Abstract
Snail and Slug are closely related transcriptional repressors involved in embryonic patterning during metazoan development. In human cancer, aberrant expression of Snail and/or Slug has been correlated with invasive growth potential, a property primarily attributed to their ability to directly repress transcription of genes whose products are involved in cell-cell adhesion, such as E-cadherin, occludin, and claudins. To investigate the molecular mechanisms of alterations in epithelial cell fate mediated by aberrant expression of Snail or Slug, we analyzed the consequences of exogenous expression of these factors in human cancer cells. Aberrant expression of either Snail or Slug led to changes in cell morphology, the loss of normal cell-cell contacts, and the acquisition of invasive growth properties. Snail or Slug expression also promoted resistance to programmed cell death elicited by DNA damage. Detailed molecular analysis revealed direct transcriptional repression of multiple factors with well-documented roles in programmed cell death. Depletion of endogenous Snail by RNA interference led to increased sensitivity to DNA damage accompanied by increased expression of the proapoptotic factors identified as targets of Snail. Thus, aberrant expression of Snail or Slug may promote tumorigenesis through increased resistance to programmed cell death.
The product of the Drosophila Snail gene represents the founding member of a superfamily of transcriptional regulators (29). Members of this family have been implicated in the formation of mesoderm and neural crest, as well as in the pathological progression of epithelial tumors (16). In mammals, the best-characterized members of the Snail superfamily, Snail and Slug, have each been implicated in pathological alterations of the epithelial cell phenotype associated with the acquisition of invasive growth properties by tumors (29). The molecular mechanisms by which aberrant expression of Snail and/or Slug lead to phenotypic alterations in epithelial cells have been a topic of intense study. A primary conclusion from this body of work asserts that pathological expression of Snail and Slug in epithelial cells leads to loss of expression of key cell-cell adhesion molecules, such as E-cadherin, claudins, and occludin (3, 6, 14, 18).
The Snail superfamily is characterized by a common protein organization. Family members contain a highly conserved region at the carboxyl terminus of the protein containing four to six zinc fingers of the C2H2 type (16, 29). The zinc fingers mediate sequence-specific interactions with DNA. Snail superfamily members bind specifically to consensus binding sites that contain the core sequence CAGGTG (16, 29). This motif constitutes a subset of the E-box (CANNTG), the conserved binding site of basic helix-loop-helix transcription factors (29). The amino termini of the superfamily are less well conserved, although most vertebrate members contain the evolutionarily conserved SNAG (for Snail/Gfi) domain at their extreme amino termini (16, 29). In addition, a conserved sequence motif near the zinc fingers differentiates Slug genes from other members of the superfamily (16, 29).
Snail superfamily members function as transcriptional repressors (16, 29). Transcriptional repression function is associated with various conserved protein domains. In vertebrates, the SNAG domain is essential to transcriptional repression function of Snail family members. Drosophila Snail lacks the SNAG domain, yet it still functions as a transcriptional repressor, through interaction with a well-characterized corepressor, CtBP (16, 29). Consensus sequences for interaction with CtBP are found in some, but not all, members of the Snail superfamily (29).
The Snail gene is essential for normal development of both Drosophila melanogaster (21) and mice (7). Homozygous null embryos in both species fail to produce mesoderm, resulting in a failure to gastrulate (7). The failure of mesodermal specification in Snail mutant embryos led to the characterization of Snail as a master regulator of epithelial to mesenchymal transitions (21). Slug, in contrast, is not essential for normal development in the mouse. Homozygous null animals are viable and fertile (23). Despite the lack of an embryonic lethal phenotype in mice, Slug has been implicated in epithelial to mesenchymal transitions during development in other vertebrates. In embryogenesis in the chick and in Xenopus sp., Slug function is required for specification of neural crest (21, 29). Thus, Snail and Slug function to promote cell fate changes during development, leading to the production of migratory, mesenchymal cells.
In addition to their roles in pattern formation and specification of mesoderm, some members of the Snail superfamily have been implicated in cell survival. The Caenorhabditis elegans CES-1 (for cell death specification) protein is a member of the Snail superfamily (most closely related to Drosophila Scratch) that can block programmed cell death of the NSM sister neurons during embryogenesis (28). In vertebrates, Slug is aberrantly upregulated by the E2A-HLF oncoprotein in certain leukemias, leading to increased cell survival (20). Further analysis of Slug during hematopoiesis has revealed a nonpathological role for Slug in promoting cell survival in hematopoietic progenitor cells (19, 33).
We investigate here the phenotypic change induced in human breast carcinoma cells after ectopic expression of Snail or of Slug. Aberrant expression of these factors has been proposed as a key determinant of pathological epithelial to mesenchymal transitions during tumor progression (29). As expected, these transcription factors induced phenotypic alterations in epithelial breast cancer cells, including loss of cell-cell contacts and acquisition of invasive growth. Surprisingly, this phenotypic change was accompanied by a dramatic alteration in the apoptotic response to the common chemotherapeutic agent, adriamycin. Cells expressing Snail or Slug were protected from apoptosis induced by this DNA-damaging agent. Analysis of apoptotic pathways revealed that ectopic expression of Snail leads to downregulation of multiple genes with known roles in programmed cell death.
MATERIALS AND METHODS
Expression vectors and adenovirus constructs.
Human Snail was cloned by PCR amplification from IMAGE clone no. 4537122 by using the following primers: Snail sense (5′-ATGCCGCGCTCTTTCCTCGTC-3′) and Snail antisense (5′-AGCGTAATCGGGGACATCGTAGGGGTAGCGGGGACATCCTGAGCAGCCGGA-3′).
Amplified Snail fragments were subcloned to pcDNA3.1 (Invitrogen). Human Slug was cloned by PCR amplification from IMAGE clone no. 3908245 by using the following primers: Slug sense (5′-CGCGGATCCGCCGCCATGCCGCGCTCCTTCCTGGTC-3′) and Slug antisense (5′-TCACTCGAGCTAAGCGTAATCGGGGACATCGTAGGGGTAGTGTGCTACACAGCAGCCAGATTCC-3′).
Amplified fragments were subcloned into pcDNA3.1 (Invitrogen). Adenovirus for expression of Snail or Slug with a hemagglutinin (HA) epitope tag was prepared as described previously (15).
To construct Snail short hairpin RNA (shRNA-snail), sense and antisense DNA oligonucleotides were designed form double-stranded RNA with a loop structure: Snail shRNA sense (5′-GGATCCCGCGAGCTGCAGGACTCTAAGAAGCTTGTTAGAGTCCTGCAGCTCGCTTTTTT-3′) and Snail shRNA antisense (5′-CTAGAAAAAAGCGAGCTGCAGGACTCTAACAAGCTTCTTAGAGTCCTGCAGCTCGCGG-3′).
After annealing, the fragment was subcloned in to pGE-1 vector (Stratagene). shRNA-snail adenovirus was constructed by excising the shRNA-snail fragment, along with the U6 promoter from shRNA-snail/pGE-1, and ligating the fragment into pAdTrack, from which the cytomegalovirus promoter used for protein expression had been deleted. Adenoviruses were purified by CsCl banding.
Cell culture, transfection, and drug treatment.
Cell lines from human breast carcinoma (MCF7) and melanoma (A375) were obtained from the American Type Culture Collection and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. MCF7 cells were treated with 3 μM adriamycin (ADR) for 2 h before culture in drug-free medium for the indicated times. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed by using commercially available reagents (In Situ Cell Death Detection Kit TMR red; Roche) according to the manufacturer's protocol after fixation as described previously (9).
Antibody and Western blotting.
Human Snail antibodies were generated in rabbits by using His6-tagged recombinant Snail protein (amino acids 1Met to 146Lys). For affinity purification, recombinant Snail was coupled to N-hydroxysuccinimide-activated Sepharose 4 Fast flow (Amersham). Other antibodies utilized included: anti-E-cadherin monoclonal antibody (MAb; Zymed), antioccludin MAb (Zymed), anti-HA MAb (Roche), antiactin MAb (Chemicon), and anti-p53 MAb (Oncogene).
Immunofluorescence staining.
Infected cells were seeded on glass coverslips, fixed with 2% paraformaldehyde in phosphate-buffered saline, and covered with blocking solution (10% fetal bovine serum-phosphate-buffered saline). Subsequently, cells were incubated with antioccludin MAb. After the cells were counterstained with goat anti-mouse antibody conjugated with Texas red, the slides were examined and photographed by using a Zeiss LSM510 laser scanning confocal microscope (Zeiss Microimaging, Inc.).
In vitro invasion analysis.
Mock and Snail adenovirus-infected cells (2.5 × 105 cells) were seeded on Matrigel invasion chambers (BD Bioscience). After 22 h of incubation in a humidified incubator, noninvading cells were removed, and the invaded cells were fixed and stained with crystal violet. Stained cell were counted by using a microscope.
RT-PCR.
Total RNA was extracted by a guanidinium thiocyanate-phenol-chloroform extraction method (11). cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase by using random hexamers. To quantify mRNA, reverse transcription-PCR (RT-PCR) with real-time quantitation was performed by using the iCycler System (Bio-Rad) as described previously (11). Primer sequences are available on request.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) was performed by using a ChIP assay kit (Upstate) as described previously (11). In brief, cells were cross-linked with 1% formaldehyde and sonicated to generate 300- to 3,000-bp DNA fragments. Immunoprecipitation was performed with anti-HA MAb (Roche) and control antibody (Upstate). Precipitated DNAs were detected by PCR by using specific primers. Primer sequences are available on request.
RESULTS
In order to investigate the physiological outcomes of aberrant expression of Snail or of Slug in human breast cancer cells, we developed a recombinant adenovirus system permitting exogenous expression of these factors (15). This system was used to assess the effects of aberrant Snail or Slug expression on epithelial architecture in MCF7 breast carcinoma cells that express endogenous Snail and Slug at very low levels. During initial characterization of this system, limiting amounts of Snail or Slug adenovirus were utilized, permitting simultaneous observation of infected and uninfected cells in single microscopic fields. Occludin, a transmembrane component of the tight junction, stained in the “chicken wire” pattern typical of epithelial cells (Fig. 1) in the absence of infection or after infection with mock virus. In contrast, in cells infected with either Snail or Slug adenovirus, occludin staining was lost, and the cells were unable to form productive contacts with their neighbors (Fig. 1). Immunoblot analysis of two well-characterized transcriptional targets of Snail (Fig. 1j) revealed a dramatic reduction of occludin (18), a finding consistent with the immunocytochemistry, and a modest reduction of E-cadherin (3, 6, 14). Slug expression resulted in qualitatively similar effects on E-cadherin and occludin albeit with slightly altered kinetics, probably relating to differences in the expression levels of Snail and Slug (Fig. 1j). The levels of repression of E-cadherin evident in this system differ somewhat from Snail/Slug-dependent repression of this molecule in MDCK cells (4) but are consistent with reports that use breast carcinoma cells (13).
FIG. 1.
Expression of Snail or Slug leads to phenotypic alterations in epithelial cells. (a to i) MCF7 cells were infected with mock (a, b, and c), Snail (d, e, and f), or Slug (g, h, and i) adenovirus. The cellular distribution of green fluorescent protein (a, d, and g) and occludin (b, e, and h) was visualized. Merged images are presented in panels c, f, and i. (j) MCF7 cells infected with Snail, Slug, or mock adenovirus were analyzed by immunoblot for Snail or Slug (by using the HA epitope), actin, E-cadherin, and occludin. Quantitation of E-cadherin abundance by densitometry revealed that Snail expression results in 35% (2 days) and 53% (4 days) reductions in E-cadherin protein, whereas Slug expression results in 63% (2 days) and 60% (4 days) reduction.
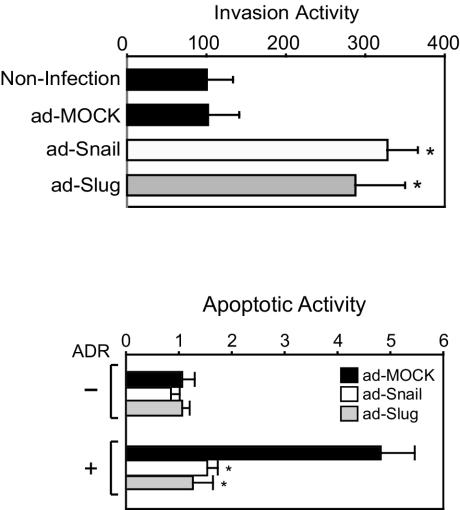
In addition to effects on junctional complex components, Snail and Slug also alter other features of epithelial cells, resulting in increases in migratory and invasive growth (16, 21, 29). In Matrigel invasion assays, Snail- and Slug-expressing cells were approximately three times more efficient at invasion than mock-infected or uninfected cells (Fig. 2, top panel). Thus, many of the phenotypic alterations attributed to aberrant Snail or Slug expression are faithfully recapitulated in cells infected with the recombinant adenoviruses. In considering the significance of this threefold increase in invasion to cancer progression, we note that a recent report documented the capacity of fibroblast growth factor 8b overexpression to alter growth characteristics of MCF7. Overexpression of this molecule resulted in a 2.5-fold increase in an in vitro invasion assay and a five- to sixfold increase in tumor volume in a xenograft model (37).
FIG. 2.
Expression of Snail or Slug leads to increased invasion and protection from programmed cell death. (a) Snail, Slug or mock adenovirus-infected MCF7 cells were seeded on Matrigel invasion chambers. Noninfected cells were included as a control. Invading cells were stained and counted. The data represent the mean value ± the standard deviation. ✽, P < 0.05 (as determined by using the Student t test). (b) After infection of MCF7 with Snail, Slug, or mock adenovirus, cells were treated with ADR, and genomic DNA cleavage was detected by TUNEL assay. For each cell line, an arbitrary value of 1 was assigned to the mock virus-infected cells in the absence of ADR treatment. All other values for a given cell line are expressed relative to this control group. The data represent the mean value ± the standard deviation. ✽, P < 0.05 (as determined by using the Student t test).
In epithelial cells, junctional complexes serve as platforms for cellular signaling (24, 34, 38). Loss of the junctional complex after transcriptional repression of E-cadherin and occludin could result in the disruption of prosurvival signaling. To investigate the response of cells expressing Snail or Slug to genotoxic stress, the topoisomerase inhibitor ADR was used to induce DNA damage. After drug exposure, the relative apoptotic activity of parental MCF7 cells increased substantially (Fig. 2b). Expression of either Snail or Slug totally prevented ADR-induced cell death in MCF7, suggesting that aberrant expression of these transcription factors in epithelial cells induces alterations in both cell morphology and in the apoptotic response. In the context of cancer progression, alterations in the in vitro response to apoptotic stimuli can be correlated to tumor growth in animal models. In MDA435 breast carcinoma cells, overexpression of the antiapoptotic molecule Bcl-xL resulted in a twofold decrease in apoptosis after transforming growth factor β treatment. In xenograft experiments, the capacity to form metastatic tumors was enhanced >5-fold (10).
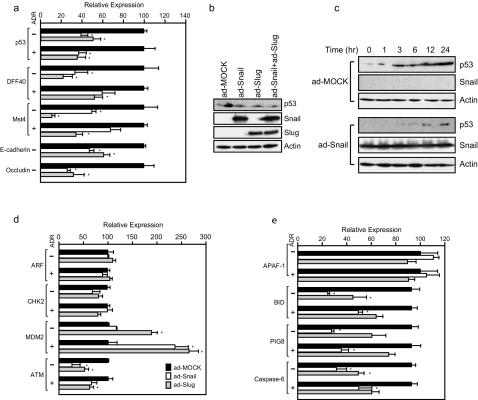
Given the function of Snail and of Slug as transcriptional repressors, their ability to interfere with the apoptotic program is predicted to result from alterations in gene expression. Since the tumor suppressor p53 plays an integral role in cellular responses to DNA damage, the role of Snail and Slug in p53 regulation was investigated. Expression of either Snail or Slug resulted in modest downregulation of steady-state levels of p53 mRNA (Fig. 3a), in keeping with the observation that the TP53 locus contains numerous potential binding sites for Snail and Slug. In addition to p53, mRNA levels for additional proapoptotic factors whose genomic loci contain potential binding sites for Snail and Slug were analyzed. Steady state mRNA levels of the apoptotic nuclease DFF40 (26), as well as Mst4, a kinase implicated in the apoptotic response (8), declined significantly in response to expression of Snail or of Slug (Fig. 3a). Occludin and E-cadherin, included as positive controls, demonstrated the expected Snail- or Slug-dependent decreases in mRNA levels (Fig. 3a). Expression of Snail also resulted in a modest decline in the steady-state level of p53 at the protein level (Fig. 3b). Similar results were obtained after expression of Slug or coexpression of Snail and Slug (Fig. 3b).
FIG. 3.
Expression of Snail or Slug alters the apoptotic response through transcriptional repression of multiple targets. (a) Total RNA was extracted from MCF7 that were infected with Snail, Slug, or mock adenovirus and treated with ADR or vehicle. Steady-state levels of the indicated transcripts were determined by using quantitative RT-PCR. All transcript levels were normalized to actin. All data represent the mean value ± the standard deviation of three independent experiments. ✽, P < 0.05 (as determined by using the Student t test). (b) Simultaneous expression of Snail and Slug does not lead to additive effects on p53 abundance. MCF7 cells were infected with mock, Snail, Slug, or Snail+Slug adenovirus. After 48 h, cells were harvested and protein lysates analyzed by immunoblotting. (c) Snail, Slug, or mock-infected MCF7 were treated with ADR, incubated for the indicated times, and analyzed by immunoblotting with the indicated antibodies. (d and e) Transcript levels for the indicated loci were analyzed as in panel a.
Regulation of the cellular p53 levels is known to result from a complicated set of regulatory steps, including posttranscriptional regulation. After DNA damage, the p53 protein is stabilized (36); therefore, the effect of Snail on p53 accumulation after ADR treatment was investigated. Mock-infected cells demonstrated the expected time-dependent accumulation of p53 (Fig. 3c). In contrast, Snail-expressing cells had reduced levels of p53 prior to treatment and had impaired accumulation of the protein following ADR treatment (Fig. 3c). Therefore, factors affecting p53 stability were examined (Fig. 3d). MDM2, an E3 ubiquitin ligase involved in p53 turnover (36), was unaffected by Snail expression in the absence of drug. However, after ADR treatment, Snail-expressing cells exhibited a statistically significant increase in MDM2 transcript levels. Slug expression resulted in increased steady-state levels of MDM2 transcript regardless of drug exposure (Fig. 3d).
Factors implicated in the stabilization of p53 showed a mixed response. ARF, the alternative reading frame product of the p16INK4a gene, stabilizes p53 independent of the DNA-damage response through inhibition of MDM2-dependent p53 turnover (36). ARF mRNA levels were unaffected by ADR treatment or by expression of either Snail or Slug (Fig. 3d). The checkpoint kinase CHK2 has been implicated in p53 stabilization through phosphorylation of the protein after DNA damage in mouse cells (17), although the relevance of this observation in human cells has recently been questioned (1, 22). CHK2 mRNA levels were not altered either by Snail expression, by Slug expression, or by ADR treatment (Fig. 3d). ATM, the gene product mutated in ataxia telangiectasia, is induced after DNA damage and is implicated in p53 modification and stabilization (2, 5). In the absence of ADR treatment, expression of either Snail or Slug results in significant repression of mRNA coding for the ATM kinase (Fig. 3d). Thus, the observed reduction in p53 protein levels after expression of Snail or Slug likely resulted from the combinatorial effects of three distinct mechanisms: a decline in p53 mRNA levels, increased MDM2-mediated turnover, and the loss of stabilizing modifications from the ATM kinase. The failure of Snail or Slug to alter the abundance of CHK2 and ARF suggests that the effects of aberrant expression of these transcriptional repressors on the p53 response are mediated through specific regulatory molecules. Since both CHK2 and ARF contain multiple E-box sequences (data not shown) and yet fail to respond to Snail or Slug, not all potential target loci respond to aberrant expression of these factors.
As levels of p53 protein were clearly affected by expression of Slug and Snail, we also analyzed transcript abundance of genes activated by p53 in response to genotoxic stress. Transcripts from a subset of these genes, including p53AIP1 (30), PIDD (25), PIG3 (35), and PUMA (40), were not detectable in MCF7 under any circumstances (data not shown). Others, including p53DINP1 (31), APAF-1 (41), and BAX (32), were unaffected by Snail (Fig. 3e and data not shown) or Slug (Fig. 3e). However, transcripts of the p53 targets BID (39), PIG8 (35), and Caspase 6 (27) were significantly repressed by ectopic expression of Snail or Slug (Fig. 3e). Thus, in addition to affects on p53 itself, aberrant Snail or Slug expression led to transcriptional effects on other genes integral to the DNA damage-induced programmed cell death pathway.
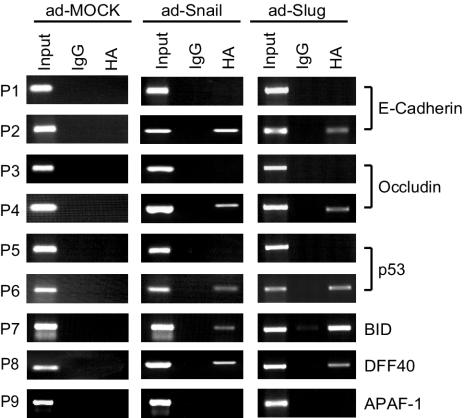
The repression at the CDH1, OCLN, TP53, BID, PIG8, and DFF40 loci could result from either direct effects or indirect effects of Snail and Slug. ChIP assays were performed to determine whether aberrantly expressed Snail and Slug bound to these loci. E-cadherin and occludin served as positive controls. After Snail or Slug expression, ChIP was performed, and coprecipitated DNA was analyzed by PCR. At all responsive loci, E-box sequences (Snail or Slug binding sites) were precipitated, whereas control sequences were not (Fig. 4). E-box DNA was amplified only after expression of the relevant repressor. Some loci containing E-boxes, such as APAF-1, are unresponsive, and the binding of Snail or Slug is not detected by ChIP. Thus, Snail and Slug can bind to the promoters of the TP53, BID, DFF40, OCLN, and CDH1 genes, the transcripts of which are all repressed after overexpression of Snail or Slug.
FIG. 4.
Snail and Slug localization at responsive genes. ChIP analyses of MCF7 cells infected with Snail, Slug, or mock adenovirus were performed. P1 to P9 indicate PCR primer sets that amplify genomic DNA from the indicated loci. Primer sets P2, P4, P6, P7, P8, and P9 amplify fragments containing Snail or Slug binding sites, whereas primer sets P1, P3, and P5 amplify nearby regions lacking Snail or Slug sites. DNA coprecipitating with Snail and control antibodies was amplified by using the indicated primer pairs.
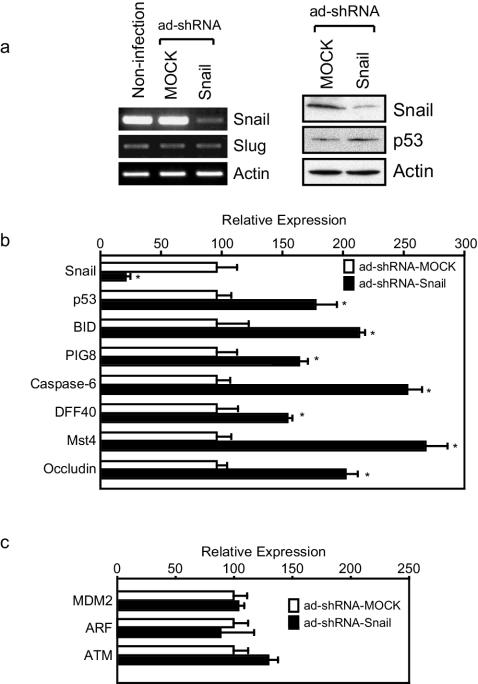
In order to confirm these results in a more physiologic context, Snail protein was depleted in A375, a human melanoma cell line that expresses Snail at moderate to high levels and is wild type at the TP53 locus, by using adenovirus-based short hairpin RNA interference (shRNA). Infection with the shRNA-Snail adenovirus resulted in depletion of endogenous Snail mRNA with no apparent effect on Slug mRNA (Fig. 5a, left panel). At the protein level, a decline in Snail protein was accompanied by moderate upregulation of p53 protein, as expected (Fig. 5a, right panel). RT-PCR was performed for selected transcripts after Snail depletion (Fig. 5b). As expected, the endogenous Snail transcript declined. Statistically significant upregulation of p53, BID, PIG8, Caspase 6, DFF40, MST4, and OCLN was observed (Fig. 5b). No significant difference was observed in MDM2, ARF, and ATM (Fig. 5c). Thus, endogenous Snail represses steady-state levels of TP53, BID, PIG8, Caspase 6, DFF40, MST4, and OCLN transcripts.
FIG. 5.
Depletion of endogenous Snail by using RNAi leads to increased p53 levels and increased sensitivity to apoptotic stimuli. (a) For the left panel, A375 cells were infected with either mock adenovirus or with shRNA-Snail adenovirus; control cells were uninfected. Total RNA was prepared from each cell population and analyzed by RT-PCR with primers for Snail, Slug, and actin as depicted in the figure. For the right panel, Snail depletion was performed by using the shRNA-Snail adenovirus, and lysates were analyzed by immunoblotting. Quantitation of steady-state levels of p53 by densitometry revealed a 1.6-fold increase in p53 levels after depletion of Snail. (b and c) Total RNA was extracted from A375 cells infected with shRNA-Snail or control adenovirus and analyzed by RT-PCR as in panel a. Transcript levels were normalized to actin. All data represent the mean value ± the standard deviation of three independent experiments. ✽, P < 0.05 (as determined by using the Student t test).
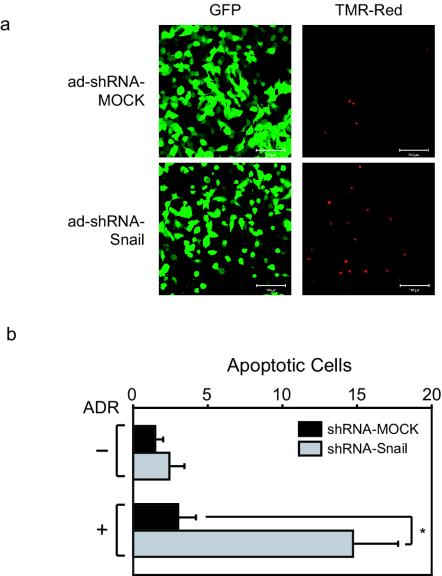
To determine whether endogenous Snail regulates DNA damage-induced programmed cell death, A375 cells were infected with shRNA-Snail or mock adenoviruses and treated with ADR. The presence of apoptotic cells (red fluorescence) was examined by an in vivo TUNEL assay in infected cells (green fluorescence) (Fig. 6a). Quantification of these data revealed a sevenfold increase in the number of apoptotic cells following adenoviral-mediated expression of the Snail-specific shRNA (Fig. 6b). These data suggest that Snail represses proapoptotic genes in the DNA damage response pathway, providing a prosurvival function.
FIG. 6.
Snail knockdown results in an increased sensitivity to programmed cell death induced by ADR treatment. (a) A375 cells infected with shRNA-Snail or mock adenovirus were treated with ADR, and TUNEL-positive cells were detected by using immunofluorescence. (b) Quantitation of the TUNEL assays described in panel a. All data represent the mean value ± the standard deviation of three independent experiments. ✽, P < 0.05 (as determined by using the Student t test).
DISCUSSION
Snail superfamily members have been identified as crucial regulators of cell fate in many biological contexts. During gastrulation, epithelial cells alter their connections to neighboring cells and acquire the characteristics necessary for migration and formation of mesoderm. This process involves major alterations in cell physiology. Previous studies have identified loss of cell-cell contact as central to the acquisition of a migratory phenotype (12), a phenotypic alteration promoted by the action of Snail and Slug. The data presented here add another property altered in epithelial cells after aberrant Snail/Slug expression: programmed cell death. Epithelial-to-mesenchymal transitions, such as those observed during development and during cancer progression, result in dramatic alterations in cell fate and in the interaction of cells with their environment. Survival in the face of such dramatic alterations in fundamental cell biology may require the development of resistance to programmed cell death. In fact, Snail superfamily members have been previously implicated in protection from programmed cell death during normal development and differentiation (19, 28, 33).
Here, we provide an extension of these findings by demonstrating that aberrant expression of Snail or of Slug in breast adenocarcinoma cells protects against apoptosis induced by genotoxic stress. We found that ectopic expression of Snail in cells with an epithelial phenotype not only promotes phenotypic transition to a less differentiated, mesenchymal pattern but also provides protection against programmed cell death. However, the mechanistic details of the protective effect of Snail or Slug in breast carcinoma cells appear to differ dramatically from those observed in hematopoietic precursors that express Slug. Specifically, p53 abundance does not appear to be altered in hematopoietic progenitors isolated from Slug knockout mice (19, 33). In the present study, regulation of p53 at both RNA and protein levels was observed. The differences in the molecular details of protection from programmed cell death likely reflect major differences in the biology of hematopoietic progenitor cells versus mammary adenocarcinoma cell lines. These findings predict that Snail superfamily members may protect cells from programmed cell death through different mechanisms in different biological contexts.
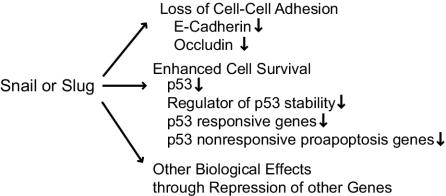
Acquisition of invasive growth is a prerequisite to metastasis during cancer progression. Aberrant expression of Snail or of Slug in breast adenocarcinomas resulted in phenotypic alterations, including loss of cell adhesion and acquisition of invasive growth accompanied by enhanced cell survival (Fig. 7). Analysis of the proapoptotic transcriptional program regulated by these factors revealed interesting overlap with the DNA damage response pathway regulated by the tumor suppressor TP53. In fact, the p53 mRNA is itself a direct transcriptional target of Snail and Slug in this system. Factors implicated in p53 turnover also represent regulatory targets. Finally, aberrant Snail/Slug expression led to transcriptional repression of other proapoptotic genes that do not fall within classical p53-dependent pathways. Thus, Snail and Slug promote cell survival after genotoxic stress through direct transcriptional repression of genes involved in many aspects of programmed cell death. In the context of cancer progression, these findings predict that increased resistance to programmed cell death induced by common chemotherapeutic agents accompany the acquisition of invasive growth induced by aberrant expression of Snail or of Slug.
FIG. 7.
Snail or Slug regulation of epithelial-to-mesenchymal transitions. The model depicts various aspects of epithelial to mesenchymal transition regulated by Snail and/or Slug. Details of the model are discussed in the text.
Acknowledgments
We gratefully acknowledge Naoyuki Fujita for technical assistance with ChIP. The manuscript was substantially improved by critical comments from Nathan Bowen, Naoyuki Fujita, Andrei Ivanov, Erwin Van Meir, Paula Vertino, and Keqiang Ye.
This study was supported by a grant from NIDDK (DK065961 to P.A.W.). K.N.M. was supported by a grant from NIGMS (T32 GM08490). P.A.W. gratefully acknowledges financial support from the Wilbur and Hilda Glenn Family Foundation.
REFERENCES
- 1.Ahn, J., M. Urist, and C. Prives. 2003. Questioning the role of checkpoint kinase 2 in the p53 DNA damage response. J. Biol. Chem. 278:20480-20489. [DOI] [PubMed] [Google Scholar]
- 2.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 3.Batlle, E., E. Sancho, C. Franci, D. Dominguez, M. Monfar, J. Baulida, and A. Garcia De Herreros. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2:84-89. [DOI] [PubMed] [Google Scholar]
- 4.Bolos, V., H. Peinado, M. A. Perez-Moreno, M. F. Fraga, M. Esteller, and A. Cano. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116:499-511. [DOI] [PubMed] [Google Scholar]
- 5.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 6.Cano, A., M. A. Perez-Moreno, I. Rodrigo, A. Locascio, M. J. Blanco, M. G. del Barrio, F. Portillo, and M. A. Nieto. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2:76-83. [DOI] [PubMed] [Google Scholar]
- 7.Carver, E. A., R. Jiang, Y. Lan, K. F. Oram, and T. Gridley. 2001. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 21:8184-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan, I., S. E. Ong, N. M. Watanabe, B. Blagoev, M. M. Nielsen, E. Kajikawa, T. Z. Kristiansen, M. Mann, and A. Pandey. 2002. Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J. Biol. Chem. 277:5929-5939. [DOI] [PubMed] [Google Scholar]
- 9.Fan, L. Z., and M. G. Cherian. 2002. Potential role of p53 on metallothionein induction in human epithelial breast cancer cells. Br. J. Cancer 87:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez, Y., L. Espana, S. Manas, A. Fabra, and A. Sierra. 2000. Bcl-xL promotes metastasis of breast cancer cells by induction of cytokines resistance. Cell Death Differ. 7:350-359. [DOI] [PubMed] [Google Scholar]
- 11.Fujita, N., D. L. Jaye, M. Kajita, C. Geigerman, C. S. Moreno, and P. A. Wade. 2003. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113:207-219. [DOI] [PubMed] [Google Scholar]
- 12.Grunert, S., M. Jechlinger, and H. Beug. 2003. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell. Biol. 4:657-665. [DOI] [PubMed] [Google Scholar]
- 13.Hajra, K. M., D. Y. Chen, and E. R. Fearon. 2002. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62:1613-1618. [PubMed] [Google Scholar]
- 14.Hajra, K. M., and E. R. Fearon. 2002. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer. 34:255-268. [DOI] [PubMed] [Google Scholar]
- 15.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemavathy, K., S. I. Ashraf, and Y. T. Ip. 2000. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene 257:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 18.Ikenouchi, J., M. Matsuda, M. Furuse, and S. Tsukita. 2003. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J. Cell Sci. 116:1959-1967. [DOI] [PubMed] [Google Scholar]
- 19.Inoue, A., M. G. Seidel, W. Wu, S. Kamizono, A. A. Ferrando, R. T. Bronson, H. Iwasaki, K. Akashi, A. Morimoto, J. K. Hitzler, T. I. Pestina, C. W. Jackson, R. Tanaka, M. J. Chong, P. J. McKinnon, T. Inukai, G. C. Grosveld, and A. T. Look. 2002. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell 2:279-288. [DOI] [PubMed] [Google Scholar]
- 20.Inukai, T., A. Inoue, H. Kurosawa, K. Goi, T. Shinjyo, K. Ozawa, M. Mao, T. Inaba, and A. T. Look. 1999. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol. Cell 4:343-352. [DOI] [PubMed] [Google Scholar]
- 21.Ip, Y. T., and T. Gridley. 2002. Cell movements during gastrulation: snail-dependent and independent pathways. Curr. Opin. Genet. Dev. 12:423-429. [DOI] [PubMed] [Google Scholar]
- 22.Jallepalli, P. V., C. Lengauer, B. Vogelstein, and F. Bunz. 2003. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J. Biol. Chem. 278:20475-20479. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, R., Y. Lan, C. R. Norton, J. P. Sundberg, and T. Gridley. 1998. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev. Biol. 198:277-285. [PubMed] [Google Scholar]
- 24.Kowalczyk, A. P., E. A. Bornslaeger, S. M. Norvell, H. L. Palka, and K. J. Green. 1999. Desmosomes: intercellular adhesive junctions specialized for attachment of intermediate filaments. Int. Rev. Cytol. 185:237-302. [DOI] [PubMed] [Google Scholar]
- 25.Lin, Y., W. Ma, and S. Benchimol. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 26:122-127. [DOI] [PubMed] [Google Scholar]
- 26.Liu, X., H. Zou, C. Slaughter, and X. Wang. 1997. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89:175-184. [DOI] [PubMed] [Google Scholar]
- 27.MacLachlan, T. K., and W. S. El-Deiry. 2002. Apoptotic threshold is lowered by p53 transactivation of caspase-6. Proc. Natl. Acad. Sci. USA 99:9492-9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzstein, M. M., and H. R. Horvitz. 1999. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol. Cell 4:309-319. [DOI] [PubMed] [Google Scholar]
- 29.Nieto, M. A. 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell. Biol. 3:155-166. [DOI] [PubMed] [Google Scholar]
- 30.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 31.Okamura, S., H. Arakawa, T. Tanaka, H. Nakanishi, C. C. Ng, Y. Taya, M. Monden, and Y. Nakamura. 2001. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8:85-94. [DOI] [PubMed] [Google Scholar]
- 32.Oltvai, Z. N., C. L. Milliman, and S. J. Korsmeyer. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74:609-619. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Losada, J., M. Sanchez-Martin, M. Perez-Caro, P. A. Perez-Mancera, and I. Sanchez-Garcia. 2003. The radioresistance biological function of the SCF/kit signaling pathway is mediated by the zinc-finger transcription factor Slug. Oncogene 22:4205-4211. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Moreno, M., C. Jamora, and E. Fuchs. 2003. Sticky business: orchestrating cellular signals at adherens junctions. Cell 112:535-548. [DOI] [PubMed] [Google Scholar]
- 35.Polyak, K., Y. Xia, J. L. Zweier, K. W. Kinzler, and B. Vogelstein. 1997. A model for p53-induced apoptosis. Nature 389:300-305. [DOI] [PubMed] [Google Scholar]
- 36.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 37.Ruohola, J. K., T. P. Viitanen, E. M. Valve, J. A. Seppanen, N. T. Loponen, J. J. Keskitalo, P. T. Lakkakorpi, and P. L. Harkonen. 2001. Enhanced invasion and tumor growth of fibroblast growth factor 8b-overexpressing MCF-7 human breast cancer cells. Cancer Res. 61:4229-4237. [PubMed] [Google Scholar]
- 38.Tsukita, S., M. Furuse, and M. Itoh. 2001. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell. Biol. 2:285-293. [DOI] [PubMed] [Google Scholar]
- 39.Wang, K., X. M. Yin, D. T. Chao, C. L. Milliman, and S. J. Korsmeyer. 1996. BID: a novel BH3 domain-only death agonist. Genes Dev. 10:2859-2869. [DOI] [PubMed] [Google Scholar]
- 40.Yu, J., L. Zhang, P. M. Hwang, K. W. Kinzler, and B. Vogelstein. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7:673-682. [DOI] [PubMed] [Google Scholar]
- 41.Zou, H., W. J. Henzel, X. Liu, A. Lutschg, and X. Wang. 1997. Apaf-1, a human protein homologous to Caenorhabditis elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90:405-413. [DOI] [PubMed] [Google Scholar]