SUMMARY
Cancer genomics have provided an unprecedented opportunity for understanding genetic causes of human cancer. However, distinguishing which mutations are functionally relevant to cancer pathogenesis remains a major challenge. We describe here a mammary stem cell (MaSC) organoid-based approach for rapid generation of somatic GEMMs (genetically engineered mouse models). By using RNAi and CRISPR-mediated genome engineering in MaSC-GEMMs, we have discovered that inactivation of Ptpn22 or Mll3, two genes mutated in human breast cancer, greatly accelerated PI3K-driven mammary tumorigenesis. Using these tumor models, we have also identified genetic alterations promoting tumor metastasis and causing resistance to PI3K-targeted therapy. Both Ptpn22 and Mll3 inactivation resulted in disruption of mammary gland differentiation and an increase in stem cell activity. Mechanistically, Mll3 deletion enhanced stem cell activity through activation of the HIF pathway. Thus, our study established a robust in vivo platform for functional cancer genomics and discovered functional breast cancer mutations.
INTRODUCTION
Breast cancer is a leading cause of death in women, with ~1.7 million new cases and more than half a million deaths globally each year (Torre et al., 2015). Complicating our efforts to develop effective treatments, breast cancers have highly heterogeneous genetic makeups. While a few genes, such as PIK3CA and TP53, are mutated at relatively high frequencies (30–40%), the numerous remaining breast cancer associated genes are only altered at intermediate to low frequencies (<10%) and their functions in cancer remain poorly understood (TCGA Network, 2012). Determining the functions of these mutations will lead to new mechanistic understandings of cancer pathogenesis and help development of targeted therapies that can be tailored specifically to each patient.
Because cancer development is a multistep process controlled by complex tissue microenvironment and systemic factors, studying a putative cancer related gene often requires in vivo models. As such, genetically engineered mouse models (GEMMs) have been a main approach for validating human cancer genes and for dissecting their mechanisms of action (Van Dyke and Jacks, 2002). However, traditional germline GEMMs are prohibitively costly and time-consuming for studying the large numbers of genes mutated in human cancers. To address this issue, we have developed a mouse mammary stem cell (MaSC) based somatic GEMM platform for functional cancer genomics studies. Using these MaSC-GEMMs, we have identified several functionally important breast cancer genes and shown that dysregulation of mammary stem/progenitor cell fate is a common mechanism of action for these genes.
RESULTS
Long-term expansion of MaSCs in vitro
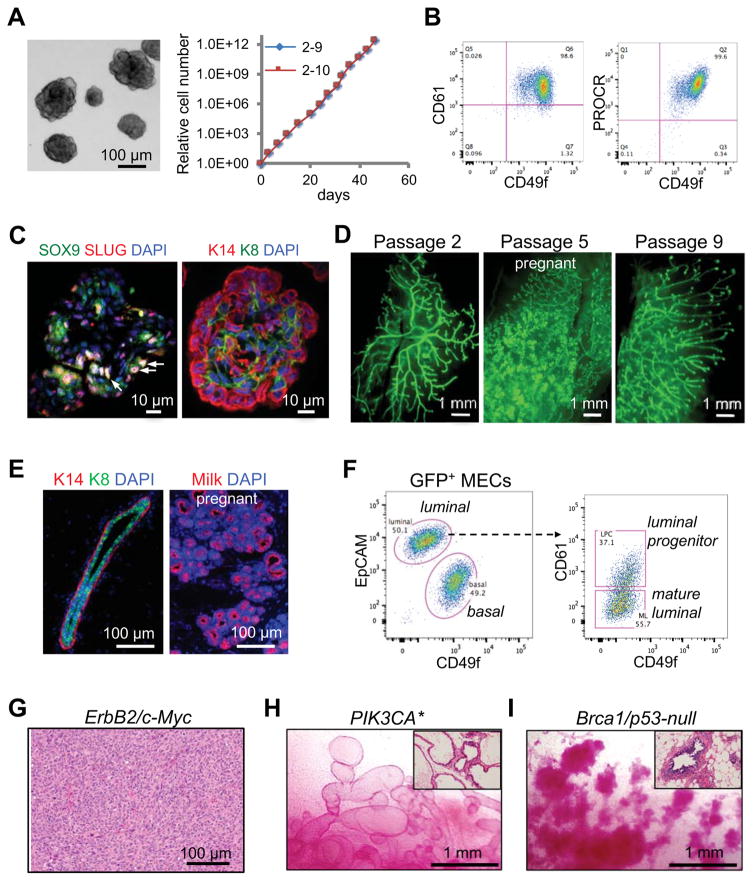
Although several methods have been established for measuring MaSC activity in vitro (Dontu et al., 2003; Guo et al., 2012; Shackleton et al., 2006; Spike et al., 2012; Stingl et al., 2006; Zeng and Nusse, 2010), robust long-term expansion of MaSCs in culture is still difficult. We previously developed an organoid assay that can be used to specifically identify bipotent MaSCs (Guo et al., 2012). Therefore, we optimized this system and tested whether it can support long-term expansion of MaSCs. To this end, we seeded CD49fhighCD61+ stem cell-enriched basal cells from CAG-EGFP transgenic mice at clonal density to generate single-cell derived organoids. The clonality of organoids formed in this condition was confirmed by mixing CAG-EGFP cells with unlabeled cells and showing that almost all individual organoids were of a single color (Figure S1A). We then dissociated individual organoids and passaged them serially. These cells multiplied rapidly with an expansion rate of ~100-fold per week even after 4 months in culture (Figure 1A). During each passage, ~10% of the cells were capable of re-initiating new organoids (Figure S1B). Additionally, the cells maintained a normal karyotype, suggesting they are genetically stable (Figure S1C).
Figure 1. Rapid generation of somatic GEMMs for breast cancer by ex vivo expansion and modification of MaSCs.
(A) Long-term expansion of MaSC organoids. Left: a representative image of organoids. Right: Growth curves of two single cell-derived organoid clones.
(B) CD49f, CD61 and PROCR flow cytometric profiles of organoids.
(C) SLUG, SOX9 and cytokeratin immunostaining in organoids. The arrows indicate examples of SLUG+SOX9+ cells.
(D) Whole-mount images of mammary ductal trees regenerated by single cell-derived GFP+ organoids at the indicated passages.
(E) Immunofluorescence images of mammary ducts (left, virgin) and alveoli (right, pregnant) regenerated by organoids (passage 2).
(F) Flow cytometric profiles of mammary ductal trees regenerated by organoids (passage 8).
(G) A representative H&E image of poorly differentiated adenocarcinoma developed in Erbb2/MYC MaSC-GEMMs (with passage 3 organoids). Organoids were transplanted into NOD-SCID mice, and the mice were treated with doxycycline for 4 months.
(H) Representative images of carmine-stained mammary fat pads reconstituted by Rosa-CreERT2; Pik3ca* organoids (passage 3–5). Mice were treated with tamoxifen 6 weeks after transplantation and then analyzed 6 weeks later. The inset shows H&E staining of the outgrowths.
(I) Representative images of carmine-stained mammary fat pads reconstituted by Blg-Cre; Brca1floxed/floxed; p53−/− organoids (passage 5). The inset shows H&E staining of the outgrowths.
See also Figure S1.
Consistent with their stem-cell origin, the organoid cells expressed the basal/MaSC markers CD49f, CD61 and PROCR (Figure 1B and Figure S1D) (Asselin-Labat et al., 2007; Stingl et al., 2006; Wang et al., 2015) and a fraction of them (~12%) coexpressed the transcription factors SLUG and SOX9 which are important for MaSC activity (Figure 1C and Figure S1E) (Guo et al., 2012). Interestingly, the organoids contained both KRT14+ basal and KRT8+ luminal cells organized in a partially polarized fashion, suggesting some degree of, albeit incomplete, differentiation (Figure 1C).
We further demonstrated that the cultured organoid cells maintained the ability to fully regenerate mammary ductal trees in vivo even after extended passaging (Figure 1D). Limiting dilution transplantation showed that as few as 25 organoid cells could fully regenerate a mammary ductal tree (Figure S1F). The frequency of mammary repopulating units was ~1/170 organoid cells, although this may underestimate the stem cell frequency due to difficulty of successfully transplanting a small number of cells. The mammary outgrowths were comprised of basal and luminal layers similar to what is observed in the endogenous mammary gland (Figure 1E). The regenerated glands also underwent robust alveologenesis to produce milk-secreting alveoli during pregnancy (Figure 1D and E). Furthermore, the outgrowths recapitulated the mammary epithelial lineages of the endogenous gland, as determined by flow cytometry (Figure 1F). These results demonstrate that our organoid culture system can sustain the long-term expansion of transplantable MaSCs, although they undergo some degree of differentiation in culture and hence only a subset of organoid cells are MaSCs.
Rapid generation of somatic GEMMs for breast cancer using genetically engineered MaSCs
We next tested whether organoid cells could be used to quickly establish somatic GEMMs for breast cancer using well-established cancer drivers (Figure S1G). We first modeled ERBB2-driven breast cancer by transducing the cells with lentiviral vectors expressing Erbb2V664E and doxycycline-inducible c-MYC, as c-MYC is often amplified or overexpressed in human ERBB2-positive cancer (Park et al., 2005). These modified cells regenerated elaborate mammary ductal trees in the absence of doxycycline (Figure S1H). Upon doxycycline treatment, the outgrowths developed palpable tumors with 100% penetrance in 5 weeks and then progressed into poorly differentiated adenocarcinomas resembling those of transgenic ERBB2 mammary tumor models (Figure 1G) (Moody et al., 2002; Muller et al., 1988).
We also modeled PI3-kinase (PI3K)-driven cancer using organoids from a Cre-inducible constitutively active PIK3CA mouse strain (Pik3ca*) (Srinivasan et al., 2009). We cultured organoids from mice carrying both Rosa26-CreERT2 and Pik3ca* alleles, then transplanted these cells into syngeneic C57BL/6 mice. The transplanted cells developed into normal mammary ductal trees (Figure S1H). However, upon administration of tamoxifen, the ductal trees developed into pronounced glandular adenosis characterized by dilated ducts (Figure 1H) and progressed into overt tumors with a median onset time of ~9 months, consistent with the median onset time of 5–16 months in previous germline PIK3CA tumor models (Adams et al., 2011; Koren and Bentires-Alj, 2013; Liu et al., 2011; Meyer et al., 2011). Additionally, we modeled Brca1 and Trp53 deletion driven neoplasia by using organoids cultured from Blg-Cre; Brca1floxed/floxed; Trp53−/− mice, a well-established basal-like tumor model (Molyneux et al., 2010). These cells developed into mammary ductal trees with numerous hyperplastic nodules 4 months post transplantation (Figure 1I). Thus, our MaSC-based somatic GEMMs (MaSC-GEMMs) provide a rapid approach for modeling tumor development of various breast cancer subtypes.
Identification of breast cancer drivers by shRNA screening in MaSC-GEMMs
To test the ability of our MaSC-GEMM platform to functionally validate the role of recently identified cancer associated mutations, we selected 4 genes (Map2k4, Cbfb, Ptprd and Ptpn22) that have frequent truncating mutations or gene deletions (TCGA Network, 2012). This class of mutations was chosen because they likely result in a loss-of-function in the gene of interest, which can be modeled by shRNA knockdown (KD). Furthermore, as they are frequently co-mutated with PIK3CA (Figure S2A), we used the aforementioned Pik3ca* model as a sensitized background to investigate their role in mammary tumorigenesis.
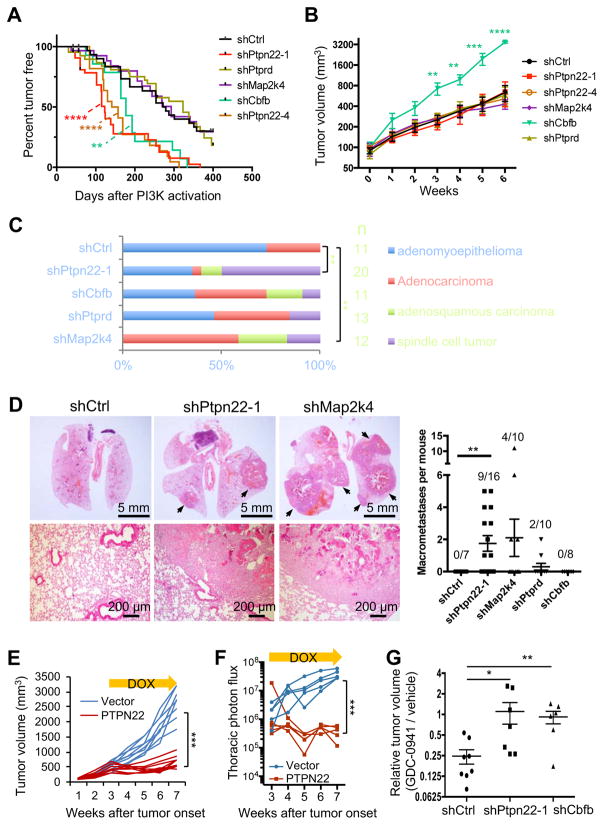
We transduced Rosa26-CreERT2; Pik3ca* organoid cells with lentiviral shRNA vectors and confirmed a KD efficiency of at least 70% for each gene (Figure S2B). The modified cells were then transplanted into C57BL/6 mice, and treated with tamoxifen to activate Pik3ca* expression 6 weeks later (Figure S2C). We found that both Ptpn22 and Cbfb KD significantly accelerated PIK3CA*-driven tumor initiation, with median onset time shortened from 276 days in the control group to 123 and 177 days in the shPtpn22 and shCbfb groups, respectively (Figure 2A). Interestingly, Cbfb KD also greatly accelerated tumor growth (Figure 2B). Similar to germline PIK3CA-driven mammary tumor models (Adams et al., 2011; Meyer et al., 2011), the control PIK3CA* MaSC-GEMM mainly developed adenomyoepithelioma and adenocarcinoma (Figure 2C and S2D). Remarkably, Ptpn22 KD led to the formation of more aggressive spindle cell tumors that resemble the ones formed by the Pik3ca and Trp53 double mutant tumor model (Figure 2C and S2D) (Adams et al., 2011).
Figure 2. Identification of cancer drivers by shRNA screening in MaSC-GEMMs.
(A) Kaplan-Meier survival analysis of tumor onset in Pik3ca* MaSC-GEMMs expressing the indicated shRNAs (n = 14–40). **P < 0.01, and ****P < 0.0001, compared to shCtrl by the log-rank test.
(B) Tumor growth rates of MaSC-GEMMs as shown in (A). Data are represented as mean ± SEM (n = 7–13).
(C) Percentages of tumors with various histological phenotypes in the indicated MaSCGEMMs (n = 11–20). **P < 0.01, Chi-square test.
(D) Lung metastases in the Pik3ca* MaSC-GEMMs expressing the indicated shRNAs. Left: representative H&E images of lung sections. Right: number of lung macrometastases per animal at the end point (mean ± SEM, n = 7–16). The frequency of mice with macrometastasis was shown on each column.
(E) Growth curve of HCC1806 xenografts expressing dox-inducible PTPN22 or the control vector. 100,000 cells were orthotopically injected into NOD-SCID mice. Three weeks after tumor onset, mice were treated with dox and measured for tumor volume. Each curve represents one tumor (n = 8, Two-way ANOVA).
(F) Lung metastasis burden of mice transplanted with Luc2-labeled HCC1806 cells, as measured by bioluminescence. Cells and mice were treated as in (E) (n = 5, Two-way ANOVA).
(G) Relative tumor volume of animals treated by GDC-0941. Each dot represents the ratio of average volume of GDC-0941-treated (for ~2 weeks) tumors to the average volume of vehicle-treated tumors that were derived from the same parental spontaneous tumor (n = 6–8). The tumors were either first or second generation allografts.
*P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
See also Figure S2.
Next we investigated the role of these genes in tumor metastasis. Although PIK3CA mutations are frequently detected in metastatic human breast cancers, the existing PIK3CA mouse mammary tumor models rarely metastasize (Koren and Bentires-Alj, 2013). We hypothesized that specific genetic alterations are required to cooperate with PIK3CA mutations in order to drive metastasis. In confirmation of our hypothesis, Ptpn22 KD significantly promoted macroscopic lung metastases (Figure 2D). Importantly, the increased rate of metastasis was not due to an increase in primary tumor burden, as no significant difference was noted among the experimental groups (Figure S2E). However, Cbfb KD had no effect on tumor metastasis even though it accelerated primary tumor growth. Taken together, our results demonstrate the power of MaSC-GEMMs as we were able to rapidly identify several functional cancer drivers that play distinct roles in tumor initiation, progression, and spread.
PTPN22 overexpression suppresses tumor growth and metastasis
To further demonstrate the tumor inhibitory function of PTPN22, we expressed it via a doxycycline-inducible vector in HCC1806 human breast cancer cells, as they lack PTPN22 expression (Figure S2F). Upon induction of PTPN22 expression, the growth of HCC1806 cells was greatly inhibited in vitro (Figure S2G). Importantly, the inhibitory effect was not due to nonspecific toxicity of PTPN22 overexpression, as similar levels of PTPN22 overexpression had no effect on the growth of immortalized normal human mammary epithelial cells (HMLE) (Figure S2F and S2G) (Elenbaas et al., 2001). Furthermore, inducing PTPN22 expression efficiently blocked the growth of HCC1806 xenografts (Figure 2E) and their metastases (Figure 2F and S2H), supporting a tumor suppressive role for PTPN22.
Ptpn22 and Cbfb knockdown confers resistance to PI3K-targeted therapy
We reasoned that our MaSC-GEMMs could also be useful as preclinical models for determining whether certain cooperating mutations alter tumor response to anti-PI3K treatment. To this end, we orthotopically transplanted small fragments of MaSC-GEMM tumors to a cohort of allograft-bearing mice for each parental tumor. As each tumor graft reached 100 mm3 in volume, mice were treated with either the vehicle control or GDC-0941, a pan class I PI3K inhibitor that is being developed clinically for breast cancer treatment (Folkes et al., 2008; Liu et al., 2011). As expected, GDC-0941 greatly diminished the growth of control shRNA-expressing tumors compared to the vehicle control (Figure 2G and Figure S2I). However, tumors derived from PIK3CA* MaSC-GEMMs expressing either shPtpn22 or shCbfb were significantly more resistant to GDC-0941. Remarkably, some of these tumors were completely refractory to the treatment (Figure 2G and Figure S2I). These results demonstrate that Ptpn22 or Cbfb KD promotes resistance to PI3K-targeted therapy.
Dysregulation of the mammary cell fate by Ptpn22 suppression
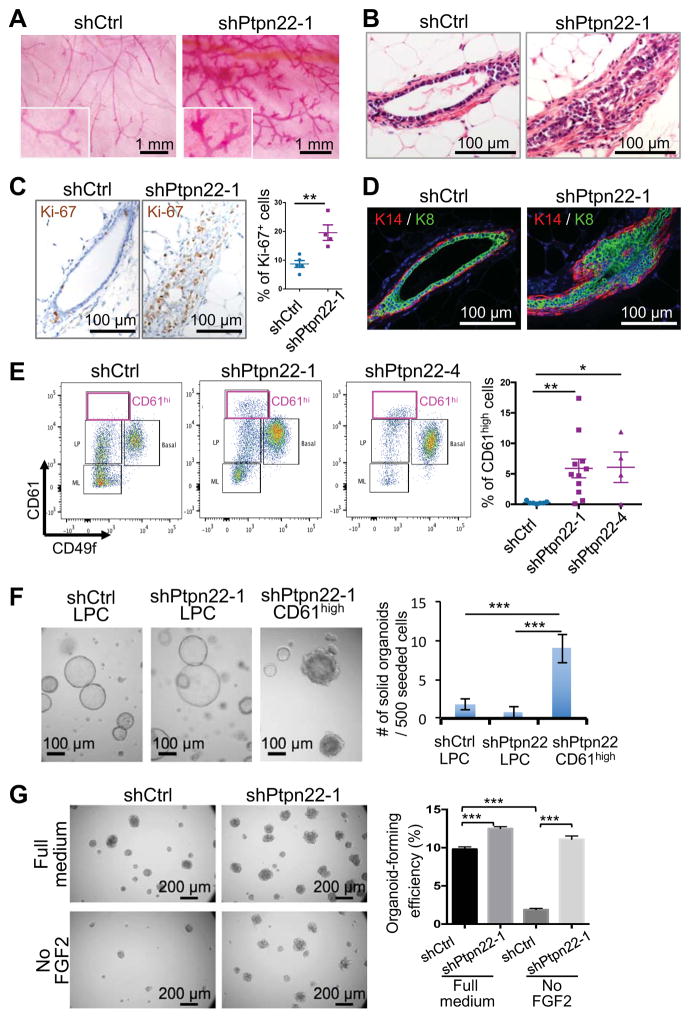
We further investigated the role of Ptpn22 in mammary gland biology, as its KD promoted the most aggressive tumor phenotypes among the candidate genes. The shPtpn22 organoids generated abnormal mammary ductal trees characterized by pronounced hyperplasia relative to estrous matched controls (Figure 3A and B). The hyperplastic ducts had significantly more proliferating cells compared to the control (Figure 3C) and showed expansion of both luminal and basal cell layers (Figure 3D). We further assessed the lineage differentiation of the outgrowths by flow cytometry (Figure S3A). Although Ptpn22 KD did not significantly alter the ratio of basal, luminal progenitor and mature luminal cells (Figure S3B), it did induce a de novo population of CD49flowCD61high cells (Figure 3E). Interestingly, these CD49flowCD61high cells generated solid organoids in vitro resembling those of bipotent MaSCs in organoid culture, although the exact nature of these cells needs to be fully characterized in future studies (Figure 3F).
Figure 3. Dysregulation of the mammary cell fate by Ptpn22 suppression.
(A) Whole-mount images of carmine-stained mammary fat pads transplanted with organoids expressing the indicated shRNAs (n = 6 for each group). Fat pads were isolated from mice in estrus ~20 weeks post transplantation.
(B) H&E images of outgrowths as generated in (A).
(C) Ki-67 IHC of outgrowths as generated in (A). Percentages of Ki-67+ cells in the mammary epithelium of individual outgrowths (minimum 6 fields/outgrowth) were shown (n = 4–5).
(D) Cytokeratin immunostaining of mammary outgrowths as generated in (A).
(E) Flow cytometric analysis of mammary outgrowths formed by organoids expressing the control or Ptpn22 shRNAs. Glands were analyzed 12 weeks post transplantation. The graph shows percentages of CD61high cells in lineage negative epithelial cells (mean ± SEM, n = 4–11).
(F) Organoid structures generated by the indicated cell populations (left) and quantification of solid organoid-forming ability (mean ± SEM) (right).
(G) Organoid-forming efficiency of the indicated cell types seeded in the full organoid medium or the medium without FGF2.
*P < 0.05, **P<0.01, ***P < 0.001.
See also Figure S3.
We next examined the effect of Ptpn22 KD on stem cell activity of the organoid cells in vitro. Ptpn22 KD significantly increased organoid-forming efficiency (Figure 3G). More strikingly, while the control organoids required FGF2 to grow, Ptpn22 KD allowed for growth in a FGF2-deficient condition, suggesting Ptpn22 KD renders organoids independent of certain niche factors (Figure 3G). Importantly, expressing human PTPN22, which is not targeted by the mouse shRNA, suppressed the FGF2-independent growth in the shPtpn22 organoids, confirming an on-target effect of shPtpn22 (Figure S3C and S3D). These results identify PTPN22 as a regulator of mammary gland differentiation and suggest that inhibition of PTPN22 promotes stem cell activity.
Generation of Mll3 mutant MaSC-GEMMs using CRISPR-Cas9
Although the shRNA screen proved effective for cancer driver discovery in MaSC-GEMMs, the incomplete inhibition of gene functions by shRNAs may potentially prevent us from identifying driver mutations that require complete gene inactivation. Therefore, we used the CRISPR technology to generate knockout MaSC-GEMMs. To accomplish this, we constructed a doxycycline-inducible Cas9 lentiviral vector and validated it by deleting Trp53 inducibly in organoid cells (Figure S4A). We then applied this approach to investigate the function of another candidate cancer gene MLL3 (gene symbol – KMT2C), a histone methyltransferase that is frequently mutated in breast cancer and other solid tumors (Cho et al., 2007; Shilatifard, 2012; TCGA Network, 2012). Intriguingly, most MLL3 mutations in breast cancer cause truncations of key functional domains, including the SET methyltransferase domain, suggesting it acts as a tumor suppressor (Figure S4B).
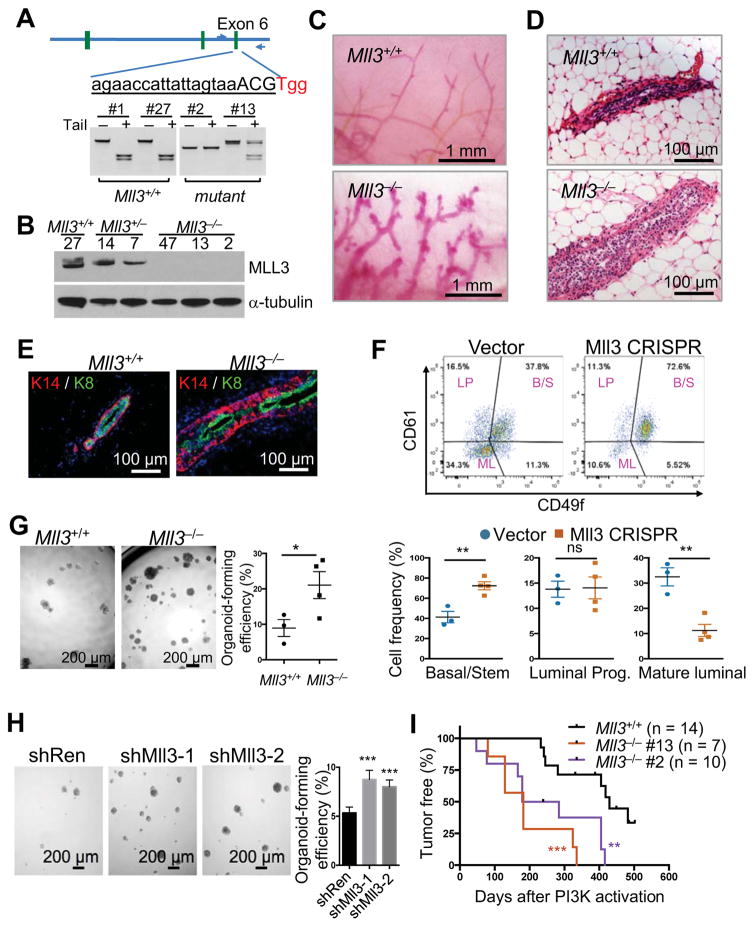
We knocked out Mll3 in Rosa26-CreERT2; Pik3ca* MaSCs, as MLL3 mutations significantly co-occur with PIK3CA mutations in human breast cancer (Figure S4B and S4C). We were able to efficiently (~10%) generate biallelic mutant organoid clones (Figure 4A). Genomic DNA sequencing and western blot revealed that these clones carried frameshift mutations resulting in the loss of the MLL3 protein (Figure 4B and S4D). We further confirmed that there were no mutations in 9 predicted low-probability off-target loci in two biallelic mutant clones (Figure S4E).
Figure 4. CRISPR-mediated MaSC-GEMMs reveal the role of Mll3 in mammary tumorigenesis and stem cell regulation.
(A) Restriction fragment length polymorphism (RFLP) assay for determining the Mll3 genotype of single cell-derived organoid clones. The wild-type amplicon can be digested by TaiI.
(B) MLL3 protein levels in organoid clones as determined by western blot.
(C) Whole-mount images of cleared mammary fat pads transplanted by Mll3+/+ (vector control) or Mll3−/− (clone #2) organoids. Glands were analyzed 12 weeks post transplantation.
(D) H&E images of outgrowths as generated in (C)
(E) KRT14 and KRT8 immunofluorescence of outgrowths as generated in (C).
(F) Flow cytometric analysis of outgrowths generated by organoids expressing the control or Mll3 lentiCRISPRv2 vector (upper). The percentages of stem/basal (B/S), luminal progenitor (LP) and mature luminal cells (ML) in lineage-negative epithelial cells were shown (mean ± SEM, n = 3–4) (lower).
(G) Organoid-forming efficiency of Mll3+/+ (vector control) and Mll3−/− organoid cells. Each data point represents an independent organoid line (mean ± SEM, n = 3–4).
(H) Organoid-forming efficiency of cells transduced by the indicated shRNAs.
(I) Kaplan-Meier survival analysis of PIK3CA*-driven tumor onset in Mll3+/+ or Mll3−/− MaSC-GEMMs. P values were determined by the log-rank test.
*P < 0.05, **P < 0.01, ***P<0.001, ns: not significant.
See also Figure S4.
Mll3 deletion causes mammary stem cell expansion and promotes PIK3CA-driven tumorigenesis
We transplanted the Mll3−/− or Mll3+/+ organoid clones into C57BL/6 mice and investigated the consequence of Mll3 deletion in mammary gland development in vivo. We found that Mll3 deletion caused severe hyperplasia (Figure 4C and 4D) and resulted in an expansion of basal cells (Figure 4E). To further rule out potential off-target effects of CRISPR, we targeted the Mll3 gene with a second sgRNA, this time by transducing pooled organoid cells with the constitutive lentiCRISPRv2 system (Sanjana et al., 2014). These pooled Mll3 mutant organoids also generated hyperplastic ductal trees similar to the Mll3 mutant organoid clones (Figure S4F).
Consistent with the basal expansion phenotype, we found that Mll3−/− mammary outgrowths had a marked increase of the CD49fhighCD61+ stem cell-enriched basal compartment and a reduction of the CD49flowCD61− mature luminal compartment (Figure 4F). In addition, compared to Mll3+/+ cells, Mll3−/− cells had significantly elevated organoid-forming ability, a property specific to bipotent MaSCs (Figure 4G). To further confirm that this phenotype is due to on-target effects of CRISPR, we knocked down Mll3 using two previously validated shRNAs (Figure S4G) (Chen et al., 2014). Consistent with the CRISPR result, Mll3 KD significantly increased in vitro organoid-forming activity (Figure 4H). This data shows that loss of Mll3 promotes stem cell self-renewal and results in a differentiation block. Interestingly, suppression of Mll3 in hematopoietic stem and progenitor cells also impairs their differentiation (Chen et al., 2014), suggesting stem cell fate regulation is likely a common function of MLL3 in various tissues.
We then examined whether Mll3 deletion cooperates with PI3K activation to drive mammary tumorigenesis. To do so, we regenerated mammary glands with pooled Mll3+/+; Pik3ca* organoids and two independent Mll3−/−; Pik3ca* organoid lines in C57BL/6 mice. Six weeks after transplantation, the mice were treated with tamoxifen to activate Pik3ca* expression and then monitored for tumor formation. We found that Mll3−/− mammary glands developed tumors significantly sooner than the control glands (Figure 4I). However, in contrast with the shPtpn22 tumors, the Mll3−/− tumors did not metastasize. Together, the above results demonstrate that Mll3 deletion leads to dysregulation of mammary stem cell fate and promotes tumorigenesis.
Activation of the HIF pathway mediates the effect of Mll3 deletion on stem cell activation
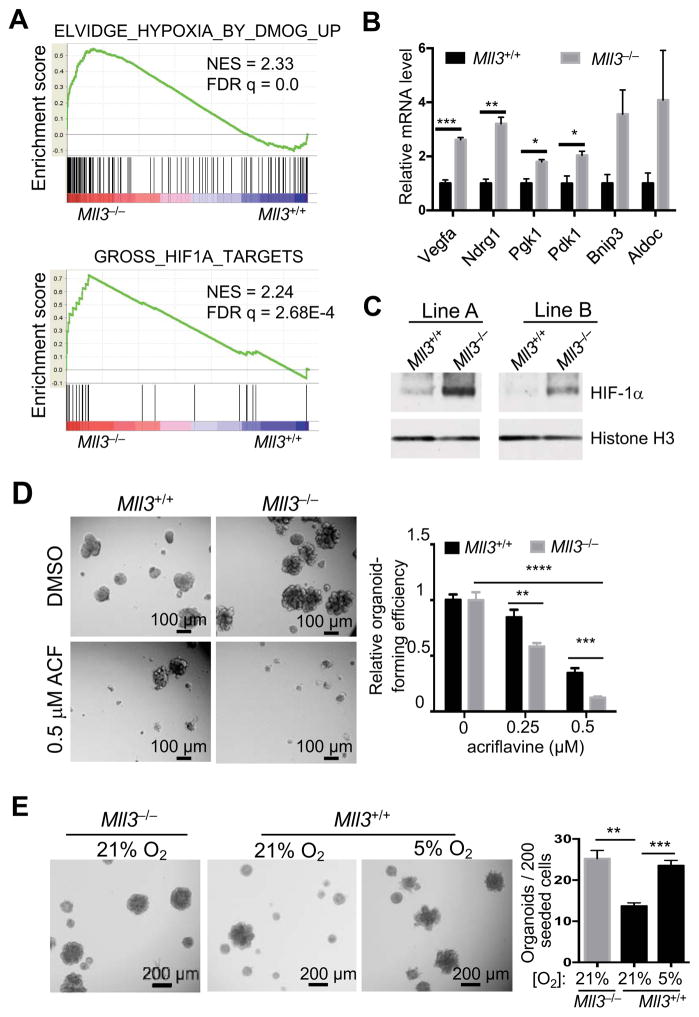
To understand mechanisms by which Mll3 deletion promotes stem cell activity, we compared transcription profiles of the vector control (Mll3+/+) and Mll3−/− organoids (3 lines for each) in regular organoid culture by microarray and then performed Gene Set Enrichment Analysis (GSEA) to identify pathways that are differentially activated between Mll3+/+ and Mll3−/− cells. Interestingly, 5 of the top 10 gene sets upregulated in Mll3−/− cells were targets of the hypoxia-inducible factor (HIF) pathway (Figure 5A and S5A). The upregulation of HIF target genes in Mll3−/− organoids was further validated by qRT-PCR (Figure 5B). Additionally, Mll3−/− organoids had markedly increased HIF-1α protein levels relative to Mll3+/+ cells in normoxia (Figure 5C and S5B). Mll3 KD by shRNAs also significantly elevated the HIF-1α protein levels (Figure S5C). Acriflavine, a specific HIF inhibitor (Lee et al., 2009), efficiently blocked organoid formation in both Mll3−/− and Mll3+/+ cells, suggesting the HIF activity is important for MaSC activity (Figure 5D). In addition, culturing WT cells in hypoxia recapitulated the effect of Mll3 deletion on stem cell activity (Figure 5E). These results suggest that Mll3 deletion activates the HIF pathway, which contributes to stem cell hyper-activation.
Figure 5. Activation of the HIF pathway mediates the effect of Mll3 deletion on stem cell activation.
(A) Two representative gene sets upregulated in Mll3−/− organoids, as determined by GSEA.
(B) Relative mRNA levels of HIF target genes in Mll3+/+ and Mll3−/− organoid lines (n=3), as measured by qRT-PCR. Gapdh and Hprt were used as internal controls.
(C) HIF-1α protein levels in 2 independent lines of Mll3+/+ (vector control) and Mll3−/− organoids as measured by western blot. Cells were transduced by the lentiCRISPRv2 vectors. Histone H3 was used as a loading control.
(D) Effect of the HIF inhibitor acriflavine (ACF) on organoid-forming ability of Mll3+/+ and Mll3−/− cells. Cells were cultured with the indicated concentration of ACF for 7 days. Organoid-forming efficiency was normalized to the respective DMSO control.
(E) Effect of hypoxia on organoid-forming ability. Mll3+/+ and Mll3−/− cells were cultured at the indicated oxygen concentrations for 7 days.
See also Figure S5.
DISCUSSION
We report here a rapid and versatile somatic GEMM approach for functional cancer genomics. Viral transduction of freshly isolated mammary epithelial cells coupled with gland reconstitution has been used to study mammary gland development and breast cancer previously (Bouras et al., 2008; McCaffrey et al., 2012; Welm et al., 2008). However, these previous methods can only maintain MaSCs transiently in culture. Hence, it is difficult to reliably generate complex modifications to model multigenic interactions, in order to recapitulate the heterogeneous genetic makeup of human cancers. To allow for complex modeling, we have developed a robust method for long-term expansion of transplantable MaSCs, which allows for precise genetic modifications by selecting and screening cells in vitro. In particular, the ability to expand single cell-derived clones will allow for CRISPR-based genome editing to generate GEMMs with defined mutations.
Using MaSC-GEMMs, we have elucidated functions of multiple candidate cancer genes at various stages of cancer pathogenesis in a single study, demonstrating the power of this approach. The data produced using our somatic mouse models suggest the loss-of-function mutations of these genes identified in human patients functionally contribute to breast cancer development. Furthermore, our tumor models driven by patient relevant mutations provide useful preclinical models for studying tumor responses to targeted therapy. We showed that inactivating Ptpn22 or Cbfb can confer resistance to PI3K inhibition in PI3K-driven tumors. Determining the genetic makeup that renders tumors sensitive or resistant to specific therapy will aid the development of effective cancer treatment. Using the rapid MaSC-GEMM approach, it is feasible to generate a large collection of preclinical tumor models that are driven by combinations of various cancer drivers. This will allow for a systematic investigation of the mechanisms of drug response and for developing combinatorial therapies.
Our results also revealed the role of these genes in cell fate regulation. Emerging evidence has suggested that breast cancer can originate from diverse lineages of mammary epithelial cells, including stem cells and committed cell types (Lim et al., 2009; Molyneux et al., 2010). However, it remains elusive whether activation of endogenous stem cell properties and de novo acquisition of such properties through cellular reprogramming contribute to breast cancer development. We found that both Ptpn22 KD and Mll3 deletion promote stem cell activity and cause expansion of stem cell-like population, although the exact identity of these stem-like cells remain to be fully characterized. Of note, recent lineage tracing studies suggest the existence of both bipotent and unipotent stem cells in the mammary gland (Rios et al., 2014; Van Keymeulen et al., 2011). The effects of these mutations in distinct stem cell populations also warrant future investigation. Interestingly, PI3K activation has also recently been shown to induce stem-like multipotency in various mammary lineages (Koren et al., 2015; Van Keymeulen et al., 2015). Together, these results suggest that stem cell fate dysregulation is likely an important mechanism of action of many cancer drivers.
EXPERIMENTAL PROCEDURES
Mice
Pik3ca* mice (JAX, #012343) were bred with Rosa26-CreERT2 mice (JAX, #008463) to generate tamoxifen inducible PIK3CA* model. Blg-Cre; Brca1flox/flox; p53−/− (#012620) and CAG-EGFP (#003291) mice were obtained from the Jackson Laboratory. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
MaSC organoid culture
Organoid medium was based on Advanced DMEM/F-12 (ThermoFisher) supplemented with 5% Matrigel (Corning, #354234), 5% heat-inactivated FBS (Sigma, F2442), 10 ng/ml EGF (Sigma, E9644), 20 ng/ml FGF2 (EMD Millipore, GF003), 4 μg/ml heparin (Sigma, H4784) and 5 μM Y-27632 (R&D, #1254). For organoid culture at clonal density, 200-500 cells/well were seeded in 96-well ultra-low attachment plates (Corning). For quantifying organoid-forming efficiency, the number of organoids (≥ 100 μm in diameter) was counted 7 days after seeding. To establish clonal lines, individual organoids were picked with P20 pipet tips and expanded. For passaging and expansion, organoids were washed with PBS, dissociated with 0.05% trypsin/EDTA and seeded at 1 × 105 cells/well in 6-well plates. Cells were then passaged every 3–4 days. Dissociated organoid cells can be frozen in calf serum containing 8% DMSO and 5 μM Y-27632, and then thawed to reestablish the culture.
Statistical analysis
P values were calculated by unpaired t-test except where otherwise specified. All analyses were done in GraphPad Prism 6. P < 0.05 was considered significant.
Additional Procedures
The details of cleared fat pad transplantation, flow cytometry, tumor analysis, tumor treatment, lentiviral vector cloning and transduction, antibody reagents, immunofluorescence, immunoblot, RFLP assay, qRT-PCR and microarray/GSEA analysis are included in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank Dulguun Amgalan for screening shRNAs and Dr. Scott Lowe for generously providing Mll3 shRNA vectors. We are grateful to the shRNA, Flow Cytometry, Histopathology, Analytical Imaging and Molecular Cytogenetic core facilities for technical assistance (supported by Einstein Cancer Center Support Grant P30 CA013330) and to the New York State Department of Health (NYSTEM Program) for shared facility grant support (C029154). This work is supported by grants from NYSTEM (C028109 and C029571), Susan Komen for the Cure (CCR12224440), and the V Foundation for Cancer Research. W.G. is a V Scholar. J.R.C. was supported by the 2T32GM007491-36 Training Program in Cellular and Molecular Biology and Genetics.
Footnotes
A CCESSION NUMBER
The microarray data is deposited in the GEO database (GSE79946).
AUTHOR CONTRIBUTIONS
Z.Z. and W.G. conceived the study. Z.Z., J.C, C.W., and W.G. performed the experiments and analyzed the data. M.O. performed tumor histology analysis. K.G. assisted with Mll3 mutant analysis. W.G. and Z.Z. wrote the paper with inputs from all other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, Wong RG, Wang W, Wright KL, Lane TF, Zacksenhaus E, et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011;71:2706–2717. doi: 10.1158/0008-5472.CAN-10-0738. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z, Shroff AS, Dickins RA, Vakoc CR, Bradner JE, et al. MLL3 Is a Haploinsufficient 7q Tumor Suppressor in Acute Myeloid Leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, Chuckowree IS, Clarke PA, Depledge P, Eccles SA, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. Journal of medicinal chemistry. 2008;51:5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Bentires-Alj M. Mouse models of PIK3CA mutations: one mutation initiates heterogeneous mammary tumors. The FEBS journal. 2013;280:2758–2765. doi: 10.1111/febs.12175. [DOI] [PubMed] [Google Scholar]
- Koren S, Reavie L, do Couto JP, De Silva D, Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina O, et al. PIK3CA induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, Yang J, Semaan DJ, Chen C, Fox EA, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DS, Brinkhaus H, Muller U, Muller M, Cardiff RD, Bentires-Alj M. Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res. 2011;71:4344–4351. doi: 10.1158/0008-5472.CAN-10-3827. [DOI] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- Park K, Kwak K, Kim J, Lim S, Han S. c-myc amplification is associated with HER2 amplification and closely linked with cell proliferation in tissue microarray of nonselected breast cancers. Hum Pathol. 2005;36:634–639. doi: 10.1016/j.humpath.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- TCGA Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Lee MY, Ousset M, Brohee S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, Zeng YA. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.