His-spb70-U03 and orp2+-three-FLAG were cultured at 36°C. Ni bead pull-down fractions from each strain's chromatin fraction were Western blotted with anti-myc and anti-FLAG for Spb70 and Orp2, respectively. The Orp2 proteins in the chromatin fraction of wild-type and mutant cells used for the Ni bead pull-down experiment are shown in the bottom panel.
The finding that spb70-U03 had strong genetic interaction with orp1-4 at 30°C (Fig. 4A) led us to analyze whether these two gene products physically associate at permissive temperature. HA-tagged Orp1 protein was readily detectable in whole-cell extracts from HA-tagged orp1+ (16) strains with or without two-myc-six-His-tagged spb70+ (Fig. 4B). Spb70 proteins were pulled down by Ni beads from whole-cell extracts of the strain containing two-myc-six-His-tagged spb70+ and HA-tagged orp1+ (16). HA-tagged Orp1 protein was detected coprecipitating with two-myc-six-His-tagged Spb70 in the Ni bead pull-down (Fig. 4B).
Fission yeast Orp proteins have been shown localizing in the chromatin-enriched fraction (30). We thus tested whether Spb70 in the chromatin-enriched fraction interacts with Orp2 (Orc2). Wild-type two-myc-six-His-spb70+ and mutant two-myc-six-His-spb70-U03 were constructed with the three-FLAG-tagged orp2+. Orp2 protein in the chromatin-enriched fraction from wild-type and mutant cells used for the Ni bead pull-down experiments are shown in Fig. 4C, bottom panel. Similar to Orp1, Orp2 was detected in the Ni bead pull-down from wild-type two-myc-six-His-spb70+ chromatin fraction. In contrast, at the restrictive temperature, when equal amounts of Spb70 protein being pulled down by Ni beads from chromatin fractions of wild type and mutant were analyzed, no Orp2 was detected in the Ni bead pull-down from two-myc-six-His-spb70-U03 chromatin fraction (Fig. 4C).
These experiments indicate that the fission yeast B-subunit of the Polα-primase complex not only genetically interacts with the ORC proteins, Orps (Orcs), but also physically associates either directly or indirectly with Orps (Orcs) in a protein complex. The compromised association of the B-subunit in the thermosensitive mutants with Orps at the restrictive temperature suggests that the association between chromatin-bound Spb70 and Orp2 is physiologically important.
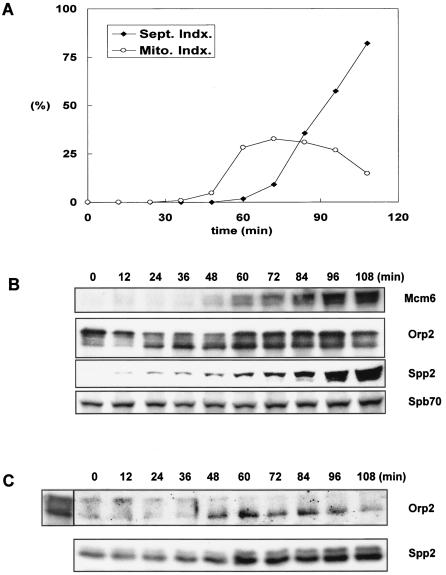
Spb70 interacts with Orp2 throughout the cell cycle.
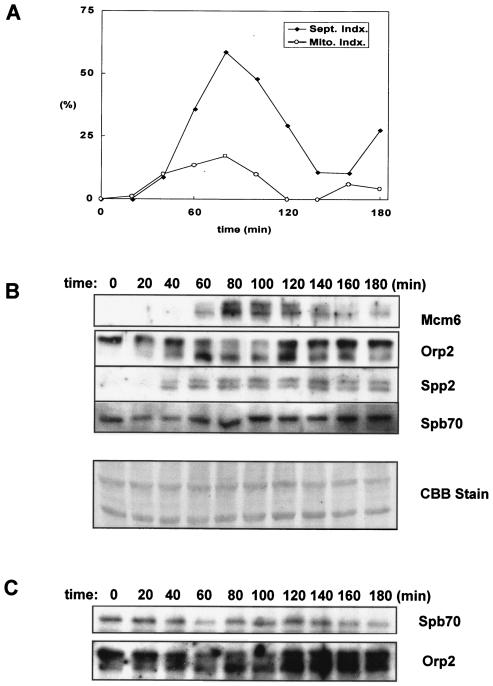
To test whether the interaction between the chromatin-bound Spb70 and Orps (Orcs) is cell cycle regulated, we constructed two-myc-six-His-tagged spb70+ and three-FLAG-tagged orp2+ at their respective genomic loci in the cdc25-22 strain. After release from temperature arrest at G2, cell samples were collected every 20 min for 3 h. Cell cycle progression was monitored by septation index and mitotic index (Fig. 5A). The septation index reached maximum at G1/S junction 80 min after release from G2 arrest. Chromatin fractions were prepared from each cell sample and probed for the presence of Mcm6, Spb70, Orp2/Orc2, and Spp2/p58 at each time point with their respective antibodies (Fig. 5B). Mcm6 protein began to appear in the chromatin fraction 60 min after release from G2 arrest and peaked at 80 min at the G1/S junction and then gradually diminished as previously reported (30). Orp2 was found in the chromatin-enriched cell fraction throughout the cell cycle and underwent cell cycle-dependent phosphorylation as evidenced by exhibiting a slower-mobility phosphorylated form of Orp2 (24, 41). The unphosphorylated Orp2 appeared 40 min after release from G2 arrest, and the abundance progressively increased from 60 to 120 min. Budding yeast Polα-primase was reported to be recruited onto chromatin during G1 independent of pre-RC formation, and the recruitment requires primase (12). We thus used the primase coupling subunit, Spp2/p58, as the representative to analyze the kinetics of Polα-primase complex chromatin association in the cell cycle. Spp2/p58 protein appeared in the chromatin fraction at 40 min after release from G2 arrest and was present from G1 to S phase, similar to the previously reported role of Polα in fission yeast (40). Interestingly, Spp2/p58 appeared in the chromatin fraction prior to the appearance of MCM6. This is consistent with the budding yeast studies that Polα chromatin binding dose not require Cdc6 and the Cdc28/Clb activation, indicating that Polα-primase is recruited onto chromatin in G1 pre-Start (12). In contrast to Spp2/p58, the Spb70 protein is present in chromatin fraction throughout the cell cycle (Fig. 5B).
FIG. 5.
Spb70 associates with Orp2/Orc2 throughout the cell cycle. The cdc25-22 strain containing two-myc-six-His-spb70+ and orp2+-three-FLAG was grown at 25°C in EMM to mid-log phase and then shifted to 36°C to arrest at G2 for 4 h. Cells were then released from G2 arrest by shift-down to 25°C and harvested every 20 min after release from G2 arrest. Chromatin fractions were prepared from each cell sample as described in Materials and Methods. Presence of Mcm6, Spb70, Orp2, and Spp2 in chromatin fraction was detected by Western blotting with anti-MCM6, anti-myc, anti-FLAG, and anti-Spp2 antibodies, respectively. (A) Cell cycle progression after the release from G2 arrest. Percentage of cells in different stages of the cell cycle was shown as the septation index or mitotic index. (B) Proteins in chromatin-enriched fractions. Kinetics of the appearance of MCM6, Orp2, primase Spp2/p58, and Spb70 in the chromatin fraction after release from G2 arrest were shown by Western blotting each protein with their respective antibody. CCB, the Coomassie brilliant blue staining of the Western blot membrane. (C) Coprecipitation of Spb70 and Orp2. Immunoprecipitates of Orp2 by anti-FLAG were Western blotted with anti-myc for Spb70 and anti-FLAG for Orp2.
As shown in Fig. 4, Spb70 protein associates with Orp2 in the chromatin fraction. We then tested whether the association of Spb70 with Orp2 (Orc2) in the chromatin fraction is cell cycle regulated. The Orp2 immunoprecipitates from the chromatin fraction were probed with anti-myc antibody for the coprecipitation of two-myc-six-His-Spb70. Spb70 proteins were found coprecipitating with Orp2 throughout the cell cycle with a moderate level of increase in cells entering G1/S junction (Fig. 5C). It is important to note that during the first 20 min of release from the G2 arrest, Spp2/p58 was not detected in the chromatin fraction by anti-Spp2 antibody (Fig. 5B), while Spb70 proteins were detected in association with Orp2 in the chromatin fraction throughout the cell cycle (Fig. 5C). These findings indicate that the Spb70 constitutively associated with Orp2p in the chromatin fraction is independent of Spp2/p58, Polα, and the primase catalytic subunit Spp1/p49.
Orp2 phosphorylation site mutants genetically interact with spb70 mutants.
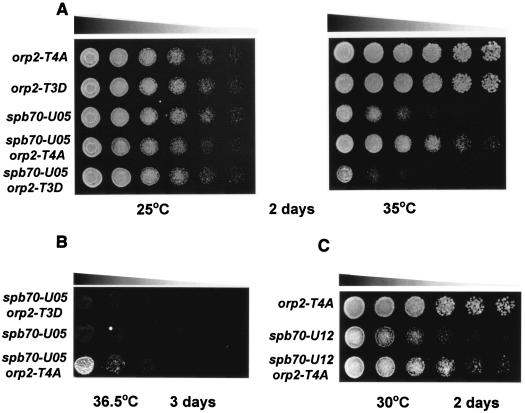
In fission yeast, Cdc2 kinase is the major kinase regulating the cell cycle progression. Phosphorylation of Orp2 by Cdc2 kinase is one of the several mechanisms by which Cdc2 regulates replication in a single cell cycle and is not required for activation of initiation of replication (41). The finding that Spb70 physically associates with Orp2 throughout the cell cycle led us to test whether phosphorylation of Orp2 by Cdc2 has an effect on spb70 mutants. We analyzed the genetic interactions of two orp2 phosphorylation site mutants, orp2-T4A and orp2-T3D (41), with spb70 mutants. Mutant Orp2-T4A protein lacks all four Cdc2 phosphorylation sites and allows rereplication, whereas Orp2-T3D mimics Orp2 phosphorylation, thus preventing rereplication from occurring (41, 45). These two orp2 phosphorylation mutant alleles were independently crossed into spb70-U05 and spb70-U12. Neither of the two orp2 mutants are thermosensitive in growth (Fig. 6A). The mutation in orp2-T4A that abolished Cdc2 phosphorylation of Orp2 suppressed the thermosensitivity of spb70-U05 at 35°C (Fig. 6A, right panel). In contrast, the mutation in orp2-T3D that mimicked constitutive phosphorylation of Orp2 exacerbated the thermosensitivity of spb70-U05 at 35°C (Fig. 6A, right panel). To ascertain the opposite effects of these two orp2 mutations on spb70-U05, the double mutants were further tested for their growth at a higher temperature of 36.5°C. The results confirmed the suppressive effect of orp2-T4A on spb70-U05 thermosensitivity (Fig. 6B). The suppressive effect is not specific to the spb70-U05 allele; a similar suppressive effect on the spb70-U12 thermosensitivity was observed at 30°C (Fig. 6C).
FIG. 6.
Phosphorylation and unphosphorylation of Orp2 affects the temperature-sensitive growth of spb70 mutants. (A) Mutants orp2-T4A and orp2-T3D exert opposite effects on spb70-U05. Single mutants orp2-T4A, orp2-T3D, and spb70-U05 and double mutants spb70-U05 orp2-T4A and spb70-U05 orp2-T3D were cultured in liquid YES medium at 25°C. Equal numbers of log-phase cells from each strain were spotted on YES plates in threefold serial dilution and incubated at 25 and 35°C for 2 days. (B) orp2-T4A, but not orp2-T3D, suppresses the temperature sensitivity of spb70-U05 at 36.5°C. Thermosensitivity of double mutant spb70-U05 orp2-T4A was compared to single mutant spb70-U05 and double mutant spb70-U05 orp2-T3D after 3 days of incubation at 36.5°C. (C) Suppression of the temperature sensitivity of spb70-U12 by orp2-T4A. Single mutants orp2-T4A and spb70-U12 and double mutant spb70-U12 orp2-T4A were incubated at 30°C for 2 days.
These genetic results support the notion that the chromatin-bound Spb70 and phosphorylation status of Orp2 have a profound physiological effect on cell growth. It is possible that the phosphorylation status of Orp2 and the chromatin-bound Spb70 affect the association of Polα-primase with Orps in G1 pre-Start. The suppression of spb70 mutants by orp2-T4A suggests that an unphosphorylated Orp2 may help to stabilize a defective Spb70 mutant protein in the Polα-primase complex onto chromatin to initiate DNA synthesis at the replication origin. The finding that the orp2-T3D mutation in spb70-U05 exacerbates the thermosensitivity supports this notion (Fig. 6A).
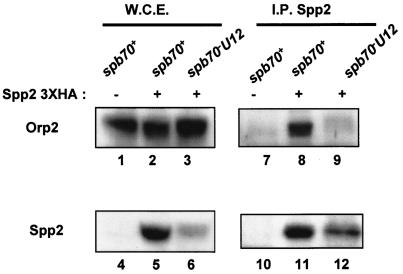
Primase subunit Spp2/p58 interacts with Orp2, and the interaction requires Spb70.
The finding that Spb70 in the chromatin fraction associates with Orp2 throughout the cell cycle prompted us to test whether other subunits of the Polα-primase complex are also able to associate with Orp2. If they do, does the association require the chromatin-bound Spb70? To this end, we used primase coupling subunit Spp2/p58 as the representative of the Polα-primase complex, since Spp2 physically links Polα/p180 and primase catalytic subunit Spp1/p49 (11, 37). Moreover, an earlier study of budding yeast has shown that Cdc6-independent association of Polα-primase with chromatin requires primase (12). We constructed strains containing spp2+-three-HA and orp2+-three-FLAG in either a wild-type spb70+ or mutant spb70-U12 background. In addition, we also constructed a wild-type spb70+ strain containing only orp2+-three-FLAG without epitope-tagged spp2+ as control. Orp2-three-FLAG proteins were expressed in whole-cell extracts from the spb70+ strain containing orp2+-three-FLAG with or without spp2+-three-HA and in cell extracts from the spb70-U12 mutant strain containing orp2+-three-FLAG and spp2+-three-HA (Fig. 7, lanes 1, 2, and 3, respectively). As expected, HA-tagged Spp2/p58 proteins were detected by Western blotting with anti-HA antibody in whole-cell extracts only from cells containing spp2+-three-HA, not from cells without spp2+-three-HA (Fig. 7, lanes 4, 5, and 6). FLAG-tagged Orp2 was only detected in the anti-Spp2/p58 immunoprecipitates from extracts of spb70+ containing orp2+-three-FLAG and the spp2+-three-HA strain (Fig. 7, lane 8) but not from extracts of the spb70+ strain containing only orp2+-three-FLAG without HA-tagged spp2+ (Fig. 7, lane 7). Importantly, FLAG-tagged Orp2 was not detected in the Spp2/p58 immunoprecipitates from cell extracts of the spb70-U12 mutant containing orp2+-three-FLAG and spp2+-three-HA (Fig. 7, lane 9). The inability of FLAG-tagged Orp2 to coprecipitate with the HA-tagged Spp2/p58 in the spb70-U12 mutant cell extract is not due to the low levels of the HA-tagged Spp2 in spb70-U12 mutant cells (Fig. 7, lane 6). HA-tagged Spp2 protein was readily detected in the HA immunoprecipitates from the cell extracts of spb70-U12 (Fig. 7, lane 12).
FIG. 7.
Spp2/p58 interacts with Orp2, and the interaction requires Spb70. Wild-type cells containing orp2+-three-FLAG either with or without spp2+-three-HA and mutant spb70-U12 containing orp2+-three-FLAG and spp2+-three-HA were cultured in EMM to mid-log phase at 25°C and then shifted to 36°C for 3.5 h. Whole-cell extracts (W.C.E.) were prepared from each strain. Presence or absence of HA-tagged Spp2/p58 and FLAG-tagged Orp2 in whole-cell extracts was shown by Western blotting (W.C.E. panel). Spp2/p58 proteins were immunoprecipitated from each strain with anti-HA antibody, and the immunoprecipitates were Western blotted with anti-HA for Spp2/p58 and anti-FLAG for Orp2 (I.P. Spp2 panel). In whole-cell extracts, Orp2-FLAG protein was detected in whole-cell extracts from spb70+ strains either with or without spp2+-three-HA (lanes 1 and 2) and from mutant spb70-U12 (lane 3), while Spp2-HA was not detected in whole-cell extracts from spb70+ cells without spp2+-three-HA (lane 4) and only detected in whole-cell extracts from spb70+ and spb70-U12 strains containing spp2+-three-HA (lanes 5 and 6). In the anti-HA immunoprecipitates (I.P. Spp2 panel), Orp2-FLAG coprecipitated with Spp2/p58 from whole-cell extracts in the spb70+ strain containing spp2+-three-HA (Lane 8) but not from mutant spb70-U12 containing spp2+-three-HA (lane 9). Anti-HA immunoprecipitates from whole-cell extracts of the spb70+ strain without spp2+-three-HA had neither detectable Orp2-FLAG nor Spp2/p58-HA (lanes 7 and 10), and Spp2-HA was detected in immunoprecipitates from whole-cell extracts of spb70+ and spb70-U12 strains containing spp2+-three-HA (lanes 11 and 12).
These results indicate that Spp2/p58 either directly or indirectly interacts with Orp2, and the interaction is significantly compromised in cells with a mutation in spb70+. These data also suggest that Polα-primase complex associates with the chromatin-bound Spb70-Orp complex (ORC), and the association involves an interaction with Spp2/p58. Importantly, the interaction requires Spb70.
Primase subunit Spp2/p58 preferentially interacts with the unphosphorylated form of Orp2 in G1.
The finding that the phosphorylation status of Orp2 can significantly affect the growth of spb70 thermosensitive mutants (Fig. 6) led us to test whether phosphorylation or unphosphorylation of Orp2 has an effect on the association between Orp2 and Spp2/p58 in the chromatin fraction. We constructed a strain containing two-myc-six-His-spb70+, orp2+-three-FLAG, and spp2-three-HA in a cdc25-22 background. After release of the cells from G2 arrest, cell samples were collected every 12 min (Fig. 8A). The kinetics of Mcm6, Orp2 (Orc2), Spp2/p58, and Spb70 appearance in the chromatin fraction were monitored by Western blotting with their respective antibodies. Mcm6 was detected in the chromatin fraction at 60 min after release from G2 arrest, and the abundance of MCM6 in chromatin fraction progressively increased as cells approached G1/S junction. Orp2 was constitutively present in the chromatin fraction throughout the cell cycle. After 24 min of release from G2 arrest, a faster-migrating unphosphorylated form of Orp2 appeared in the chromatin fraction. Abundance of the unphosphorylated Orp2 peaked prior to the increase of MCM6 and gradually decreased as cells approached G1/S junction. After 12 min of release from G2 arrest, a very faint Spp2/p58 protein band began to be detectable by anti-HA antibody. The abundance of Spp2/p58 in the chromatin fraction progressively increased as cells progressed into S phase (Fig. 8B).
FIG. 8.
Spp2/p58 coprecipitates with unphosphorylated Orp2/Orc2. The cdc25-22 strain containing two-myc-six-His-spb70+, orp2+-three-FLAG, and spp2+-three-HA was cultured at 25°C in EMM to mid-log phase and shifted to 36°C for 4 h to arrest at G2. Cells were then released from G2 arrest by shift to 25°C and harvested every 12 min. Chromatin fractions were prepared as described in Materials and Methods. (A) Cell cycle progression was monitored by septation index and mitotic index. (B) Kinetics of the appearance of MCM6, Orp2, Spp2/p58, and Spb70 in the chromatin fractions prepared from each time point were detected by Western blotting with their respective antibodies as described for Fig. 5B. (C) Coprecipitations of the unphosphorylated Orp2-FLAG with the HA-tagged Spp2/p58 were shown by Western blotting the anti-HA immunoprecipitates of Spp2 with anti-FLAG and anti-HA antibodies. A demonstration of phosphorylated and unphosphorylated Orp2 is shown next to the 0-min time point.
Having established that Spb70 associates with Orp2 in the chromatin fraction throughout the cell cycle (Fig. 5C) and Spp2/p58 associates with Orp2 depending on a functional Spb70 (Fig. 7), we tested whether the interaction between Spp2/p58 and Orp2 is cell cycle regulated. The HA-tagged Spp2/p58 was immunoprecipitated and probed for coprecipitation of FLAG-tagged Orp2 (Fig. 8C). Consistent with a progressive increase of Spp2/p58 abundance in chromatin fraction as cells approach G1/S boundary, the abundance of Spp2/p58 in the anti-HA immunoprecipitates progressively increased (Fig. 8C, bottom panel). Nominal levels of Orp2 proteins were detectable in the Spp2/p58 immunoprecipitates when most of the Orp2 was in the phosphorylated form during the first 36 min after release from G2 block. After 36 min of release from G2 arrest, increased amounts of unphosphorylated Orp2 appeared in the chromatin fraction (Fig. 8B), and increased amounts of unphosphorylated Orp2 were also detected in the Spp2/p58 immunoprecipitates (Fig. 8C), indicating that Spp2/p58 preferentially associates with the unphosphorylated Orp2 during G1 pre-Start. As cells approached G1/S junction, Cdc2 began to phosphorylate Orp2; progressively decreased amounts of unphosphorylated Orp2 were found to coprecipitate with Spp2/p58 (Fig. 8C). These results indicate that Spp2/p58 preferentially associates with the unphosphorylated Orp2 in G1 pre-Start prior to Orp2 being phosphorylated by Cdc2.
DISCUSSION
DNA Polα-primase is unique among all the replicative DNA polymerases for its role in initiation at the replication origin and in elongation during Okazaki fragment synthesis on the lagging strand. Among the four subunits of DNA Polα-primase complex, the biological function of B-subunit is largely unknown. In vitro experiments have shown that a reconstituted three-subunit Polα-primase without the B-subunit is fully catalytic competent in either polymerase or primase enzymatic activity (11). The B-subunits in budding yeast and human cells are not phosphorylated in G1 and early S phase and are progressively phosphorylated when cells approach late S phase and G2 (14, 29). These findings suggest that the B-subunit phosphorylation might play a regulatory role in late S phase and G2; however, how the cell cycle-dependent phosphorylation of B-subunit regulates replication is not clear. In this study, we generated nine fission yeast thermosensitive mutants of B-subunit. By genetic and biochemical approaches, we have found that (i) a fraction of the fission yeast B-subunit is bound to chromatin-enriched cellular fraction independent of the other subunits, (ii) the B-subunit genetically interacts and physically coexists with ORC proteins Orp1 (Orc1) and Orp2 (Orc2) in a protein complex; the phosphorylation status of Orp2 has a profound effect on the thermosensitive growth of the spb70 mutants, and (iii) primase coupling subunit Spp2/p58 interacts with both B-subunit and Orp2; the interaction between Spp2/p58 and Orp2 requires a functional B-subunit and the unphosphorylated form of Orp2.
These findings led us to propose a hypothesis: there are two populations of B-subunits in proliferating cells. The main bulk of the B-subunit is in complex with Polα/p180 and the primase subunits, which are involved in initiation of DNA synthesis during elongation of Okazaki fragments. Another fraction of the B-subunit constitutively associates with the ORC, Orps (Orcs), on the chromatin at replication origins throughout the cell cycle. The chromatin-bound Spb70 in complex with Orps recruits a population of Polα-primase complex to the origin by interacting with the primase during G1 pre-Start, prior to the events of CDK activation, Orp2 phosphorylation, and loading of MCM onto the chromatin. The Polα-primase complexes that are recruited onto the replication origins by Spb70-Orps during G1 pre-Start are to set the stage for initiation at the replication origins. We discuss this hypothesis below.
The B-subunit-Orp complex recruits Polα-primase for initiation at the replication origins.
Studies of SV40 in vitro replication have proposed that the B-subunit of Polα-primase complex is involved in tethering Polα-primase to interact with T antigen for initiation at the SV40 replication origin (10). A study of budding yeast B-subunit has hypothesized that the B-subunit might have a regulatory role in loading the Polα-primase complex at replication origin by either directly interacting with the ORC or indirectly via some yet to be identified factors (15). However, thus far, no experimental evidence has been reported to support this hypothesis. Association of budding yeast Polα-primase with chromatin in G1 has been shown to be independent of Cdc6. This has been interpreted as Polα-primase being involved in a Cdc6-independent mitotic resetting event (12). A fission yeast study has also shown that Polα-primase is loaded onto chromatin in G1 pre-Start (40). Thus, both budding and fission yeasts studies have demonstrated that Polα-primase associates with chromatin in early G1 (12, 40). However, how and why Polα-primase associates with chromatin in G1 is not known.
In this study, we have found that although the main bulk of the four-subunit Polα-primase complex is in the soluble cellular fraction, a fraction of the Polα-primase is chromatin bound (Fig. 1D). Notably, the B-subunit is chromatin bound and associates with Orp2 protein throughout the cell cycle (Fig. 5B and C). In contrast, primase Spp2/p58 does not associate with chromatin in G2 (at 0 min; Fig. 5B and 8B) and begins to associate with chromatin 20 to 24 min after release from G2 arrest (Fig. 5B and 8B). These findings indicate that there is a distinct population of B-subunit that constitutively associates with Orps in the chromatin-enriched fraction throughout the cell cycle independent of Spp2/p58, Polα/p180, and Spp1/p49.
We show in this study that in fission yeast, there is a three-way interplay of the chromatin-bound B-subunit (Spb70), ORC (Orps), and primase coupling subunit Spp2/p58 in the Polα-primase complex. A mutation in spb70+ not only compromises the interaction between Orp and Spb70 protein but also compromises the association between Orps and primase Spp2/p58 (Fig. 4C and 7). In early G1 prior to CDK activation, the unphosphorylated Orp2 coprecipitates with primase subunit Spp2/p58 in the Polα-primase complex (Fig. 8B and C). Together, these data suggest that the B-subunit Spb70 associating with Orps on the chromatin mediates the recruitment of the four-subunit Polα-primase complex onto the chromatin during G1 prior to CDK activation. The recruitment may occur through either a direct or indirect interaction of the unphosphorylated Orp2 with primase Spp2/p58 in the four-subunit Polα-primase complex. The Spb70 in the Orp2-Spb70 complex may either enhance and/or stabilize the interaction between the unphosphorylated Orp2 and the primase subunit Spp2/p58 in the four-subunit Polα-primase complex. Thus, a mutation in spb70+ that compromises the stability of Spb70-Orp2 complex formation may cause either a weakening or an inability of the unphosphorylated Orp2 to associate with Polα-primase, resulting in thermosensitive cell growth (Table 2 and Fig. 2). In this study, we used the primase coupling subunit Spp2/p58 as the representative of the Polα-primase complex to analyze the association with Orp2. Previous studies of Polα-primase complex from fission yeast and human cells have shown that Polα/p180 can form a complex with the two-primase subunits independent of the B-subunit, and the trimeric Polα-primase complex is enzymatically active in vitro (11, 31, 37). Spp2/p58 couples the Polα/p180 and primase catalytic subunit Spp1/p49 (11, 37). A previous study of budding yeast has shown that the Cdc6-independent association of Polα-primase complex with chromatin requires the primase (12). Thus, it is reasonable to assume that the interaction of chromatin-bound Orp-Spb70 complex with Spp2/p58 is a recruitment of the Polα-primase complex.
Orp2 is known to be unphosphorylated during G1 prior to Start (41). Mutations in orp2+ that abolish all of its Cdc2 phosphorylation sites suppress the spb70 mutants' thermosensitivity, whereas a mutation in orp2+ that mimics constitutive phosphorylation of Orp2 exacerbates the thermosensitivity of the spb70 mutant (Fig. 6). Mutations in orp2+ that abolish its CDK phosphorylation site have been show to promote rereplication (41). Because unphosphorylation of Orp2 allows rereplication (41), the unphosphorylated Orp2 might enhance the efficiency of origin firing, resulting in the offsetting of a compromised interaction between a Polα-primase complex containing a mutated B-subunit with Orps. It is also possible that an unphosphorylated Orp2 may enhance the tethering of a defective Polα-primase complex to the origin. Thus, a constitutively unphosphorylated Orp2 could suppress the thermosensitivity of spb70 mutants, whereas a mutant orp2 that mimics constitutive phosphorylation could exacerbate the thermosensitivity of the spb70 mutant (Fig. 6).
The timing of recruiting Spp2/p58 onto the chromatin coincides with the appearance of the unphosphorylated Orp2 (Fig. 5B) and prior to the loading of MCMs, indicating that the recruitment occurs in G1 pre-Start before the chromatin is licensed. There are two possible roles of the Polα-primase recruited onto chromatin at this stage of the cell cycle. It is possible that the fraction of Polα-primase recruited onto chromatin by Orp-Spb70 in G1 pre-Start is for some type of repair during G1. It is also possible that the fraction of Polα-primase recruited onto chromatin by Orp-Spb70 at this stage of G1 is for initiation of DNA synthesis at the origin of replication. The findings that a mutation in budding yeast B-subunit arrests the cells prior to HU arrest point (15), the human B-subunit interacts with SV40 T antigen at the SV40 origin (10), and initiation of SV40 replication requires the B-subunit (31) support the second proposed role of Polα-primase complex recruited in G1 pre-Start; we thus favor the hypothesis that the chromatin-bound Spb70 in complex with Orps mediates recruitment of the four-subunit Polα-primase complex onto replication origins in G1 phase before Start to get ready for Polα-primase to initiate both leading and lagging strand synthesis.
Chromatin association of Polα-primase at G1/S junction and during S phase requires Cdc45.
Studies of budding yeast and other organisms have established that pre-RCs are assembled at replication origin during late mitosis or early G1 phase in a period when S-phase CDK are inactive (7). Upon activation of CDK and DDK at G1/S junction, pre-RC is transformed to preinitiation complex by loading Cdc45 onto pre-RC (7, 38, 47). In Xenopus egg extracts, association of Polα onto chromatin requires Cdc45 (25, 26). In budding yeast, DNA Polα-primase is loaded onto preinitiation complex after a mutual dependent loading of Cdc45, MCMs, and RPA (40, 46-48). Studies of budding yeast cdc45 mutants have suggested that Cdc45 has a critical role in initiation and associates with origins during G1 or early S phase (1, 2, 46-48). By using a cdc45 thermolabile degron mutant and density shift analysis in budding yeast, it has been shown that Cdc45 is required for both entry of S phase and elongation of DNA replication during S-phase progression (38). Studies of fission yeast Cdc45 (also named Sna41) have shown that Cdc45 and Polα-primase interacts throughout the cell cycle and Cdc45 interacts with MCMs during S phase. At G1/S boundary, fission yeast Cdc45 and Polα-primase complex are both loaded onto chromatin, and Polα-primase complex associates with MCMs in a Cdc45-dependent manner (40). The fission yeast results suggest that loading of Polα-primase onto replication origin has two distinct modes. Polα-primase is loaded onto chromatin in early G1 during pre-Start. At G1/S boundary, additional Polα-primase is loaded onto chromatin to associate with MCMs in pre-RC in a Cdc45-dependent manner (40).
Cdc45 was not detected in chromatin fraction until the S-phase CDK were activated (48). Upon activation of CDK and DDK, Cdc45 is then loaded into the pre-RC at G1/S junction (38, 47). We propose that at the G1/S junction, additional Polα-primase is loaded onto the chromatin unwound by MCMs in the Cdc45-dependent manner; thus, the unwinding of chromatin by MCMs and initiation of lagging synthesis by Polα-primase are carried out in a coordinated manner. Since Cdc45 is required for both entry of S phase and elongation of replication during S phase (38), this could explain the finding that Polα-primase associates with Cdc45 throughout the S phase in fission yeast (40). Hence, the Cdc45-dependent chromatin association of Polα-primase at the G1/S junction and during S-phase progression is required for initiation of Okazaki fragments during lagging strand synthesis. As we proposed above, in this study, the fraction of Polα-primase recruited onto chromatin during G1 pre-Start via the interaction with the chromatin-bound B-subunit-Orps complex initiates DNA synthesis at the replication origins for both leading and lagging strands.
Acknowledgments
We thank members of our lab for helpful discussion, Rose Borbely for her excellent technical help, Janet Leathewood for the orp2 mutants, and Hisao Masukata for the antibody of MCM6.
This work is supported by grant CA14835 from the National Cancer Institute of the National Institutes of Health, and M.U. is a recipient of the Stanford University Medical School Dean's Postdoctoral Fellowship.
REFERENCES
- 1.Aparicio, O. M., A. M. Stout, and S. P. Bell. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. USA 96:9130-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 3.Arezi, B., and R. D. Kuchta. 2000. Eukaryotic DNA primase. Trends Biochem. Sci. 25:572-576. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 6.Baker, T. A., and S. P. Bell. 1998. Polymerases and the replisome: machines within machines. Cell 92:295-305. [DOI] [PubMed] [Google Scholar]
- 7.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 8.Bhaumik, D., and T. S.-F. Wang. 1998. Mutational effect of fission yeast Polα on cell cycle events. Mol. Biol. Cell 9:2107-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke, J. D., and K. L. Gould. 1994. Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol. Gen. Genet. 242:169-176. [DOI] [PubMed] [Google Scholar]
- 10.Collins, K. L., A. A. Russo, B. Y. Tseng, and T. J. Kelly. 1993. The role of the 70 kDa subunit of human DNA polymerase α in DNA replication. EMBO J. 12:4555-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copeland, W. C., and T. S.-F. Wang. 1993. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268:26179-26189. [PubMed] [Google Scholar]
- 12.Desdouets, C., C. Santocanale, L. S. Drury, G. Perkins, M. Foiani, P. Plevani, and J. F. X. Diffley. 1998. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J. 17:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, D. L., and P. Nurse. 1996. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 15:850-860. [PMC free article] [PubMed] [Google Scholar]
- 14.Foiani, M., G. Liberi, G. Lucchini, and P. Plevani. 1995. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol. Cell. Biol. 15:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foiani, M., F. Marini, D. Gamba, G. Lucchini, and P. Plevani. 1994. The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grallert, B., and P. Nurse. 1996. The ORC homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 10:2644-2654. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths, D. J. F., V. F. Liu, P. Nurse, and T. S.-F. Wang. 2001. Role of fission yeast primase catalytic subunit in the replication checkpoint. Mol. Biol. Cell 12:115-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm, C., J. Kohli, J. Murray, and K. Maundrell. 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selective marker. Mol. Gen. Genet. 215:81-86. [DOI] [PubMed] [Google Scholar]
- 19.Hayles, J., D. Fisher, A. Woodlard, and P. Nurse. 1994. The temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78:813-822. [DOI] [PubMed] [Google Scholar]
- 20.Jallepalli, P. V., and T. J. Kelly. 1997. Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? Curr. Opin. Cell Biol. 9:358-363. [DOI] [PubMed] [Google Scholar]
- 21.Kamimura, Y., Y. S. Tak, A. Sugino, and H. Araki. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukimoto, I., H. Igaki, and T. Kanda. 1999. Human Cdc45 protein binds to minichromosome maintenance 7 protein and the p70 subunit of DNA polymerase alpha. Eur. J. Biochem. 265:936-943. [DOI] [PubMed] [Google Scholar]
- 23.Leatherwood, J., A. Lopez-Girona, and P. Russell. 1996. Interaction of cdc2 and cdc18 with a fission yeast ORC2-like protein. Nature 379:360-363. [DOI] [PubMed] [Google Scholar]
- 24.Lygerou, Z., and P. Nurse. 1999. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 112:3703-3712. [DOI] [PubMed] [Google Scholar]
- 25.Mimura, S., T. Masuda, T. Matsui, and H. Takisawa. 2000. Control role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5:439-452. [DOI] [PubMed] [Google Scholar]
- 26.Mimura, S., and H. Takisawa. 1998. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J. 17:5699-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondesert, O., C. H. McGowan, and P. Russell. 1996. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol. Cell. Biol. 16:1527-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 29.Nasheuer, H.-P., A. Moore, A. F. Wahl, and T. S.-F. Wang. 1991. Cell cycle-dependent phosphorylation of human DNA polymerase α. J. Biol. Chem. 266:7893-7903. [PubMed] [Google Scholar]
- 30.Ogawa, Y., T. Takahashi, and H. Masukata. 1999. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 19:7228-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott, R. D., C. Rehfuess, V. N. Podust, J. E. Clark, and E. Fanning. 2002. Role of the p68 subunit of human DNA polymerase alpha-primase in simian virus 40 DNA replication. Mol. Cell. Biol. 22:5669-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, H., S. Francesconi, and T. S.-F. Wang. 1993. Cell cycle expression of two replicative DNA polymerases α and δ from Schizosaccharomyces pombe. Mol. Biol. Cell 4:145-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piatti, S., T. Bohm, J. H. Cocker, J. F. Diffley, and K. Nasmyth. 1996. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 10:1516-1531. [DOI] [PubMed] [Google Scholar]
- 34.Saha, P., K. C. Thome, R. Yamaguchi, Z. Hou, S. Weremowicz, and A. Dutta. 1998. The human homolog of Saccharomyces cerevisiae CDC45. J. Biol. Chem. 273:18205-18209. [DOI] [PubMed] [Google Scholar]
- 35.Sazer, S., and S. W. Sherwood. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97:509-516. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, D. A., and S. L. Forsburg. 1998. Schizosaccharomyces pombe Mcm3p, an essential nuclear protein, associates tightly with Nda4p (Mcm5p). Nucleic Acids Res. 26:3955-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan, S., and T. S.-F. Wang. 2000. Analysis of fission yeast primase defines the checkpoint responses to aberrant S phase initiation. Mol. Cell. Biol. 20:7853-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tercero, J. A., K. Labib, and J. F. Diffley. 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 19:2082-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchiyama, M., K. Arai, and H. Masai. 2001. Sna41goa1, a novel mutation causing G1/S arrest in fission yeast, is defective in a CDC45 homolog and interacts genetically with polalpha. Mol. Genet. Genomics 265:1039-1049. [DOI] [PubMed] [Google Scholar]
- 40.Uchiyama, M., D. Griffiths, K. Arai, and H. Masai. 2001. Essential role of Sna41/Cdc45 in loading of DNA polymerase α onto minichromosome maintenance proteins in fission yeast. J. Biol. Chem. 276:26189-26196. [DOI] [PubMed] [Google Scholar]
- 41.Vas, A., W. Mok, and J. Leatherwood. 2001. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell. Biol. 21:5767-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 43.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 44.Wang, T. S.-F. 1996. Cellular DNA polymerases, p. 461-493. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Wuarin, J., V. Buck, P. Nurse, and J. B. Millar. 2002. Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111:419-431. [DOI] [PubMed] [Google Scholar]
- 46.Zou, L., J. Mitchell, and B. Stillman. 1997. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol. Cell. Biol. 17:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou, L., and B. Stillman. 1998. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 280:593-596. [DOI] [PubMed] [Google Scholar]