Abstract
The effects of estrogens, particularly 17β-estradiol (E2), are mediated by estrogen receptor α (ERα) and ERβ. Upon binding to E2, ERs homo- and heterodimerize when coexpressed. The ER dimer then regulates the transcription of target genes through estrogen responsive element (ERE)-dependent and -independent pathways that constitute genomic estrogen signaling. Although ERα and ERβ have similar ERE and E2 binding properties, they display different transregulatory capacities in both ERE-dependent and -independent signaling pathways. It is therefore likely that the heterodimerization provides novel functions to ERs by combining distinct properties of the contributing partners. The elucidation of the role of the ER heterodimer is critical for the understanding of physiology and pathophysiology of E2 signaling. However, differentially determining target gene responses during cosynthesis of ER subtypes is difficult, since dimers formed are a heterogeneous population of homo- and heterodimers. To circumvent the pivotal dimerization step in ER action and hence produce a homogeneous ER heterodimer population, we utilized a genetic fusion strategy. We joined the cDNAs of ERα and/or ERβ to produce single-chain ERs to simulate the ER homo- and heterodimers. The fusion ERs interacted with ERE and E2 in a manner similar to that observed with the ER dimers. The homofusion receptors mimicked the functions of the parent ER dimers in the ERE-dependent and -independent pathways in transfected mammalian cells, whereas heterofusion receptors emulated the transregulatory properties of the ERα dimer. These results suggest that ERα is the functionally dominant partner in the ERα/β heterodimer.
Estrogen hormones, particularly 17β-estradiol (E2), exert their effects through a complex array of convergent and divergent signaling pathways that mediate genomic and nongenomic events, resulting in target tissue-specific responses (11, 31). The E2 information is conveyed by the transcription factors, estrogen receptor α (ERα) and ERβ (11, 31), which are encoded by distinct genes and are expressed in different tissues as well as in the same tissue at various levels (11, 31).
Upon binding to E2, ER dimerizes and interacts with permutations of a palindromic DNA sequence separated by three nonspecific nucleotides: 5′-GGTCAnnnTGACC-3′, the consensus estrogen responsive element (ERE) (11, 18, 31). The E2-ER-ERE complex subsequently recruits coactivators/regulators to promote local chromatin remodeling and to bridge with general transcription factors for the initiation of transcription (11, 31). This pathway is called ERE-dependent ER signaling. The E2-ER complex also regulates gene expression through functional tethering to a transcription factor bound to its cognate regulatory element on DNA. This is the DNA-dependent and ERE-independent signaling pathway (22, 36). Furthermore, E2 elicits effects through the membrane and cytoplasmic ERs (24, 39).
ERα and ERβ share high amino acid identity (96%) in their DNA-binding domains (DBDs) (11, 31), which is reflected in the abilities of ERs to bind to the same spectrum of ERE sequences with similar affinities (27, 45). The carboxyl-terminal ligand-binding domains (LBDs) also show considerable homology (53%) responsible for similar ligand-binding affinities. Despite comparable biochemical properties, ERs differ in their transregulation potencies in a ligand, promoter, and cell context-dependent manner (11, 31). Studies have indicated that the distinct amino-terminal A/B domains, a lesser conserved region between ERs (30% homology), play critical roles in the manifestation of transregulatory activity of the receptor subtype (5, 12, 28, 44).
In addition to acting as a homodimer, ERβ heterodimerizes with ERα in vitro and in situ when coexpressed (4, 34, 40). However, the role of the ERα/β heterodimer in E2 signaling remains largely unknown due to the presence of a heterogeneous population of ER dimers. An innovative approach was introduced to address the functions of ERα/β heterodimer in the ERE-dependent signaling pathway (40). This approach changes the DNA-binding specificity of an ER subtype to that of the glucocorticoid hormone receptor (GR) (10). Coexpression of a wild-type (WT) ER with a consensus glucocorticoid (GRE)-binding ER allows the measurement of transcriptional properties of the ERα/β heterodimer from a hybrid response element composed of ERE and GRE half-sites without an interference from the ER homodimers. Using this system, it was shown that ERα/β heterodimer generates new attributes to E2 signaling by combining functional properties of both contributing partners (40). Although a powerful approach for structure-function analysis, this system could produce not only ERα/β heterodimer but also ERα and ERβ homodimers. Furthermore, since the approach utilizes a hybrid DNA response element, analysis of responses from natural EREs would be precluded. In addition, in the presence of both homo- and heterodimer ERs, examination of DNA-dependent and ERE-independent regulation of estrogen-responsive genes by a specific ER dimer is difficult.
We recently sought an alternative approach to generate a homogeneous population of homo- or heterodimers of ERα by circumventing the pivotal dimerization step in receptor action (30). We engineered a single-chain α-α protein by genetically conjugating two ERα cDNAs in tandem. Although a monomer, the homofusion α-α exhibited biochemical and functional properties that mimicked those of the ERα dimer. Using this genetic conjugation approach, we have now generated single-chain α-β and β-α receptors to study the role of the ERα/β heterodimer in genomic estrogen signaling pathways. The homofusion α-α and β-β simulated the functional properties of the parent ERα and ERβ dimers. The heterofusions α-β and β-α, on the other hand, emulated the functions of the ERα dimer in both ERE-dependent and ERE-independent genomic signaling pathways. These results suggest that ERα dictates the transregulatory properties of the ERα/β heterodimer.
MATERIALS AND METHODS
Construction of fusion ER receptors and glutathione S-transferase (GST) fusion cofactors.
The human WT ERα and ERβ cDNAs with or without the Flag epitope were described previously (30, 38, 44). The WT ERβ cDNA encodes a 530-amino-acid protein. Construction of fusion ERs was accomplished as described previously (30). ERs defective in DNA binding were constructed by changing two Cys residues in the first zinc finger of the C domain to His residues at positions 202 and 205 in ERα and at positions 166 and 169 in ERβ. Dimerization defective ERs were constructed by changing three Leu residues at positions 504, 508, and 511 to Glu in ERα and at positions 455, 459, and 462 in ERβ. Mutant fusion receptors were engineered by exchanging mutant ERα and/or ERβ cDNAs in the WT fusion receptor cDNAs.
The generation of GST-cofactor fusion proteins was described previously (30, 38, 44). GST fusion steroid receptor cofactor 1 (SRC-1) and transcription intermediary factor 2 (TIF-2) polypeptides contain nuclear interacting signature motifs within the region encompassing residues 219 to 399 (SRC-1219-399) and 623 to 986 (TIF-2623-986), respectively. The GST fusion TIF2-Q polypeptide contains a glutamine-rich region that encompasses residues 1125 to 1325.
Synthesis and detection of fusion ERs in vitro and in situ.
The linearized pBluescript II KS bearing no cDNA or a receptor cDNA was transcribed by using T3 RNA polymerase and translated by using a rabbit reticulocyte lysate as directed by the manufacturer (Promega, Madison, Wis.). For quantification of ERs synthesized in vitro, we used l-[methyl-3H]methionine (72 Ci/mmol; NEN Life Sciences, Boston, Mass.), followed by fluorography as described previously (30). Equal aliquots of reaction mixtures (5 μl/50-μl reaction mixture) were also subjected to Western blot analysis. Proteins were probed with a polyclonal ERα (HC-20; Santa Cruz Biotechnology, Santa Cruz, Calif.)- or ERβ (PA1-313; Affinity Bioreagents, Golden, Colo.)-specific antibody directed to the carboxyl terminus. Proteins were visualized by using the ECL-Plus Western blotting system (Amersham Pharmacia). In Western blot analysis with a monoclonal Flag antibody (M2; Sigma-Aldrich, St. Louis, Mo.), we used a Flag Western detection kit as directed by the manufacturer (Stratagene, La Jolla, Calif.).
EMSA.
Both strands of oligomers were synthesized and purified by the Integrated DNA Technologies (Coralville, Iowa). Regions flanking test sequences were identical. The oligomers were annealed, 32P end labeled, and subjected to electrophoretic gel mobility shift assay (EMSA) as described previously (30). We used fivefold greater molar concentrations of ERβ to obtain similar amounts of ERE binding as described below. Equal amounts (10 μg) of total proteins from whole-cell extracts were used for EMSA. For the recruitment of cofactors, equal molar concentrations of in vitro-synthesized receptors were processed as described previously (45).
Estimation of relative DNA- and ligand-binding abilities of receptors.
The affinity of ER for various EREs was determined by electrophoretic gel mobility shift competition assays as described previously (45). Ligand binding was carried out as described previously (20).
A hydroxyl radical assay was done as previously described (45).
Cell culture and transfections.
A receptor cDNA was excised from pBluescript II KS(+) with XhoI and BamHI and inserted into a mammalian expression vector (pM2-AH). For transient transfections, Chinese hamster ovary (CHO-K1), human cervical carcinoma HeLa, and breast adenocarcinoma MDA-MB-231 (American Type Culture Collection, Rockville, Md.) cells were used. Cells in 48-well tissue culture plates were transfected as described previously (30) by using 75 ng of expression vector bearing no receptor or a receptor cDNA, together with 125 ng of a reporter plasmid driving the expression of the firefly luciferase enzyme cDNA. A reporter plasmid bearing the Renilla luciferase cDNA (Promega; 2 ng/well) was used to monitor transfection efficiency. Cells were then treated without or with 10−9 M of E2 (Sigma-Aldrich) for 24 h. Luciferase assays were performed with a dual luciferase assay kit (Promega). The reporter vectors containing a TATA box promoter without or with one or two EREs in tandem, the human complementary 3 (C3), and pS2 gene promoters were described previously (30, 45). Reporter plasmids bearing collagenase (Col) promoter and the RARα gene promoter were described recently (15). To assess the effects of ligands on ER-induced transcriptional responses from Col and RARα promoters, we used 125 ng of expression and reporter vectors/well. In these series of experiments, cells were treated for 40 h in the absence or presence of various concentrations of E2 or of ICI 182,780 (ICI; Tocris, Inc., Ballwin, Mo.). The amount of ICI was based on the preliminary studies in which 10−7 M ICI was the optimal concentration to induce a response without eliciting cell toxicity.
For Western blotting and EMSA, cells were plated onto six-well plates and transiently transfected with 3 μg of the expression vector bearing no cDNA or an ER cDNA/well as described above. Cells were lysed in buffer containing 20 mM Tris-HCl (pH 7.5), 400 mM KCl, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20% glycerol, and 1% protease inhibitor cocktail (Sigma-Aldrich) by a freeze-thaw cycle of three times. Then, 10 μg of total protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In cotransfections, a constant amount of an expression vector bearing a receptor cDNA was cotransfected with increasing concentrations of another expression vector as described in the text.
For immunocytochemistry, cells were processed as described previously (30).
RESULTS
Heterodimerization of ERs.
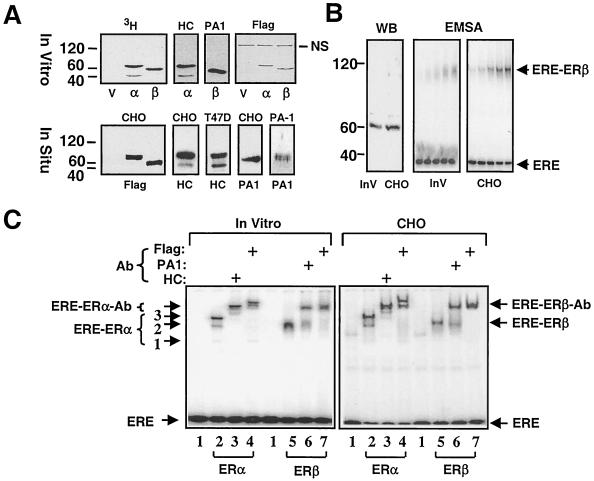
Previous studies showed that coexpression of the human “short” ERβ of 477-amino-acid with the human WT ERα leads to the formation of the ERα/β heterodimer in vitro and in situ (4, 34, 40). We wanted to address whether the WT ERβ (ERβ) that contains an additional 53 amino acids at the amino terminus also heterodimerizes with WT ERα (ERα) when cosynthesized. In order to facilitate the examination of biochemical and functional properties of heterodimer receptors, we initially examined the abilities of ER homodimers synthesized in vitro and in situ to interact with a 32P-end-labeled DNA fragment bearing the consensus ERE derived from the vitellogenin A2 gene by EMSA. ERα and ERβ cDNAs contain sequences that encode a Flag epitope at the amino terminus. [3H]methionine was used to quantify in vitro-synthesized ERs. Fluorography indicated that ERα migrates at molecular masses of 67 and 46 kDa, the latter being a minor species synthesized at various levels among experiments and not always observable (Fig. 1, 3H). ERβ displayed an electrophoretic migration of 60 kDa. The detection of proteins by Western blot analysis by using ERα (HC-20 and HC)- or ERβ (PA1-313 and PA1)-specific antibody or the M2 (Flag) antibody for both receptors confirmed the identity of receptors. The absence of detection of the minor species of ERα migrating at 46 kDa by the Flag antibody either in vitro or in cell extracts from the transiently transfected ER-negative CHO cells suggests that this is an amino-terminally truncated ERα isoform. Two distinct ERα species were also detected by HC in Western blots of transfected CHO or ERα-positive T47-D cells derived from breast ductal carcinoma. ERβ showed a 60-kDa band by the PA antibody in cell extracts from transfected CHO or ERβ-positive PA-1 cells of ovarian teratocarcinoma.
FIG. 1.
In vitro and in situ synthesis of ERα and ERβ. (A) For in vitro synthesis of ERs, 1.5 μg of linearized pBS-KS bearing no cDNA or a cDNA for an ER was transcribed by T3 polymerase and translated by rabbit reticulocyte lysate in the presence of 4 μl of [3H]methionine in a total reaction volume of 50 μl. Equal amounts (5 μl) of reaction mixtures were subjected to SDS-7.5% PAGE, visualized by fluorography (3H) or by Western blotting with an antibody directed to the carboxyl terminus of ERα (HC), of ERβ (PA1) or the amino-terminal Flag epitope (Flag) of both ERs. For in situ synthesis of ERs, CHO cells were transiently transfected with 3 μg of expression vector bearing none (V) or a cDNA for an ER. After 24 h, cells were lysed, and equal amounts (10 μg) of total cellular proteins were resolved by SDS-7.5% PAGE, followed by Western blotting with Flag, HC, or PA1 antibody. Endogenously expressed ERα in T47D cells (T47D) and ERβ in PA-1 cells (PA-1) were also analyzed by Western blotting with HC and PA1 antibodies, respectively. NS, nonspecific protein. (B) In vitro (InV, 10 μl) and in situ (CHO, 10 μg) synthesized ERβ were analyzed by Western blotting with Flag antibody. The ERE binding of ERβ synthesized in vitro (InV) or in CHO cells (CHO) was assessed by EMSA. Briefly, 1, 2, 4, 7, and 10 μl of transcription-translation mixture and 1, 2, 4, 7, and 10 μg of total protein from cell extract were incubated with 62.5 pmol of 32P-labeled ERE-containing DNA fragment for 30 min at 4°C and then subjected to 5% nondenaturing PAGE. The gel is dried and exposed to a PhosphorImager. Unbound ERE (ERE) and ERE-ERβ complexes are indicated. (C) The binding of ERα (lanes 2, 3, and 4) and ERβ (lanes 5, 6, and 7) synthesized in vitro (In Vitro) or in transiently transfected CHO cells (CHO) to the consensus ERE was assessed by EMSA. In this assay, 10 μl of in vitro-synthesized ERβ and 2 μl of ERα were used to obtain similar amounts of ERE-bound receptors. Reactions were incubated in the absence or presence (+) of the Flag (lanes 4 and 7), HC (lane 3), or PA1 (lane 6) antibody. ER-ERE complexes representing truncated-ERα homodimer (complex 1; the least prominent ER-ERE complex), truncated-ERα-WT-ERα heterodimer (complex 2), and WT-ERα homodimer (complex 3) are indicated. Lane 1 indicates reactions with the parent expression vector.
Despite similar amounts of receptors as assessed by the Flag antibody in Western blots (Fig. 1B, WB), the extent of interaction of the in vitro-synthesized ERβ with an equal concentration of ERE (Fig. 1B, EMSA) was significantly lower than the in situ-synthesized ERβ whether or not a saturating concentration (10−7 M) of E2 was present (data not shown). Moreover, the ERβ synthesized in vitro showed a diffuse ERE-binding pattern that gradually formed a distinct complex as a result of increasing receptor concentration. In contrast, the ERβ synthesized in situ interacted with ERE efficiently. ERβ synthesized in S2 (data not shown) cells or in Sf9 insect cells (45) also binds efficiently to DNA. Although it is not clear, differential processing of ERβ species in vitro versus in situ could lead to differences in the extent of homodimerization of ERβ and/or stability of the ERβ homodimer. This, in turn, could be manifested as differences in the amount and pattern of ERβ interaction with DNA.
ERα synthesized in vitro formed three distinct ERE-bound complexes as detected by EMSA in the absence or presence of the HC antibody: a major complex and two minor complexes (Fig. 1C, In Vitro), as reported previously (4, 29). The slowest-migrating complex (complex 3) likely represents the WT ERα homodimer. The fastest-migrating band, the least prominent complex, corresponds to the amino-terminally truncated ERα homodimer (complex 1). The intermediate ER-ERE complex appears to be the heterodimer of the WT and the truncated ERα species (complex 2). This conclusion is suggested because the Flag antibody specific to the amino-terminal Flag epitope fails to retard the migration of the complex 1 (lane 4). A similar ERE-binding pattern was also observed with the ERα synthesized in situ (Fig. 1C, CHO). The ERβ synthesized in vitro, which we used fivefold more compared to ERα to obtain similar amount of binding, forms a single ER-ERE complex that migrates somewhat faster than the complex 2 of the ERα, whereas the ERE-bound in situ-synthesized ERβ migrates slightly faster than the complex 3 of ERα.
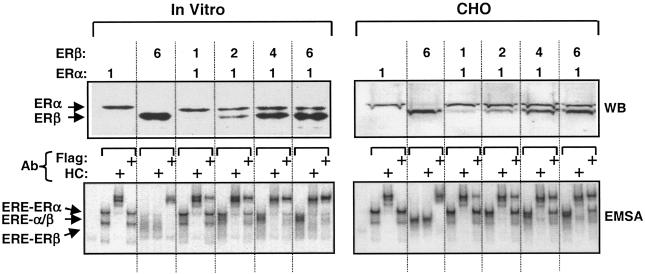
To examine the formation of a functional ERα/β heterodimer, we used a constant amount of the ERα expression vector, together with increasing amounts of ERβ expression vector in transcription-translation reactions or in transfection into CHO cells. The relative amounts of ER proteins were assessed in Western blots of equal aliquots of the in vitro reactions (Fig. 2, WB, In Vitro) or of CHO cell extracts (CHO) with the Flag antibody. The results indicated that increasing amounts of ERβ were cosynthesized with a constant amount of ERα. To assess the formation of a functional heterodimer, we used ERα without the Flag epitope to distinguish ER subtypes with the use of the Flag antibody. The cosynthesis of ERs in vitro or in situ led to the formation of a functional ERα/β heterodimer that showed an electrophoretic migration between ERα-ERE complex 3 representing the ERE-bound ERα homodimer and the ERβ homodimer in EMSA (Fig. 2, EMSA). The extent of heterodimer formation was correlated with a gradual decline in the ERα-ERE complex. It is also evident that residual ER homodimers were still present.
FIG. 2.
Coexpression of ERα and ERβ in vitro and in situ leads to formation of ERα/β heterodimer. For Western blotting of in vitro-synthesized ERs, 0.3 μg of pBS-KS vector bearing the Flag ERα cDNA was coexpressed with 0.3, 0.6, 1.2, or 1.8 μg of vector bearing the Flag-ERβ cDNA that correspond to 1-, 2-, 4-, or 6-fold-greater amounts, respectively, of the expression vector. Equal amounts (5 μl) of reaction mixtures were subjected to SDS-7.5% PAGE, followed by Western blotting with the Flag antibody. Migration of ERα and ERβ are indicated. The dimerization of ERα and ERβ was assessed by EMSA with equal amounts of transcription-translation mixture (5 μl). For Western blotting of in situ-synthesized ERs, 0.5 μg of the mammalian expression vector bearing the Flag-ERα cDNA was cotransfected with 0.5, 1.0, 2.0, or 3.0 μg (corresponding to 1, 2, 4, or 6, respectively) of the expression vector carrying the Flag-ERβ cDNA in CHO cells. Equal amounts of cell extracts (10 μg of total protein) were subjected to SDS-7.5% PAGE, followed by Western blotting with the Flag antibody. For EMSA, the cosynthesis of ERs was accomplished as described above, except that the cDNA for ERα does not contain sequences for the Flag epitope to distinguish ER species by the use of the antibody. Equal amounts of total protein (10 μg) of CHO cell extracts were subjected to EMSA. ERE-ER complexes were analyzed by using the HC or Flag antibody. ERE-bound ERα and ERβ homodimers and the ERα/β heterodimer are indicated. Unbound ERE is not shown.
Thus, the coexpression of WT ERβ, as shown previously for the “short” ERβ (4, 34), with ERα leads to the formation the ERα/β heterodimer, the extent of which is dependent upon the relative amount of the contributing partners.
Construction of fusion ERs.
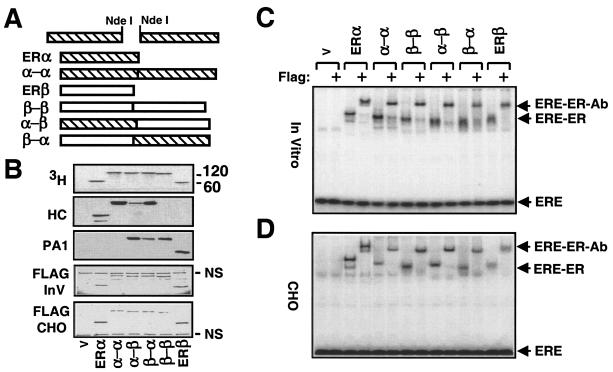
To study the effects of ERα/β heterodimer in genomic estrogen signaling, we used a genetic conjugation approach (30) to circumvent the pivotal dimerization step in receptor action. Single-chain ERs were produced by joining the 3′ end of the coding sequence of one ERα or ERβ cDNA to the 5′ coding sequence of another to encode two ER monomers in tandem (Fig. 3A) without or with a Flag epitope at the amino terminus. Analysis of radiolabeled proteins by fluorography revealed that the homofusion α-α and β-β and the heterofusion α-β and β-α displayed electrophoretic migrations with molecular masses of 120 to 136 kDa that correspond to two ER molecules (Fig. 3B, 3H). The detection of proteins by Western blot analysis with ERα (HC)-, ERβ (PA1)-, or Flag epitope-specific antibody (Flag-InV) confirms the identity of receptor species. Since radiolabeled counterparts and the Flag antibody show synthesis at comparable levels, differences in the extent of detection of α-β and β-α by the receptor-specific antibodies in Western blotting are likely due to a position-dependent alteration in the epitope recognition site.
FIG. 3.
Construction and synthesis of fusion ERs. (A) Schematics of ER fusion receptor cDNAs. A PCR-generated NdeI restriction enzyme site at the 5′ or 3′ end of an ER was used to genetically fuse two ER cDNAs in tandem. (B) Synthesis of fusion ERs in vitro with the parent vector bearing no cDNA (V) or a cDNA for an ER was accomplished as described for Fig. 1. Equal amounts of in vitro reaction mixtures (5 μl) or of (10 μg of total protein) of cell extracts from transfected CHO cells (Flag-CHO) were subjected to SDS-7.5% PAGE. The in vitro samples were visualized by fluorography (3H). The same samples were also probed with the HC, the PA1, or the Flag antibody (Flag-InV). (C) ERE binding of in vitro or in situ (CHO)-synthesized fusion receptors. Equal amounts (2 μl) of reaction mixtures, with the exception of the mixture containing ERβ (10 μl), were incubated with radiolabeled ERE in the absence or presence of the Flag antibody. Reaction mixtures were electrophoresed by 5% nondenaturing PAGE. (D) Equal amounts of CHO cell extracts (10 μg) were subjected to EMSA. The results from a representative experiment of two independent experiments are shown.
Similarly, the fusion receptors synthesized in transiently transfected CHO cells migrated with molecular masses ranging from 120 to 136 kDa assessed by the Flag antibody, whereas ERα and ERβ migrated with molecular masses of 67 and 60 kDa, respectively (Fig. 3B, Flag-CHO).
Characterization of biochemical properties of the fusion ERs. (i) DNA binding.
We examined by EMSA whether single-chain receptors synthesized in vitro (Fig. 3C) or in situ (Fig. 3D) interact with the consensus ERE. Equal amounts of the in vitro-synthesized ERα and fusion receptors retarded the electrophoretic migration of labeled ERE. Due to inefficient ERE binding of the ERβ synthesized in vitro, we used a fivefold greater concentration of receptor to obtain similar amounts of ERE-bound receptors. The ERE-bound homo- and heterofusion ERs showed migrations comparable to those of the ER dimers. The interaction of receptor species with ERE is specific, because the protein-ERE complex was further retarded by the Flag antibody. Similarly, equal amounts of cell extracts of transfected CHO cells synthesizing an ER or a single-chain receptor interacted efficiently with the consensus ERE in vitro.
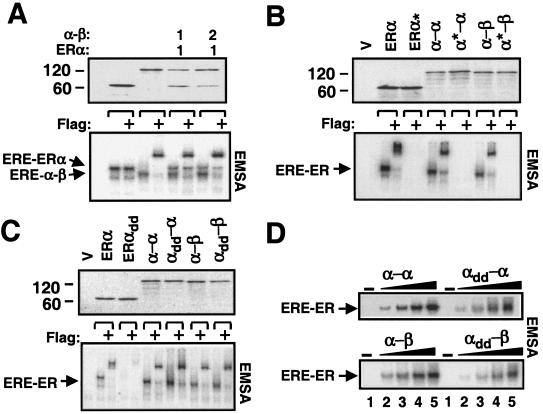
Since ERs bind to EREs as dimers, similar migration among ERE-bound WT ERs and fusion species suggests that the fusion receptors interact with DNA as monomers. We also observed that the ERE-bound homo- and heterofusion proteins showed slightly faster electrophoretic migrations than ERα, possibly reflecting differences in size and in conformation among ER species. We took advantage of these differences in the electrophoretic mobility to test the conclusion that the fusion receptors indeed bind to ERE as monomers and, implicitly, do not dimerize. We used the ERα expression vector together with the same (1:1) or a twofold-greater (1:2) amount of the expression vector bearing the α-β or the α-α (data not shown) cDNA in transcription-translation reactions with radiolabeled Met (Fig. 4A, upper panel). The ERα cDNA lacks the amino-terminal Flag epitope to discern ERE-bound receptor species by the use of Flag antibody. Reactions were subjected to EMSA (Fig. 4A, lower panel). The Flag antibody shifted only the ERE-bound α-β without altering the migration of ERα. This result demonstrates that the fusion receptors bind to ERE as monomers.
FIG. 4.
(A) The fusion ERs bind to ERE as monomers. The expression plasmids bearing ERα and the same (1:1) or a twofold-greater (1:2) concentration of the expression vector bearing the heterofusion Flag-α-β were cosynthesized in the presence of [3H]methionine in vitro. Equal amounts (5 μl) of reaction mixtures were subjected to SDS-7.5% PAGE or EMSA in the absence or presence (+) of the Flag antibody. (B) Intact DBDs are required for binding to ERE. Radiolabeled WT or DNA-binding defective (✽) ERα, the homofusion α-α, or the heterofusion α-β were subjected to SDS-PAGE and EMSA in the absence or presence (+) of Flag antibody. (C) Effect of dimerization surfaces in the LBDs on the ability of fusion receptor to bind to ERE. Equal amounts (5 μl) of WT or variant, with mutations in dimerization interface (indicated by the “dd” subscript), ERα, α-α, or α-β synthesized in vitro were subjected to SDS-PAGE. Then, 100 pM concentrations of each construct were electrophoresed on 5% nondenaturing PAGE (EMSA) in the absence or presence (+) of Flag antibody. (D) Dimerization interfaces in LBDs are required for an efficient binding of the fusion receptors to ERE. Equal molar concentrations (0, 6, 12, 25, and 50 pM [lanes 1, 2, 3, 4, and 5, respectively]) of WT and mutant (subscript dd) α-α or α-β were subjected to EMSA. For all experimental series, a representative experiment from at least two independent experiments is shown.
Each DBD of ER consists of two zinc finger-like modules that form a single functional domain. Each module contains a zinc ion with tetrahedral coordination by four Cys residues (8). Substitution of two of the zinc-coordinating Cys with His residues of the first zinc finger in the DBD of ERα prevents the receptor from interacting with DNA (21). If the fusion receptors bind to ERE as monomers, analogous mutations in the DBD of one α monomer should prevent the interaction of the variant fusion ERs with an ERE. Although synthesized at comparable levels, as assessed by fluorography (Fig. 4B, upper panel), the mutant homofusion α*-α and heterofusion α*-β failed, just as the DNA-binding defective ERα* dimer had failed, to bind to ERE in EMSA in contrast to the WT counterparts (Fig. 4B, lower panel). This result is consistent with the conclusion that fusion receptors bind to DNA as monomers.
The dimerization of ERα is primarily mediated by a surface located within the LBD of each monomer (21, 23). The absence of dimerization also suggests that two ER monomers in a single-chain receptor fold into a compact structure whose formation may or may not be dependent upon interactions between LBD dimerization surfaces. To address this point, we generated dimerization defective ERα. A previous study showed that changing of three Leu residues to Glu in the helix 11 of the LBD of the murine ERα prevents dimerization (41). We made analogous mutations by changing three Leu residues at positions 504, 508, and 511 to Glu. Similarly, we produced variant homofusion α-α that bears these same mutations in either (αdd-α or α-αdd) or both (αdd-αdd) ERα monomers, and we produced the heterofusion αdd-β that has mutations in the ERα monomer. The synthesis of mutant and WT fusion receptors was assessed by fluorography (Fig. 4C, upper panel). EMSA revealed that the dimerization defective ERαdd, as expected, did not significantly interact with an ERE (Fig. 4C, lower panel). Although the mutant fusion receptors (αdd-α is shown in Fig. 4), bound to an ERE, the interaction of these receptors with the ERE was also compromised (∼3-fold) compared to the WT counterparts when the binding was assessed at low concentrations of receptors (Fig. 4D). This indicates that the interactions between dimerization surfaces of both ERα monomers of the single-chain species increase the efficiency with which the single-chain receptor interacts with ERE.
Moreover, the αdd-β heterofusion, or homofusion αdd-αdd, as its WT counterpart, did not dimerize with the cosynthesized ERα (data not shown). Thus, fusion receptors fold to allow complementary interactions of the dimerization interfaces in the two LBDs, despite the head-to-tail fusion of two ER monomers in a single-chain configuration.
(ii) DNA-binding specificity.
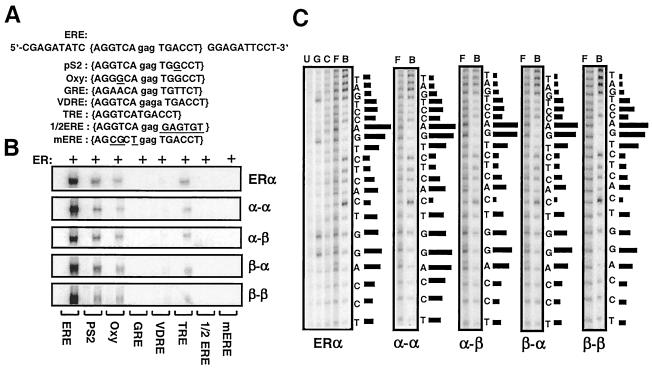
Efficient binding of single-chain receptors to ERE in a manner similar to the ER dimers implies that the DNA-binding specificity of ER is also preserved in fusion receptors. Therefore, we assessed the interaction of receptor species with DNA fragments that contain various test sequences (Fig. 5A) by EMSA (Fig. 5B). Previously, we (45) and others (27) have shown that ERα and ERβ bind to various ERE sequences with similar preference and affinity. Likewise, WT and fusion receptors bound the consensus ERE and ERE sequences derived from the estrogen-responsive pS2, complementary 3 (C3), oxytocin (Oxy) genes, and the thyroid hormone responsive element (TRE). Receptors had very little, if any, interaction with GRE and vitamin D responsive element (VDRE). There was no observable interaction with half-ERE (1/2ERE) or a mutant ERE sequence that bears three nucleotide substitutions in the consensus (mERE).
FIG. 5.
(A) Upper strand of the sequence containing the consensus ERE (in brackets), pS2, and Oxy ERE, GRE, VDRE, and TRE responsive elements and 1/2 ERE and mERE with the underlined five- and three-nucleotide changes from the ERE, respectively. The central base spacer is shown in lowercase. (B) Fusion ERs bind to the same spectrum of response elements. Equal molar concentrations of in vitro-synthesized ERα dimer or a fusion receptor were subjected to EMSA. A representative phosphorimage of two independent experiments is shown. (C) Fusion receptors bind to ERE in a manner similar to ERα. Critical nucleosides for ER-ERE interaction were identified by missing nucleoside hydroxyl radical assay. End-labeled DNA fragment containing consensus ERE was randomly cleaved by hydroxyl radical, incubated with 50 μl of transcription-translation mixtures, and subjected to 5% nondenaturing PAGE. Radioactive bands containing bound and free DNA were excised, eluted, and subjected to 15% sequencing gel electrophoresis. The intensities of individual DNA bands from sequencing gels were quantified by using a PhosphorImager. The ratio of free (F) to bound (B) DNA at each base was plotted as horizontal bars, the length of which approximates the strength of nucleoside contact with the protein. Uncut (U), Maxam-Gilbert G reaction-cut (G), and randomly cut (C) DNA fragments were also electrophoresed. A representative phosphorimage from at least two independent experiments is shown.
Conservation of the DNA-binding specificity of ERα in single-chain receptors also suggests that the fusion receptors utilize both half-sites in an ERE for binding. We tested this prediction by using the missing nucleoside hydroxyl radical assay that allows the analysis of DNA-protein interaction at a single-nucleotide resolution. It is expected that the protein binding to DNA would be adversely affected if a nucleoside important for binding were missing (13, 45). A low-intensity or missing band in the lane containing ERE-bound ERs (B) or, conversely, a high-intensity band in the lane containing free ERE (F), identifies a nucleoside important for the formation of the ER-ERE complex. A high ratio of free to bound ERE is represented by a long horizontal bar in Fig. 5C; the length of a bar represents the strength of contacts with ERs. The results revealed that the fusion receptors, just as for the ERα dimer, occupied both half-sites of the consensus ERE by contacting the same nucleosides with similar strength (Fig. 5C).
Moreover, the fusion receptors bound to the consensus, pS2 and Oxy ERE sequences with affinities similar to those of the ERα dimer as assessed by DNA competition assay (http://dbb.urmc.rochester.edu/labs/muyan/figure.htm). Likewise, all receptor species bound to E2 with comparable affinities (http://dbb.urmc.rochester.edu/labs/muyan/figure.htm). We also found that stoichiometry of E2 binding among ER species was primarily conserved. For example, as we reported previously (30), 1 mol of ERα binds 1 mol of E2, whereas 1 mol of α-α or α-β binds 1.86 ± 0.11 or 1.77 ± 0.19 (n = 3) mol of E2, respectively. Thus, the folding of the fusion ERs allows the receptors to bind to ERE and E2 efficiently.
(iii) Cofactor interactions.
The p160 family of coregulators interacts with the agonist-nuclear receptor complex through a LXXLL nuclear receptor interacting motif (NRIM) (14). Upon E2 binding, the LBD undergoes a conformational change by which the helix 12 located at the carboxyl terminus of the LBD is realigned over the ligand-binding pocket (3, 35). Since the conjugation approach uses a fusion between the amino terminus of one monomer and the carboxyl terminus of the other, the critical alignment of helix 12 could be compromised despite the fusion receptors bind to E2 efficiently.
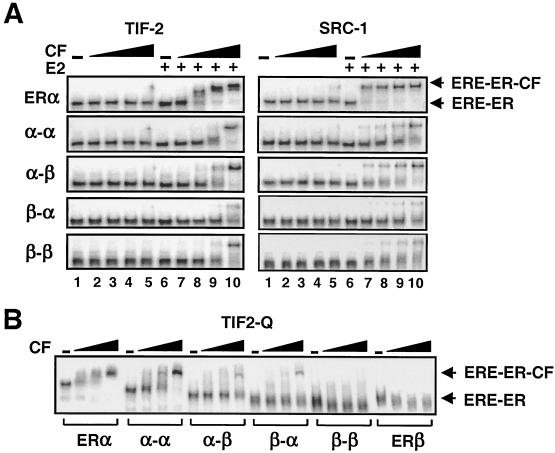
To address this issue, we examined the abilities of the fusion receptors to interact with cofactors by using EMSA. We (45) and others (1, 19, 26, 33, 42, 43) showed that the p160 family of cofactors, including TIF-2 and SRC-1, interact with ERα and ERβ in an agonist-dependent manner. Equal molar concentrations of fusion receptors synthesized in vitro and the ERα dimer were preincubated without or with a saturating concentration (10−7 M) of E2. Samples were then incubated with an end-labeled ERE. GST alone or the GST fusion polypeptide of TIF-2623-986 containing NRIM was added into the reaction mixtures at increasing concentrations. Reactions were subjected to EMSA (Fig. 6A). GST alone at any concentration had no effect on the electrophoretic mobility of the ERE-ER complexes in the absence or presence of E2 (data not shown). The electrophoretic mobility of ERE-bound ERα was quantitatively retarded by TIF-2 in response to E2 (lanes 7 to 10). E2 also enhanced the ability of the fusion receptors to recruit TIF-2 with affinities about twofold lower than that of the ERα dimer. SRC-1219-399 (SRC-1) interacted with the E2-bound ERα-ERE complex. The SRC-1 was also recruited by the fusion receptors, however, with efficiencies ∼5-fold lower than that observed with the ERα dimer. Apparent differences in the extent of SRC-1 recruitment compared to TIF-2 by the fusion receptors also suggest differences in the binding affinities of cofactors for fusion receptors.
FIG. 6.
(A) Interactions of cofactors (CF) with fusion ERs as assessed by EMSA. Equal molar concentrations of in vitro-synthesized receptors were preincubated in the absence (−, lanes 1 to 5) or presence (+, lanes 6 to 10) of 10−7 M E2 (E2), followed by the addition of the end-labeled consensus ERE. The reactions were then incubated with 0 (−),1.56, 6.25, 25, or 100 ng (lanes 1 to 5 and 6 to 10, respectively) of GST fusion TIF-2623-986 (TIF-2) or 0, 0.125, 0.25, 0.5, and 1 μg (lanes 1 to 5 and 6 to 10, respectively) of SRC-1219-399 (SRC-1). Reactions were resolved on 5% nondenaturing PAGE. A representative phosphorimage of two independent experiments is shown. Unbound ERE is not shown. (B) Interaction of increasing concentrations (0.3, 0.6, and 1 μg) of TIF21125-1325 (TIF2-Q)-GST fusion protein with ERE-bound ERs. A representative phosphorimage of two independent experiments is presented. Free DNA is not shown.
In addition to NRIM, TIF-2 through a glutamine-rich region (Q) also interacts efficiently with the amino terminus of ERα, but not of ERβ, independent of E2 (38, 43, 45). To examine whether this receptor-subtype specific interaction is preserved in the heterofusion ERs, the interaction of the GST fusion TIF-2 containing residues 1125 to 1325 (TIF2-Q) was tested in EMSA with receptors synthesized in vitro (Fig. 6B). The results revealed that increasing concentrations of the cofactor gradually retarded the electrophoretic migration of the EREbound ERα and α-α in the absence or presence (data not shown) of 10−7 M E2. The α-α bound to the cofactor with ∼2-fold-lower affinity than the dimer ERα. The TIF-Q, on the other hand, did not affect the electrophoretic migration of ERβ and β-β. TIF2-Q interacted quantitatively with the heterofusion receptors, however, with affinities twofold lower than those observed with the homofusion α-α.
Overall, these results suggest that cofactor interacting surfaces in the fusion receptors are altered.
Biological activities of the fusion ERs. (i) ERE-dependent ER signaling.
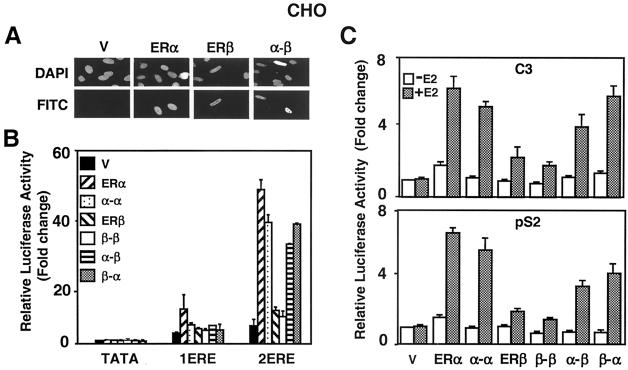
We next examined whether the homofusion α-α and β-β simulate the transactivation properties of the parent ER dimers. Furthermore, we attempted to determine whether the heterofusions α-β and β-α display novel functions by incorporating functional properties of both contributing partners in genomic estrogen signaling pathways. ERβ has considerably less transcription potency than ERα in ERE-dependent signaling pathways independent of the promoter and cell context (11, 31). The expression vector without or with cDNA for an ER was cotransfected into CHO cells, together with a reporter plasmid bearing no (TATA), one (1ERE), or two (2ERE) consensus EREs placed upstream of a simple TATA box (TATA) promoter that drives the expression of the firefly luciferase cDNA as reporter. Normalized activity from each reporter construct was compared to the basal activity from the reporter bearing no ERE in response to the parent expression vector (V) in the absence of E2, with the latter value set to one. Proteins were synthesized at similar amounts (Fig. 3B, Flag-CHO) and localized to the nucleus in the absence (Fig. 7A) or presence (data not shown) of a physiological concentration (10−9 M) of E2. As we showed previously (44), ERα, but not ERβ, induced transcription synergistically from two ERE-containing reporter constructs in CHO cells (Fig. 7B). Although α-α increased luciferase activity synergistically in response to E2, albeit to a lesser extent than that observed with ERα, β-β had only an additive effect on luciferase activity. On the other hand, heterofusion α-β or β-α induced synergy at levels comparable to that observed with the α-α. These results show that the homofusion α-α and β-β emulate the activities of the parent receptors and suggest, as proposed previously (40), that ERα is the dominant partner in the ER heterodimer when tested from a simple TATA box promoter construct bearing only tandem consensus EREs.
FIG. 7.
(A) Intracellular localization of fusion ERs was examined by immunocytochemistry. The expression vector without (V) or with a cDNA for Flag ERα, ERβ, or fusion α-β was transiently transfected into CHO cells. The proteins were probed with the Flag antibody and visualized by using fluorescein-conjugated secondary antibody (FITC). DAPI (4′,6′-diamidino-2-phenylindole) staining indicates the nucleus. There was no protein detectable in cells transfected with the parent vector. The absence or presence (data not shown) of 10−9 M E2 did not affect the intracellular localization of the ERs. (B) CHO cells were transiently transfected with 75 ng of expression vector bearing a cDNA for an ER, together with 125 ng of reporter plasmid bearing no (TATA), one (1ERE), or two copies of the consensus ERE (2ERE). The TATA box promoter drives the expression of the firefly luciferase cDNA. The transfection efficiency was monitored by determining the coexpression of 2 ng of reporter plasmid bearing the Renilla luciferase cDNA. Cells were treated without (data not shown) or with 10−9 M E2 (E2) for 24 h. The data represent the means ± the standard errors of the mean (SEM) of three independent experiments performed in duplicate. (C) CHO cells transiently transfected as described above by using a reporter plasmid bearing the C3 or pS2 promoter that drive the expression of the firefly luciferase enzyme cDNA. Cells were treated in the absence (−E2) or presence (+E2) of 10−9 M E2 for 24 h. The results from three independent experiments in duplicate are represented as the means ± the SEM.
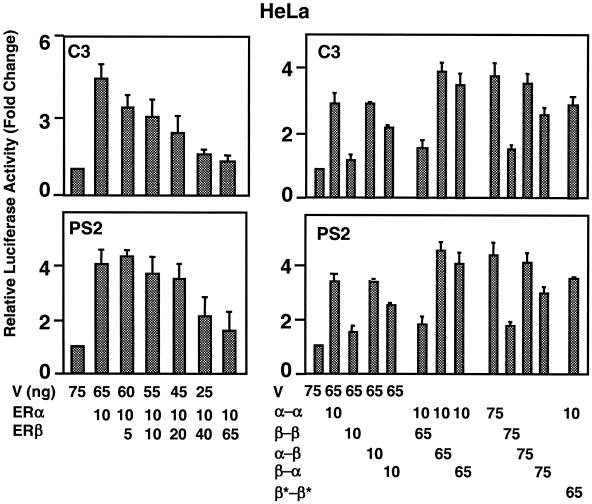
The expression of estrogen-responsive genes is the result of integrated effects of various trans-acting factors that are critical for gene expression. We examined the biopotencies of the fusion ERs from constructs bearing the enhancer-promoter region of estrogen-responsive pS2 and C3 genes that drive the expression of the luciferase enzyme cDNA. To accomplish this, we transiently transfected CHO cells with an expression vector bearing a receptor cDNA (Fig. 7C). Normalized activity from each reporter was compared to the promoter activity in response to the parent expression vector (V) in the absence of E2, with the latter value set to one. ERα and the homofusion α-α in response to 10−9 M E2 augmented the luciferase activity from both the pS2 and C3 promoters ∼6-fold, whereas ERβ and β-β increased transcription ∼2-fold. The heterofusion ERs showed biopotencies that were similar to those observed with ERα and α-α from the C3 promoter. The biopotencies of the heterofusions were, on the other hand, intermediate between the homofusion receptors that simulated the transcription activities of the parent ERα and ERβ when tested with the pS2 promoter construct.
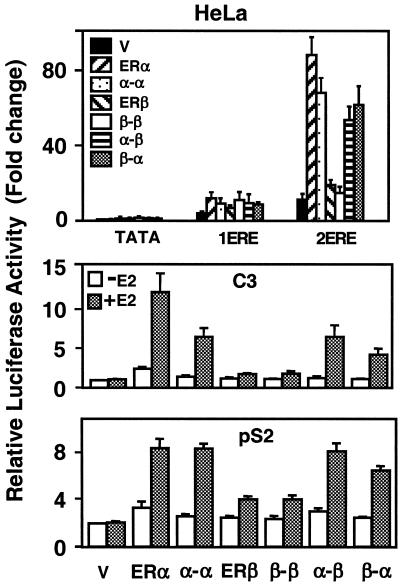
In HeLa cells, the heterofusion ERs also induced synergy with biopotencies that simulate that of α-α, which showed less potency than the ERα dimer (Fig. 8). The ERβ dimer and β-β had little effect on luciferase activity. The magnitude of transcription from the C3 promoter by the heterofusion ERs in response to 10−9 M E2 was similar to that of α-α, which had significantly lower potency than the ERα dimer (Fig. 8). Both ERβ and β-β, on the other hand, had minimal effects on luciferase activity. The heterofusion receptors showed transcription capacities comparable to those of α-α and ERα when tested from the PS2 promoter.
FIG. 8.
HeLa cells were transiently transfected as described for Fig. 7. The data represent the means ± the SEM of three experiments performed in duplicate.
These results suggest that, although the biopotencies of the fusion receptors are dependent upon the promoter and cell context, the heterofusions α-β and β-α emulate the activity of α-α rather than the β-β when tested with estrogen-responsive gene promoters. Thus, it appears that ERα dictates the mode of transcription of the ERα/β heterodimer in the ERE-dependent ER signaling pathway.
(ii) Repression of ERα transactivity by ERβ.
Previous reports indicated that ERβ functions as a dominant inhibitor of ERα transcriptional activity in the ERE-dependent signaling pathway in transfected mammalian cells (12). Indeed, cotransfections of an expression vector bearing ERα cDNA, together with increasing amounts of ERβ cDNA in transiently transfected HeLa cells, revealed that ERβ effectively inhibits the E2-ERα induced transcription from the C3 or the pS2 gene promoter (Fig. 9, left panels). At the concentration that induced maximal repression, the inhibition of ERα-induced transcription by ERβ was independent of the E2 amount regardless of the promoter-type (data not shown).
FIG. 9.
Effects of coexpression of ERs and fusion receptor on transcriptional responses. (Left) HeLa cells were transiently transfected with a constant amount (in nanograms) of ERα expression vector and increasing amounts of the ERβ expression vector. Cells were cotransfected with a reporter plasmid bearing the C3 or pS2 promoter that drives the expression of the firefly luciferase enzyme cDNA. Cells were treated without (data not shown for simplicity) or with 10−9 M E2 for 24 h. (Right) HeLa cells were transiently transfected with an expression vector bearing a fusion ER cDNA alone or together with another fusion ER cDNA. Cells were also transfected with the C3 or pS2 reporter plasmid. After transfection, cells were incubated in the absence (data not shown) or presence of 10−9 M E2 for 24 h. V, parent expression vector. The data are the means ± the SEM of three independent experiments performed in duplicate.
This inhibitory capacity of ERβ on ERα-induced transcription in coexpression systems could occur at multiple levels. These include heterodimerization, competition for binding to DNA, and/or transcriptional silencing through squelching. Since the fusion receptors do not dimerize, hence generating homogenous populations of receptor species, we reasoned that coexpression of the fusion receptors would allow us to address the underlying mechanism. To accomplish this, we performed a cotransfection assay using an expression vector bearing the α-α cDNA with an expression vector carrying the β-β or heterofusion α-β or β-α cDNA (Fig. 9, right panels). The results revealed that the cotransfected β-β repressed the transregulatory activity of α-α. On the other hand, the heterofusion α-β or β-α augmented the luciferase activity induced by α-α. This finding also suggests that the repression of the α-α activity by β-β is independent of sequestration of factors required for transactivation under the condition we are testing. Since the heterofusion ERs simulate the activities of the ERα dimer and α-α, these results also imply that ERβ as a homodimer represses the ERα function in the ERE-dependent signaling pathway. Indeed, the DNA-binding-defective β-β (β*-β*) bearing two mutant ERβ monomers did not affect luciferase levels induced by α-α. Thus, an effective repression of ERα-mediated transcriptional responses by ERβ occurs through competition for ERE binding.
(iii) ERE-independent and DNA-dependent ER signaling.
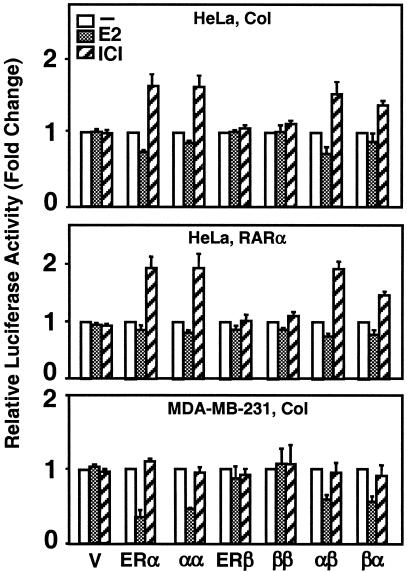
The ligand-ER complex regulates the expression of the Col and RARα genes through the DNA-dependent and ERE-independent signaling pathway. The functional tethering of the AF-1 and/or AF-2 domains of both ERs to the Jun/Fos family of proteins bound to the AP-1 element in the promoter of the Col gene provides the basis for gene responsiveness to ligand-ER signaling (22). Similarly, the functional interaction of the activation domains of ERα, but not ERβ, with the Sp-1 transcription factor bound to the GC box provides responsiveness for the RARα gene expression (36). Using a reporter plasmid that contains the Col or RARα promoter as a model for the DNA-dependent and ERE-independent signaling, we examined whether or not the heterofusion ERs emulate the ligand-mediated effects of ERα. The expression vector with cDNA for an ER was cotransfected into cells with a reporter plasmid with the Col or RARα promoter that drives the expression of the firefly luciferase cDNA. Ligand-mediated responses from the reporter plasmid by an expression vector were compared to the activity in the absence of ligand, the latter value being set to one.
The ligand-mediated responses from reporter vectors were cell type dependent. Although ligand ER did not alter luciferase activity from either the Col or the RARα promoter in transiently transfected CHO cells, the homofusion ERs simulated the transactivation abilities of the parent dimers in response to ER ligands in HeLa cells (Fig. 10). ERα and α-α induced the transactivation from both the Col and the RARα promoters in response to 10−7 M ICI (see also Materials and Methods), whereas ERβ or β-β had no effect on luciferase activity in the presence of ICI. The heterofusion ERs induced transcription in response to ICI in a manner similar to the ERα dimer and α-α. E2 at any concentration tested, shown at 10−9 M, had little effect on luciferase activity from the Col or the RARα promoter.
FIG. 10.
Transcriptional responses to fusion ERs from the ERE-independent signaling pathway. HeLa or MDA-MB-231 cells were transiently transfected with 125 ng of expression vector bearing no cDNA (V) or a cDNA for an ER, together with 125 ng of reporter vector that contained the Col or the RARα promoter driving the expression of firefly luciferase enzyme cDNA. Cells were treated without (−) or with 10−9 M E2 (+E2) or 10−7 M ICI (ICI) for 40 h. The means ± the SEM of four independent experiments are shown.
E2, on the other hand, suppressed reporter enzyme activity mediated by ERα and α-α in the ER-negative MDA-MB-231 cell line derived from a breast adenocarcinoma; whereas ICI had little effect on luciferase activity (Fig. 10, lower panel). Both the α-β and the β-α heterofusions emulated the E2-mediated effects of ERα and α-α on transcription from the Col promoter. The ligand-receptor complexes had no effect on responses from the RARα promoter in this cell line (data not shown). Thus, as observed with the ERE-dependent signaling, it appears that ERα dictates the mode of transcriptional properties of the heterofusion proteins in the DNA-dependent and ERE-independent signaling pathway as well.
DISCUSSION
In addition to acting as homodimers, the coexpression of ERs leads to heterodimerization, the extent of which, as we show here, is dependent upon the relative amount of each subtype. However, the role of the ERα/β heterodimer in E2 signaling has remained largely unknown. This is because coexpression leads to the formation of not only the ERα/β heterodimer but also the ERα and ERβ homodimers that prevent examination of the transcriptional ability of the heterodimer and elucidation of its role in E2 signaling. Using a genetic conjugation approach, we generated monomeric α-β and β-α receptors to integrate the functions of ERα and ERβ into a single-chain protein in order to study the role of ERα/β heterodimer in genomic signaling pathways. The homofusion α-α and β-β simulated the functions of the parent ERα and ERβ dimers, respectively. The heterofusions α-β and β-α, on the other hand, mimicked the function of the ERα dimer. Thus, ERα defines the transregulatory mode of the ERα/β heterodimer.
Single-chain receptors are in a dimer-like configuration.
We show here that single-chain receptors as monomers interacted with the consensus ERE by utilizing the same nucleosides as the parent ER dimers. Moreover, single-chain receptors bound to the same spectrum of responsive elements as the ER dimers with binding affinities similar to those of ERs. Likewise, the single-chain receptors bound to E2 with high affinity. These results suggest that the two ER monomers in fusion proteins fold to a configuration that juxtaposes two DBDs and LBDs to interact with EREs and E2 in a manner comparable to the ER dimers. These results suggest that despite the tail-to-head joining of two receptor monomers in the single-chain receptor, monomers fold to associate intramolecularly in a manner comparable to the ER dimers that dimerize through an intermolecular assembly.
ERα dictates the functions of the ERα/β heterodimer in genomic signaling pathways.
Conformational changes induced by the binding of E2 to ER realign helix 12 of the LBD over the ligand-binding pocket to form a cofactor interacting surface, together with the contributions from helices 3 and 4/5 (3, 35). Previous studies indicated that distinct classes of coactivators recognize distinct but overlapping sites on the agonist occupied LBD of ERs (7). It was also shown that one molecule of SRC-1 through two LXXLL motifs interacts with both cofactor interacting surfaces of LBDs in an agonist-bound ERα dimer (9, 17), as reported for the PPARγ homodimer (32). The genetic conjugation fuses the amino terminus of one monomer and the carboxyl terminus of the other, domains of the protein that are critical for interactions with cofactors. Although single-chain ERs bind to EREs and E2 with high affinities, the E2-dependent recruitment of the AF2-dependent cofactors by fusion ERs was found to be less efficient than that of the ER dimers. This suggests that the cofactor interacting surfaces are altered. Remarkably, however, two ER monomers in fusion proteins apparently form a ligand-dependent cofactor surface sufficient for interaction with the p160 family of cofactors.
In addition to NRIMs, TIF-2 through a glutamine-rich region (TIF2-Q) interacts preferentially with the amino-terminal AF-1 domain of ERα independently from ligands (2, 30, 38, 43, 45). We also observed here that the homofusion α-α, but not β-β, recruited TIF2-Q with a lower affinity than that observed with the ERα dimer. These results indicate that the receptor subtype-specific interaction of TIF2-Q, although not at the natural extent, is preserved in single-chain receptors. Similarly, the heterofusions α-β and β-α interacted with the TIF2-Q with efficiencies twofold lower than that observed with the homofusion α-α. This indicates that the presence of one ERα monomer is sufficient for the ability of the heterodimer to recruit ligand-independent cofactors.
ERβ has considerably less transcription potency than ERα in heterologous expression systems that utilize EREs. Studies indicated that receptor-specific AF-1 defines the transcriptional strength of ERα and ERβ. It appears that the ability of ERα to recruit the p160 family of cofactors through the AF-1 domain (2, 30, 38, 43, 45) and to subsequently integrate the AF-1 and AF-2 functions (2, 45) is critical for the biopotency of the receptor. TIF2-Q is also recruited preferentially by the homofusion α-α and the heterofusions α-β and β-α, in contrast to β-β, independently from E2. This implies that the abilities of these fusion receptors to interact with distinct surfaces of a cofactor likely underlie their mimicry of ERα functions in the ERE-dependent signaling pathway. We also observed that the extent of transcription induction by fusion receptors was dependent upon the promoter and cell context. This could be due to combinatorial effects of many trans-acting components specific to each gene and, consequently, differences in the composition or concentration of coregulatory proteins, which vary among E2 target tissues. Since the interaction of fusion receptors with SRC-1 was more severely affected compared to TIF-2, differences in the binding affinities of cofactors for fusion receptors could also contribute to differences in a promoter and cell type responses to single-chain receptors.
Our observations that the ligand-mediated responses of the ER dimers are preserved in the corresponding homofusion receptors from the Col and RARα promoters representing ERE-independent genomic signaling are consistent with the suggestion that single-chain receptors display configurations that mimic the parent ER dimers. Since heterofusion receptors emulated the functions of the ligand-ERα complex, ERα appears to dictate the ligand-mediated functions of ERα/β heterodimer in the ERE-independent DNA-dependent pathway as well. We therefore suggest that the ERα/β heterodimer mimics the mode of transcription of ERα in the genomic estrogen signaling.
Implications for E2 signaling.
Although the consequences of ERα and ERβ cosynthesis in vivo are unknown, a fine tuning of tissue responses to E2 is likely mediated by the integrated effects of the ER dimers and the ERα/β heterodimer. It is certain that ERβ mediates the expression of ERE-bearing responsive genes in response to E2, albeit less potently than ERα, when expressed alone. Coexpression of ERβ could attenuate the transregulatory potential of ERα in the ERE-dependent genomic signaling pathway by heterodimerization and could also antagonize the effects of the ERα dimer as a homodimer. Due to the absence of significant transcriptional responses to ligand-ERβ in experimental approaches we used, assessing the cross-effects of the ligand-ER complexes on transcriptional responses from the Col or the RARα promoter was difficult. It remains possible that in addition to the ERE-dependent signaling pathway, the ligand-ERβ complex could also modulate the transcriptional responses mediated by the ERα dimer from the ERE-independent genomic signaling pathway. Therefore, an alteration in the regulatory balance resulting from aberrant synthesis of either or both ER subtypes could lead to a malignancy in E2 target tissues wherein both subtypes are synthesized. Studies, for example, suggest that ERα and ERβ can be coexpressed in normal breast tissue (6, 16, 25, 37). However, the ratio of ERα to ERβ appears to be altered in samples obtained from breast cancer patients; while ERα is expressed at higher levels, the expression of ERβ is decreased (25).
The genetic conjugation approach precludes the limitation of monomer association into biologically active dimers and the dissociation of dimers into inactive monomer induced by destabilizing mutations. This offers an opportunity to address the roles of ER dimers as homogeneous populations in the physiology and pathophysiology of estrogen signaling. Moreover, single-chain ERs could be utilized to further the understanding of nongenomic effects of ER dimers. Permitting functional analysis of unique symmetrical or asymmetrical mutations that simulate variant homo- and heterodimers, genetic conjugation would also allow us to address the effects of various signaling pathways that cross talk with ER and fundamentally alter ligand-ER functions. The fusion protein approach could be extended further to other members of multisubunit complexes of the steroid/thyroid hormone receptor superfamily, and to non-ligand-dependent transcription factors that act primarily as dimers.
Acknowledgments
We thank Jay Reeder for allowing us to access the fluorescence microscopy facilities. We are grateful to Cheeptip Benyajati, Jeffrey J. Hayes, and Mark Sowden for critical reading of the manuscript.
This study was supported by National Institutes of Health grant HD 24459 (R.H., R.A.B., and M.M.) and an American Cancer Society Institutional Research Grant (M.M.).
REFERENCES
- 1.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 2.Benecke, A., P. Chambon, and H. Gronemeyer. 2000. Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 1:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brzozowski, A. M., A. C. Pike, Z. Dauter, R. E. Hubbard, T. Bonn, O. Engstrom, L. Ohman, G. L. Greene, J. A. Gustafsson, and M. Carlquist. 1997. Molecular basis of agonism and antagonism in the estrogen receptor. Nature 389:753-758. [DOI] [PubMed] [Google Scholar]
- 4.Cowley, S. M., S. Hoare, S. Mosselman, and M. G. Parker. 1997. Estrogen receptors α and β form heterodimers on DNA. J. Biol. Chem. 272:19858-19862. [DOI] [PubMed] [Google Scholar]
- 5.Cowley, S. M., and M. G. Parker. 1999. A comparison of transcriptional activation by ER α and ERβ. J. Steroid Biochem. Mol. Biol. 69:165-175. [DOI] [PubMed] [Google Scholar]
- 6.Dotzlaw, H., E. Leygue, P. H. Watson, and L. C. Murphy. 1999. Estrogen receptor-β messenger RNA expression in human breast tumor biopsies: relationship to steroid receptor status and regulation by progestins. Cancer Res. 59:529-532. [PubMed] [Google Scholar]
- 7.Eng, F. C., A. Barsalou, N. Akutsu, I. Mercier, C. Zechel, S. Mader, and J. H. White. 1998. Different classes of coactivators recognize distinct but overlapping binding sites on the estrogen receptor ligand binding domain. J. Biol. Chem. 273:28371-28377. [DOI] [PubMed] [Google Scholar]
- 8.Freedman, L. P. 1992. Anatomy of the steroid receptor zinc finger region. Endocrinol. Rev. 13:129-145. [DOI] [PubMed] [Google Scholar]
- 9.Gee, A. C., K. E. Carlson, P. G. Martini, B. S. Katzenellenbogen, and J. A. Katzenellenbogen. 1999. Coactivator peptides have a differential stabilizing effect on the binding of estrogens and antiestrogens with the estrogen receptor. Mol. Endocrinol. 13:1912-1923. [DOI] [PubMed] [Google Scholar]
- 10.Green, S., V. Kumar, I. Theulaz, W. Wahli, and P. Chambon. 1988. The N-terminal DNA-binding ‘zinc finger' of the estrogen and glucocorticoid receptors determines target gene specificity. EMBO J. 7:3037-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, J. M., J. F. Couse, and K. S. Korach. 2001. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 276:36869-36872. [DOI] [PubMed] [Google Scholar]
- 12.Hall, J. M., and D. P. McDonnell. 1999. The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ER alpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 140:5566-5578. [DOI] [PubMed] [Google Scholar]
- 13.Hayes, J. J., and T. D. Tullius. 1989. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry 28:9521-9527. [DOI] [PubMed] [Google Scholar]
- 14.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J., P. Yi, X. Li, R. Hilf, R. A. Bambara, and M. Muyan. 2004. Targeting estrogen responsive elements (EREs): design of potent transactivators for ERE-containing genes. Mol. Cell Endocrinol. 218:65-78. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, E. V., G. Cheng, C. Palmieri, S. Saji, S. Makela, S. Van Noorden, T. Wahlstrom, M. Warner, R. C. Coombes, and J. A. Gustafsson. 2001. Estrogen receptors and proliferation markers in primary and recurrent breast cancer. Proc. Natl. Acad. Sci. USA 98:15197-15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the estrogen receptor. EMBO J. 17:232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinge, C. M. 2001. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29:2905-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraichely, D. M., J. Sun, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2000. Conformational changes and coactivator recruitment by novel ligands for estrogen receptor-α and estrogen receptor-β: correlations with biological character and distinct differences among SRC coactivator family members. Endocrinology 141:3534-3545. [DOI] [PubMed] [Google Scholar]
- 20.Kuiper, G. G., B. Carlsson, K. Grandien, E. Enmark, J. Haggblad, S. Nilsson, and J. A. Gustafsson. 1997. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138:863-870. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, V., and P. Chambon. 1988. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell 55:145-156. [DOI] [PubMed] [Google Scholar]
- 22.Kushner, P. J., D. A. Agard, G. L. Greene, T. S. Scanlan, A. K. Shiau, R. M. Uht, and P. Webb. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74:311-317. [DOI] [PubMed] [Google Scholar]
- 23.Lees, J. A., S. E. Fawell, R. White, and M. G. Parker. 1990. A 22-amino-acid peptide restores DNA-binding activity to dimerization-defective mutants of the estrogen receptor. Mol. Cell. Biol. 10:5529-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin, E. R. 2002. Cellular functions of plasma membrane estrogen receptors. Steroids 67:471-475. [DOI] [PubMed] [Google Scholar]
- 25.Leygue, E., H. Dotzlaw, P. H. Watson, and L. C. Murphy. 1998. Altered estrogen receptor α and β messenger RNA expression during human breast tumorigenesis. Cancer Res. 58:3197-3201. [PubMed] [Google Scholar]
- 26.Loven, M. A., V. S. Likhite, I. Choi, and A. M. Nardulli. 2001. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor β conformation. J. Biol. Chem. 276:45282-45288. [DOI] [PubMed] [Google Scholar]
- 27.Loven, M. A., J. R. Wood, and A. M. Nardulli. 2001. Interaction of estrogen receptors α and β with estrogen response elements. Mol. Cell Endocrinol. 181:151-163. [DOI] [PubMed] [Google Scholar]
- 28.McInerney, E. M., K. E. Weis, J. Sun, S. Mosselman, and B. S. Katzenellenbogen. 1998. Transcription activation by the human estrogen receptor subtype β (ERβ) studied with ERβ and ERα receptor chimeras. Endocrinology 139:4513-4522. [DOI] [PubMed] [Google Scholar]
- 29.Metzger, D., M. Berry, S. Ali, and P. Chambon. 1995. Effect of antagonists on DNA binding properties of the human estrogen receptor in vitro and in vivo. Mol. Endocrinol. 9:579-591. [DOI] [PubMed] [Google Scholar]
- 30.Muyan, M., P. Yi, G. Sathya, L. J. Willmert, M. D. Driscoll, R. Hilf, and R. A. Bambara. 2001. Fusion estrogen receptor proteins: toward the development of receptor-based agonists and antagonists. Mol. Cell Endocrinol. 182:249-263. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, S., and J. A. Gustafsson. 2002. Estrogen receptor action. Crit. Rev. Eukaryot. Gene Expr. 12:237-257. [DOI] [PubMed] [Google Scholar]
- 32.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 33.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 34.Pace, P., J. Taylor, S. Suntharalingam, R. C. Coombes, and S. Ali. 1997. Human estrogen receptor β binds DNA in a manner similar to and dimerizes with estrogen receptor α. J. Biol. Chem. 272:25832-25838. [DOI] [PubMed] [Google Scholar]
- 35.Pike, A. C., A. M. Brzozowski, R. E. Hubbard, T. Bonn, A. G. Thorsell, O. Engstrom, J. Ljunggren, J. A. Gustafsson, and M. Carlquist. 1999. Structure of the ligand-binding domain of estrogen receptor β in the presence of a partial agonist and a full antagonist. EMBO J. 18:4608-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safe, S. 2001. Transcriptional activation of genes by 17 β-estradiol through estrogen receptor-Sp1 interactions. Vitam. Horm. 62:231-252. [DOI] [PubMed] [Google Scholar]
- 37.Sasano, H., T. Suzuki, Y. Matsuzaki, T. Fukaya, M. Endoh, H. Nagura, and M. Kimura. 1999. Messenger ribonucleic acid in situ hybridization analysis of estrogen receptors α and β in human breast carcinoma. J. Clin. Endocrinol. Metab. 84:781-785. [DOI] [PubMed] [Google Scholar]
- 38.Sathya, G., P. Yi, S. Bhagat, R. A. Bambara, R. Hilf, and M. Muyan. 2002. Structural regions of ER α critical for synergistic transcriptional responses contain cofactor interacting surfaces. Mol. Cell Endocrinol. 192:171-185. [DOI] [PubMed] [Google Scholar]
- 39.Simoncini, T., A. Hafezi-Moghadam, D. P. Brazil, K. Ley, W. W. Chin, and J. K. Liao. 2000. Interaction of estrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature 407:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tremblay, G. B., A. Tremblay, F. Labrie, and V. Giguere. 1999. Dominant activity of activation function 1 (AF-1) and differential stoichiometric requirements for AF-1 and -2 in the estrogen receptor α-β heterodimeric complex. Mol. Cell. Biol. 19:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valentine, J. E., E. Kalkhoven, R. White, S. Hoare, and M. G. Parker. 2000. Mutations in the estrogen receptor ligand binding domain discriminate between hormone-dependent transactivation and transrepression. J. Biol. Chem. 275:25322-25329. [DOI] [PubMed] [Google Scholar]
- 42.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160-kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 43.Webb, P., P. Nguyen, J. Shinsako, C. Anderson, W. Feng, M. P. Nguyen, D. Chen, S. M. Huang, S. Subramanian, E. McKinerney, B. S. Katzenellenbogen, M. R. Stallcup, and P. J. Kushner. 1998. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol. Endocrinol. 12:1605-1618. [DOI] [PubMed] [Google Scholar]
- 44.Yi, P., S. Bhagat, R. Hilf, R. A. Bambara, and M. Muyan. 2002. Differences in the abilities of estrogen receptors to integrate activation functions are critical for subtype-specific transcriptional responses. Mol. Endocrinol. 16:1810-1827. [DOI] [PubMed] [Google Scholar]
- 45.Yi, P., M. D. Driscoll, J. Huang, S. Bhagat, R. Hilf, R. A. Bambara, and M. Muyan. 2002. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ERα and ERβ. Mol. Endocrinol. 16:674-693. [DOI] [PubMed] [Google Scholar]