Abstract
Telomere synthesis in most organisms depends on the action of the telomerase enzyme, which contains an RNA subunit that is stably associated with the reverse transcriptase subunit as well as additional telomerase proteins. In the budding yeast Saccharomyces cerevisiae, several structural domains that are responsible for mediating protein interactions with the telomerase RNA TLC1 have been identified. We report here the identification and characterization of a TLC1 stem-loop that is required for its interaction with the Est2 reverse transcriptase protein. This hairpin, which does not contain any bulges in the duplex stem that commonly mediate protein-RNA interaction, appears to be a part of a larger structure, as nucleotides immediately to either side of this stem-loop contribute to the interaction of TLC1 with the Est2 protein. Surprisingly, replacement of a 95-nucleotide region of the yeast telomerase RNA that is required for Est2 interaction with a 39-nucleotide pseudoknot from a distantly related telomerase RNA results in a functional telomerase enzyme. These findings suggest that the ability of the budding yeast reverse transcriptase to associate with the telomerase RNA depends on a highly structured region rather than specific sequence elements.
Telomeres, the specialized protein-DNA complexes that cap chromosome ends, make essential contributions to both genome stability and cellular proliferation. End-to-end fusions between chromosomes, a fatal event for most genomes, are prevented by a suite of telomere-bound proteins (18, 24), which also protect these termini from unregulated degradation due to nucleolytic processing or intrachromosomal deletions of telomeric tracts (22, 23, 37, 40, 57, 62). Linear chromosomes also employ a specialized mechanism(s) to counter the sequence loss that would occur if such chromosomes were replicated solely by semiconservative replication (31). The consequence of degradative activities or incomplete replication is progressive telomere shortening, which eventually provides a barrier to proliferation of cells in culture (4, 36, 60). Such telomere erosion can also have consequences at the organismal level. In humans, genetically inherited syndromes that result in telomere shortening are characterized by bone marrow failure (42, 58, 59), and even within the general population, telomere length decline shows a correlation with the onset of age-dependent mortality (5).
In most organisms, the enzyme telomerase is the primary factor responsible for countering telomere shortening (25). It does so by elongating the 3′ terminus of one strand of chromosome ends, with the complementary strand filled in by the lagging-strand DNA machinery. Telomerase uses a mode of synthesis akin to that employed by the reverse transcriptases: a tightly associated telomerase RNA subunit provides a short template that is iteratively reverse transcribed by the catalytic protein subunit (called TERT, for telomerase reverse transcriptase). Not surprisingly, the telomerase enzyme is subject to regulation at multiple levels. For example, expression of the TERT subunit is tightly controlled in human cells by both positive and negative transcriptional regulators (13, 30). Access of telomerase to its substrate is also regulated both positively and negatively (17). At fully replicated telomeres, telomerase accessibility to chromosome termini is inhibited, in cis, by proteins bound to the duplex telomeric tract RNP (1, 34, 38, 39, 51). Substrate access is also controlled as the result of an interaction between telomerase and the telomere-bound Cdc13 protein (15, 44). In Saccharomyces cerevisiae, this recruitment activity depends on a direct interaction between the Est1 telomerase holoenzyme subunit and the Cdc13 protein (44; R. Cervantes, S. Post and V. Lundblad, unpublished data). Yeast strains that bear specific est1 or cdc13 missense mutations that disrupt this interaction are no longer able to maintain their telomeres, with eventual consequences for long-term cell proliferation. Identification of orthologs of the Est1 protein as telomerase-associated proteins in Candida albicans, Schizosaccharomyces pombe, and humans also suggests that this recruitment mechanism may be widely utilized (35).
Telomerase is a multisubunit holoenzyme, and increasing evidence suggests that individual components may make multiple contributions to its enzyme action. The RNA subunit is an example of a subunit that has more than one role in telomerase function: it not only provides the template for the elongation reaction, it also modulates enzyme activity and contributes a scaffold for the assembly of the telomerase RNP (6). Dissection of RNA-protein interactions that contribute to RNP assembly and enzyme activity has been greatly facilitated by the elucidation of the secondary structures of the ciliate and mammalian telomerase RNAs (8, 32, 46). Strikingly, common structural features have been observed even among telomerase RNA molecules recovered from evolutionarily distant species that exhibit essentially no primary sequence conservation. The most prominent conserved feature is a pseudoknot structure located immediately adjacent to the template domain (8, 32, 46, 52, 56). Chemical mapping and nuclear magnetic resonance structural studies suggest that the vertebrate telomerase RNA pseudoknot is dynamic and in balance between different conformational states, potentially triggered in response to telomeric repeat synthesis (2, 12, 54). Analysis of the ciliate and vertebrate RNA secondary structures also revealed the presence of multiple sites that are required for association of TERT with its RNA partner (3, 9, 28, 41, 53).
Determination of a detailed telomerase RNA secondary structure for the ciliate and vertebrate RNAs has been aided, at least in part, by their small size (148 to 209 and 382 to 559 nucleotides, respectively). In contrast, elucidation of the structure of the budding yeast telomerase RNA (called TLC1 in S. cerevisiae and TER1 in Kluyveromyces lactis) has lagged behind, in part due to its much larger size (≈1,300 nucleotides) and substantial sequence divergence. Recent observations from several different laboratories have nevertheless begun to reveal discrete structural features of the yeast telomerase RNA (6). One such structure is a base-paired element immediately adjacent to the template, which provides a template boundary element and directs template usage (49, 55). In addition, two stem-loop structures provide protein binding sites that mediate association of the Est1 telomerase subunit and the Ku heterodimer with the telomerase RNP. The Est1 protein binds to a bulged stem just downstream of the template (48) and does so in a manner that is independent of the assembly of other proteins into the telomerase complex (21, 61). Association of Est1 with this TLC1 structure has been proposed to provide a bridge between the catalytic core of telomerase and the telomere-bound Cdc13 protein, thereby mediating an essential step in telomere replication (15, 48, 61). The Ku heterodimer also interacts directly with yeast telomerase through a second RNA structural element (45, 50). The biochemical consequence of this Ku-TLC1 interaction has not yet been elucidated, but it may either promote the association of telomerase with its substrate or influence enzyme activity subsequent to localization of telomerase at the telomere.
A third phylogenetically conserved stem-loop structure, immediately adjacent to the Est1-interacting structure, was recently characterized in K. lactis, a budding yeast closely related to S. cerevisiae (56). We report here that an analogous stem-loop forms in the S. cerevisiae telomerase RNA and is required for association of the yeast TERT subunit (called Est2 in S. cerevisiae) with the telomerase RNP. This stem-loop does not contain any bulges that would dictate protein-RNA interaction, arguing that additional structural information must help mediate the association of Est2 with TLC1. Both deletion analysis and the construction of chimeric telomerase RNAs implicate additional nucleotides immediately to either side of this stem-loop that are also required for the interaction of TLC1 with Est2. Surprisingly, replacement of a 95-nucleotide Est2-interacting region with a pseudoknot structure from a distantly related telomerase RNA still results in a functional telomerase RNP. These findings suggest that a highly structured region, rather than specific sequence elements, may dictate the ability of the budding yeast reverse transcriptase to associate with the telomerase RNA.
MATERIALS AND METHODS
Yeast strains and plasmids.
Two isogenic haploid S. cerevisiae strains, YVL1008 and YVL1009 (containing tlc1-Δ and tlc1-Δ rad52-Δ mutations, respectively), were used for all senescence assays and telomere length analysis described in this study. Both YVL1008 (MATa tlc1-Δ::LEU2 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ/pSD120) and YVL1009 (MATα tlc1-Δ::LEU2 rad52-Δ::LYS ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ/pSD120) were descended from YPH275 (36). These two strains are maintained with a TLC1 covering plasmid (pSD120, URA3 CEN TLC1; generously provided by Dan Gottschling), which was shuffled out prior to experiments (see below). The strains used for biochemical analysis contain tagged versions of Est1, YVL2183 (MATa HA3-EST1 tlc1-Δ::LEU2 leu2 trp1 ura3-52 prb prc pep4-3/pA258) and YVL2434 (MATa HA3-EST1 tlc1-Δ148-440 leu2 trp1 ura3-52 prb prc pep4-3), or Est2, YVL2192 (MATa ProA-EST2 tlc1-Δ::LEU2 leu2 trp1 ura3-52 prb prc pep4-3/pA258) and YVL2435 (MATa ProA-EST2 tlc1-Δ148-440 leu2 trp1 ura3-52 prb prc pep4-3). These tagged versions of Est1 and Est2 have been used previously for telomerase immunoprecipitations (21, 33). These strains also contain tlc1-Δ148-440, a deletion of TLC1 shown to be proficient for Est1 and Est2 association (33), either present on a low-copy-number plasmid (pA258; see Fig. 3 and 4) or integrated into the endogenous TLC1 locus by standard two-step gene replacement (with the integrating vector pVL2527), to generate YVL2434 and YVL2435 (used for the experiments in Fig. 5, 6, and 7).
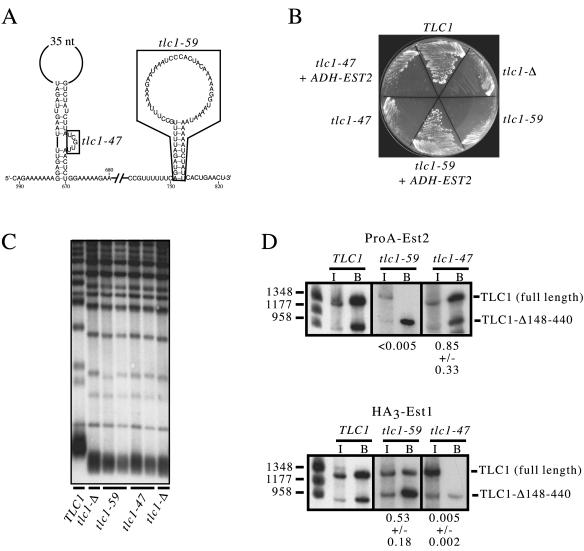
FIG. 3.
Defining a region of TLC1 required for interaction with Est2. (A) Sequences deleted in tlc1-47 and tlc1-59 are indicated by boxes. (B) Growth after ≈75 generations of a tlc1-Δ strain harboring plasmids expressing TLC1, tlc1-47, tlc1-59, or vector, with or without an ADH-EST2 plasmid (pVL369). (C) Telomere length of a tlc1-Δ strain transformed with TLC1 (lane 1), vector (lanes 2 and 7), tlc1-59 (lanes 3 and 4), or tlc1-47 (lanes 5 and 6). (D) Coimmunoprecipitation of TLC1 RNAs with either ProA-Est2 (top) or HA3-Est1 (bottom). The tlc1-59 and tlc1-47 mutant RNAs as well as the control tlc1-Δ148-440 RNA were expressed on CEN plasmids that were introduced into a tlc1-Δ strain. A representative experiment is shown, as are the averaged values of three independent experiments.
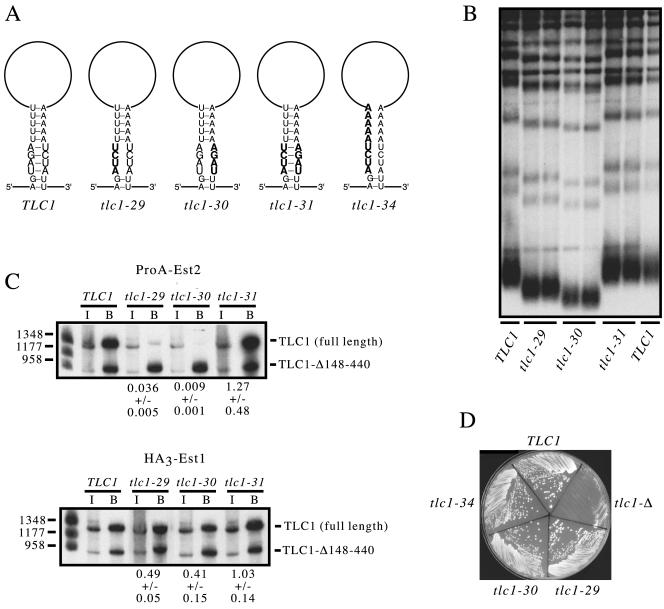
FIG. 4.
The Est2 protein interacts with a stem-loop structure. (A) Diagram of the nucleotide changes introduced into the duplex portion of the stem: tlc1-29 and tlc1-30 (predicted to disrupt base pairing in a portion of the stem), tlc1-31 (which should restore base pairing as the result of compensating sets of mutations), and tlc1-34 (predicted to disrupt base pairing of the entire stem). A second set of mutations introduced into this stem are shown in the supplemental material (Fig. S2). (B) Telomere length of a tlc1-Δ rad52-Δ strain transformed with plasmids encoding wild-type TLC1 (lanes 1 and 8), tlc1-29 (lanes 2 and 3), tlc1-30 (lanes 4 and 5), and tlc1-31 (lanes 6 and 7) and grown for ≈60 generations. (C) Northern blot of tlc1-29, tlc1-30, and tlc1-31 after immunoprecipitation of ProA-Est2 or HA3-Est1 from a tlc1-Δ strain harboring a tlc1-Δ148-440 plasmid. (D) Growth of a tlc1-Δ strain harboring either vector or plasmids that express TLC1, tlc1-29, tlc1-30, or tlc1-34.
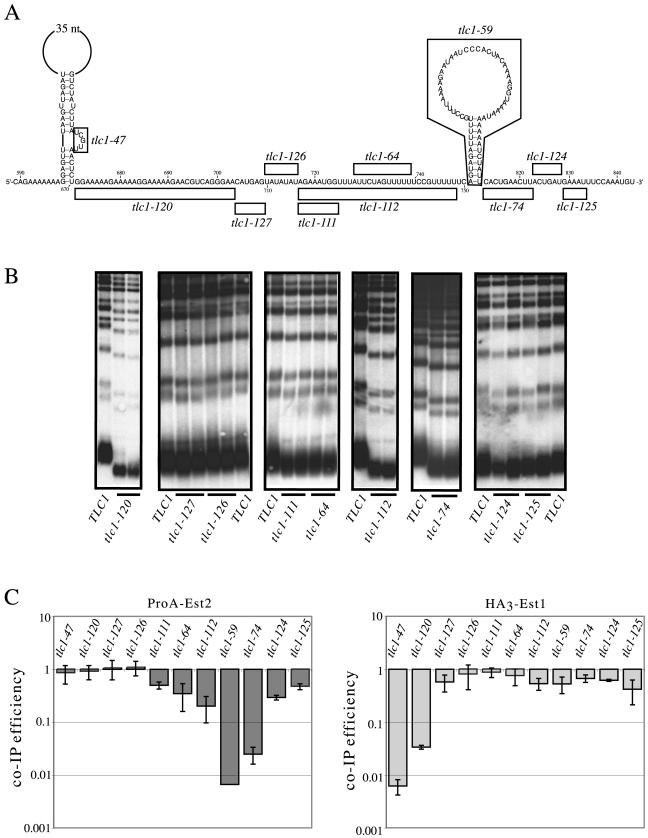
FIG. 5.
Regions adjacent to the stem-loop contribute to the interaction of the Est2 protein with TLC1. (A) Construction of a panel of deletion mutations on either side of the Est2-interacting stem-loop. Nucleotides that were deleted for each allele are indicated. (B) Telomere lengths of tlc1 mutants shown in panel A. Plasmids were transformed into a tlc1-Δ rad52-Δ strain and grown for ≈60 generations prior to analysis; data for tlc1-47 and tlc1-59 are shown in Fig. 3C. (C) Efficiency of coimmunoprecipitation of each tlc1 allele shown in part A with either ProA-Est2 or HA3-Est1; the relative efficiency, normalized to tlc1-Δ148-440, was determined from three independent experiments.
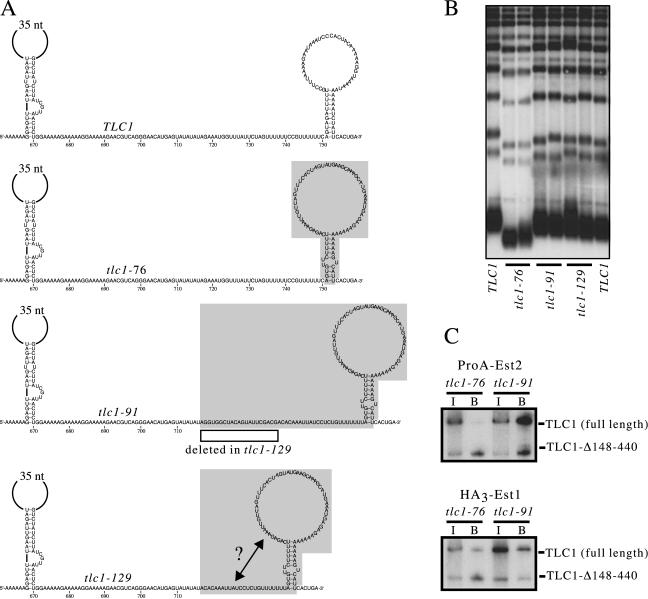
FIG. 6.
Chimeric TLC1 RNAs are functional in S. cerevisiae. (A) Diagram of three chimeric TLC1 molecules, constructed by replacing sequences in the S. cerevisiae TLC1 RNA with corresponding sequences from the S. kluyveri RNA. (B) Telomere length of a tlc1-Δ strain transformed with S. cerevisiae TLC1 (lanes 1 and 8), tlc1-76 (lanes 2 and 3), tlc1-91 (lanes 4 and 5), or tlc1-129 (lanes 6 and 7). (C) Coimmunoprecipitation of tlc1-76 and tlc1-91 with ProA-Est2 or HA3-Est1 in parallel with tlc1-Δ148-440.
FIG. 7.
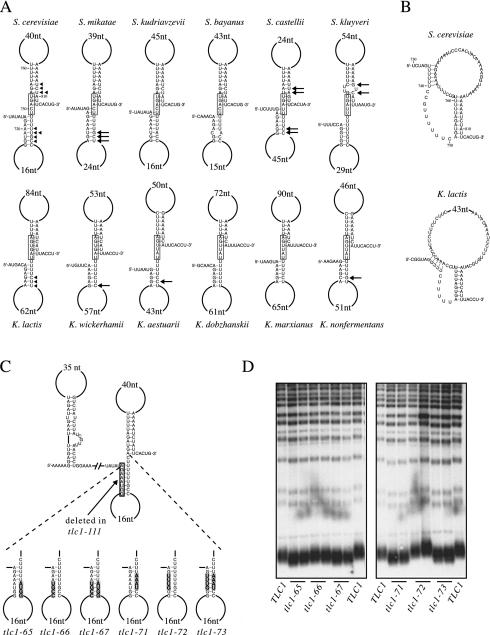
Two alternative models for an additional Est2-interacting structure. (A) Proposed second stem-loop structure, shown for six Saccharomyces and six Kluyveromyces RNAs; the larger stem-loop corresponds to that depicted in Fig. 2B. In order to identify base pairs that covary, thetwo sets of six RNAs were coaligned on a block of conserved residues, which is boxed. Arrows indicate base pairs that covary among the Saccharomyces homologues compared to analogous base pairs in the S. cerevisiae RNA (top) and among the Kluyveromyces homologues compared to the K. lactis RNA (bottom). Arrowheads alongside the S. cerevisiae and K. lactis RNAs summarize the number of covarying base pairs identified within each set of six RNAs. The sequence alignments that support these proposed structures are shown in supplemental Fig. S1. (B) A pseudoknot structure that has been previously proposed for the K. lactis telomerase RNA (56) and which may also form in the S. cerevisiae RNA; the regions that base-pair to form the proposed pseudoknot are also highlighted in grey in Fig. 2B. (C) Diagram of the nucleotide changes introduced into the duplex portion of the proposed second stem-loop (see panel A). The position of the tlc1-111 deletion mutation, which removes one strand of the predicted duplex stem, is also indicated. (D) Telomere blot of strains bearing plasmids containing the tlc1-65, tlc1-66, tlc1-67, tlc1-71, tlc1-72, and tlc1-73 mutant RNAs. Note that predicted compensatory mutations (tlc1-67 and tlc1-73), which should restore base pairing of the predicted duplex structure, do not restore the telomere length defect.
Mutations in TLC1 (Table 1) were introduced into pVL795 (TLC1 TRP1 CEN) by oligonucleotide-directed single-stranded mutagenesis (27) or with the QuikChange site-directed mutagenesis kit (Stratagene), and the mutational change was confirmed by sequencing. For tlc1 mutants that exhibited a telomere replication defect (tlc1-29, tlc1-30, tlc1-47, tlc1-59, tlc1-74, tlc1-76, tlc1-94, tlc1-96, and tlc1-112), the entire TLC1 gene plus an additional 300 nucleotides upstream and 300 nucleotides downstream of the gene, was sequenced to eliminate the possibility of unlinked mutations and confirm that the defect was the consequence of the introduced mutation.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Mutations or comments | Source or reference |
|---|---|---|---|
| pSD120 | TLC1 CEN URA3 | D. Gottschling | |
| pA258 | tlc1-Δ148-440 CEN URA3 | Δ148-440 | 33 |
| pVL2527 | tlc1-Δ148-440 URA3 | tlc1-Δ148-440 subcloned into YIp211 | pA258, YIp211 |
| pVL369 | ADH-EST2 2μm URA3 | 32a | |
| pVL795 | TLC1 CEN TRP1 | 2.6-kb TLC1 gene in pRS414 | This work |
| pVL1221 | CDC13-EST1 CEN HIS3 | In-frame fusion between CDC13 and EST1 in pRS414 | 15 |
| pVL1881 | TLC1S. castellii 2μm TRP1 | 7.2-kb genomic insert from S. castellii in YEp112 | This work |
| pVL1882 | TLC1S. bayanus 2μm TRP1 | 2.6-kb genomic insert from S. bayanus in YEp112 | This work |
| pVL1883 | TLC1S. kluyveri 2μm TRP1 | 6.9-kb genomic insert from S. kluyveri in YEp112 | This work |
| pVL2090 | tlc1-29 CEN TRP1 | TAGA753-756 → ATCT | pVL795 |
| pVL2091 | tlc1-30 CEN TRP1 | TCTA807-810 → AGAT | pVL795 |
| pVL2099 | tlc1-31 CEN TRP1 | TAGA753-756 → ATCT + TCTA807-810 → AGAT | pVL795 |
| pVL2122 | tlc1-34 CEN TRP1 | TAGATTTTT753-761 → ATCTAAAAA | pVL795 |
| pVL2175 | tlc1-43 CEN TRP1 | GATTT755-759 → CTAAA | pVL795 |
| pVL2126 | tlc1-44 CEN TRP1 | AAATC804-808 → TTTAG | pVL795 |
| pVL2176 | tlc1-45 CEN TRP1 | GATTT755-759 → CTAAA + AAATC804-808 → TTTAG | pVL795 |
| pVL2128 | tlc1-47 CEN TRP1 | Δ660-664 | pVL795 |
| pVL2192 | tlc1-59 CEN TRP1 | Δ751-812 | pVL795 |
| pVL2195 | tlc1-62 CEN TRP1 | Δ762-795 | pVL795 |
| pVL2428 | tlc1-64 CEN TRP1 | Δ728-739 | pVL795 |
| pVL2208 | tlc1-65 CEN TRP1 | CCGT741-744 → GGTA | pVL795 |
| pVL2209 | tlc1-66 CEN TRP1 | ATGG721-725 → TGCC | pVL795 |
| pVL2210 | tlc1-67 CEN TRP1 | CCGT741-744 → GGTA + ATGG721-725 → TGCC | pVL795 |
| pVL2218 | tlc1-71 CEN TRP1 | GTTT743-746 → TAAA | pVL795 |
| pVL2214 | tlc1-72 CEN TRP1 | AAAT719-722 → TTTG | pVL795 |
| pVL2215 | tlc1-73 CEN TRP1 | GTTT743-746 → TAAA + AAAT719-722 → TTTG | pVL795 |
| pVL2216 | tlc1-74 CEN TRP1 | Δ813-822 | pVL795 |
| pVL2204 | tlc1-76 CEN TRP1 | Replacement of nt 751-812 with analogous sequences from TLC1S. kluyveri | pVL795 |
| pVL2245 | tlc1-91 CEN TRP1 | Replacement of nt 717-812 with analogous sequences from TLC1S. kluyveri | pVL795 |
| pVL2249 | tlc1-94 CEN TRP1 | Replacement of nt 717-812 with nt 59-98 of O. nova telomerase RNA | pVL795 |
| pVL2251 | tlc1-96 CEN TRP1 | Replacement of nt 717-812 with nt 90-183 of hTR | pVL795 |
| pVL2430 | tlc1-111 CEN TRP1 | Δ717-724 | pVL795 |
| pVL2339 | tlc1-112 CEN TRP1 | Δ717-748 | pVL795 |
| pVL2426 | tlc1-120 CEN TRP1 | Δ671-703 | pVL795 |
| pVL2611 | tlc1-122 CEN TRP1 | Δ717-812 | pVL795 |
| pVL2431 | tlc1-124 CEN TRP1 | Δ823-828 | pVL795 |
| pVL2432 | tlc1-125 CEN TRP1 | Δ829-833 | pVL795 |
| pVL2435 | tlc1-126 CEN TRP1 | Δ710-716 | pVL795 |
| pVL2434 | tlc1-127 CEN TRP1 | Δ704-709 | pVL795 |
| pVL2497 | tlc1-129 CEN TRP1 | Derived from pVL2245 with AGGTGGCTACAGTATTCGACG deleted | pVL2245 |
Senescence assay and telomere length analysis.
Senescence and telomere length analyses were conducted with either YVL1008 or YVL1009 after plating on 5-fluoroorotic acid-containing medium to counterselect for loss of the TLC1 covering plasmid. A single 5-fluoroorotic acid-resistant colony was inoculated into 50 ml of YPD for subsequent transformation with relevant tlc1 mutant plasmids. Transformants were propagated by successive single-colony streak-outs on medium that maintained selection for the tlc1 mutant plasmid to observe senescence, as described previously (16, 29). For telomere length analysis, colonies from the first streak-out were inoculated into 10 ml of medium and grown to saturation. Genomic DNA was prepared by bead-beating and phenol-chloroform extraction (20) and digested with XhoI, and telomere length was examined by Southern blot analysis (29).
Isolation of TLC1 genes from other Saccharomyces species.
Genomic libraries of S. bayanus (ATCC 8741), S. castellii (generously provided by Ted Young), and S. kluyveri (ATCC 22512) were constructed by Sau3AI partial digestion of genomic DNA, followed by purification of DNA fragments in the ≈6- to 10-kb size range and ligation into BamHI-digested YEp112 (TRP1 2μm). S. bayanus and S. castellii TLC1 homologs were recovered by functional complementation by transforming a freshly generated S. cerevisiae tlc1-Δ rad52-Δ strain with the relevant genomic library. Candidate colonies that exhibited healthy growth were recovered, and plasmids were rescued and retransformed into an S. cerevisiae tlc1-Δ rad52-Δ strain to confirm rescue of senescence, followed by sequence analysis to identify the TLC1 gene in each clone.
The S. kluyveri TLC1 gene was recovered by screening for clones that hybridized to an oligomer that was complementary to the predicted 26-nucleotide S. kluyveri TLC1 template sequence (11). The S. kluyveri genomic library was transformed into Escherichia coli DH10B, and ≈35,000 colonies were plated. Colonies were transferred, lysed, and fixed to 137-mm transfer membranes (Biotechnology Systems). Membranes were prehybridized in SSC prehybridization buffer [6× SSC (3 M NaCl, 0.3 M trisodium citrate, pH 7.0), 5× Denhardt's solution (0.5% Ficoll 400, 0.5% polyvinylpyrrolidone, 0.5% bovine serum albumin), 0.05% sodium pyrophosphate, 100 μg of salmon sperm DNA per ml and 0.5% sodium dodecyl sulfate] at 37°C for 90 min and probed at 54°C overnight in SSC hybridization solution (6× SSC, 1× Denhardt's solution, 100 μg of yeast tRNA per ml, and 0.05% sodium pyrophosphate) with a 5′-end-labeled oligonucleotide (5′-TACTGTGAGGTCTGGGTG-3′). Membranes were washed twice in 6× SSC plus 0.05% sodium pyrophosphate at 65°C for 20 min and exposed on film. Candidate colonies were recovered, and sequence analysis was used to identify the S. kluyveri TLC1 gene present in the genomic insert; this sequence matched that of the TLC1 gene in the S. kluyveri genome (10).
TLC1 sequences were aligned with the Dialign program (http://bibiserv.techfak.uni-bielefeld.de/dialign/), and secondary structures were predicted with RNA Mfold version 3.1 (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/), aided by previously published structures that had been elucidated for portions of the S. cerevisiae and K. lactis telomerase RNAs (45, 48, 49, 55, 56).
Association of Est1 and Est2 with mutant tlc1 RNAs.
Coimmunoprecipitation experiments were performed as previously described (21). Briefly, strains were grown in 500 ml of selective medium to an optical density at 600 nm of 1.0, and cells were pelleted and resuspended in TMG buffer [10 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 10% (vol/vol) glycerol] plus 200 mM NaCl. Extracts were prepared by grinding cell pellets in the presence of liquid nitrogen and clarified twice by centrifugation for 10 min at 14,000 rpm. For HA3-Est1 immunoprecipitations, 0.5 to 2 mg of extract was incubated with 1 μl of antihemagglutinin (HA) antibody (3F10; Roche Molecular) for 1 h at 4°C, rotating, followed by a 2-h incubation at 4°C with 40 μl of protein G plus protein A-agarose beads (Oncogene). For ProA-Est2 immunoprecipitations, 0.5 to 2 mg of extract was incubated with 20 μl of immunoglobulin G-Sepharose (Amersham Biosciences) at 4°C for 3 h. Beads were collected by gentle centrifugation, washed three times for 5 min with TMG-200 mM NaCl-0.5% Tween 20-phenylmethylsulfonyl fluoride-RNasin, and washed once for 5 min with TMG plus 50 mM NaCl, phenylmethylsulfonyl fluoride, and RNasin. RNA was prepared by phenol-chloroform extraction, separated on a 7 M urea-4% polyacrylamide gel, transferred to a Hybond-N nylon membrane (Amersham Biosciences), and hybridized with a 470-bp fragment (nucleotides 451 to 921) of the TLC1 gene.
The relative efficiency of coimmunoprecipitation of various tlc1 mutations with Est1 or Est2 was determined by detection of both the tlc1-Δ148-440 and the tlc1 mutant RNA in question with PhosphorImager detection (Molecular Dynamics Storm 860) and subsequent analysis with ImageQuant software. The relative immunoprecipitation efficiency of the various tlc1 mutants was calculated as (bound tlc1 mutant RNA/bound tlc1-Δ148-440)/(input tlc1 mutant RNA/input tlc1-Δ148-440). A minimum of three independent immunoprecipitations were conducted for each mutant. The absolute coimmunoprecipitation efficiency of the Tlc1-Δ148-440 RNA with either ProA-Est2 or HA3-Est1 was generally around 20%.
Nucleotide sequence accession number.
A 1.6-kb region encompassing the S. bayanus TLC1 gene was sequenced and has been deposited in GenBank (accession number AY547299). The sequence of the S. castellii TLC1 gene matched that of the TLC1 gene in the sequenced S. castellii genome (10), which was completed while this project was ongoing.
RESULTS
Identification of TLC1 genes from several Saccharomyces species.
In order to identify phylogenetically conserved structural elements in the budding yeast telomerase RNA, we analyzed TLC1 genes from six Saccharomyces species. TLC1 genes from S. bayanus and S. castellii were cloned by complementation of the senescence phenotype of a S. cerevisiae tlc1-Δ rad52-Δ strain, whereas the S. kluyveri telomerase RNA was recovered by screening a genomic library for clones containing the predicted template sequence (see Materials and Methods for details). The TLC1 genes from S. mikatae and S. kudriavzevii were subsequently identified from the completed sequences of these two genomes (10, 26).
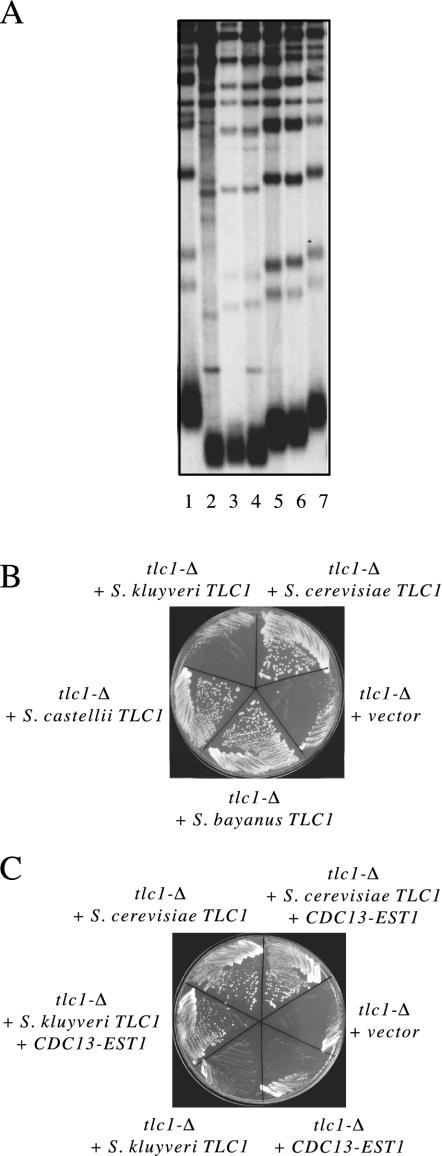
These candidate TLC1 genes exhibited a surprising degree of primary sequence divergence, even though all were recovered from Saccharomyces species. The S. bayanus and S. castellii genes exhibited 65 and 55% identity, respectively, compared to the S. cerevisiae RNA, whereas the S. kluyveri and S. cerevisiae TLC1 RNAs shared so little sequence identity that it was difficult to entirely align these two sequences. In order to confirm that bona fide TLC1 genes had been cloned, the ability of the S. bayanus, S. castellii, and S. kluyveri genes to complement an S. cerevisiae tlc1-Δ strain was examined. As shown in Fig. 1, the S. bayanus TLC1 gene (expressed by the S. bayanus promoter) substantially rescued the telomere replication defect of a tlc1-Δ strain. In the tlc1-Δ/pTLC1S. bayanus strain, telomere length was stably maintained at an intermediate length (Fig. 1A, lanes 5 and 6), and this strain exhibited a growth phenotype comparable to that of a TLC1 strain (Fig. 1B). The S. castellii TLC1 gene was somewhat less functional in this cross-species complementation experiment: telomeres in a tlc1-Δ/pTLC1S. castellii strain were extremely short (Fig. 1A, lanes 3 and 4), although not so short as to confer senescence (Fig. 1B).
FIG. 1.
TLC1 genes from S. bayanus, S. castellii, and S. kluyveri are partially functional in S. cerevisiae. (A) Telomere length of a tlc1-Δ strain transformed with S. cerevisiae TLC1 (lanes 1 and 7), vector (lane 2), S. castellii TLC1 (lanes 3 and 4), or S. bayanus TLC1 (lanes 5 and 6). Telomere length was examined after ≈75 to 100 generations of growth, at a time when telomeres in the tlc1-Δ strain (lane 2) had begun to undergo rearrangements indicative of a recombination-dependent pathway for telomere maintenance. (B) Growth of a tlc1-Δstrain transformed with either vector or plasmids encoding the S. cerevisiae, S. bayanus, S. castellii, or S. kluyveri TLC1 gene. (C) Growth of a tlc1-Δ strain transformed with plasmids encoding S. kluyveri TLC1, S. cerevisiae TLC1, or vector in the presence or absence of a CDC13-EST1 fusion plasmid.
The S. kluyveri TLC1 gene failed to rescue either telomere length (data not shown) or the senescence defect of a S. cerevisiae tlc1-Δ strain (Fig. 1B). Since this RNA showed substantial sequence divergence, the limited cross-species complementation might be due to a reduced interaction with S. cerevisiae telomerase protein subunits and/or a less functional telomerase enzyme, once assembled. If so, either defect might be alleviated by increasing the interaction of this telomerase RNP, composed of heterologous components, with chromosome termini. To test this, we introduced the S. kluyveri TLC1 gene into a tlc1-Δ strain containing a fusion between a subunit of telomerase and the Cdc13 telomere binding protein. Previous work has shown that such fusions increase telomere replication, presumably as a consequence of increasing the local concentration of telomerase at chromosome termini (15). In the presence of a Cdc13-Est1 fusion protein, the S. kluyveri TLC1 gene was capable of complementing the senescence defect of a tlc1-Δ strain (Fig. 1C), although telomeres still remained extremely short (data not shown). This result supports the conclusion that the S. kluyveri sequence, recovered on the basis of a predicted template sequence, encodes a telomerase RNA.
Phylogenetically conserved stem-loop structures present in six Saccharomyces RNAs.
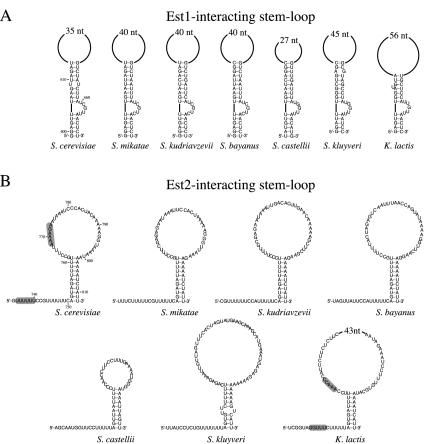
As discussed above, alignment of the six Saccharomyces RNAs revealed a surprisingly high level of sequence divergence. Despite this, two previously characterized secondary structures located upstream of the template region, the template boundary element (49) and a stem-loop that mediates association with the Ku heterodimer (50), were readily identified in all six Saccharomyces RNAs (7). Two stem-loop structures downstream of the template were also detected (Fig. 2). One of these has been shown to mediate association with the Est1 telomerase subunit (48). Interaction of the Est1 protein with TLC1 has been shown to depend on a 5-nucleotide bulge present within the stem of the Est1-interacting structure. Comparison of the sequence of this bulge among 12 Saccharomyces and Kluyveromyces RNAs indicates that three of the five nucleotides in this bulge are conserved, suggesting that both structural elements and sequence specificity may contribute to Est1 binding (Fig. 2) (48).
FIG. 2.
Two previously identified stem-loops are conserved among the budding yeast telomerase RNAs. Predicted secondary structures for the Est1-interacting (A) and Est2-interacting (B) structures of the telomerase RNAs from six Saccharomyces species and from K. lactis. Determination of structures was based on primary sequence alignments and comparison to previously determined structural data (48, 56), aided by RNA Mfold version 3.1. The duplex portion of the Est1-interacting structure is extended by an additional 6 bp relative to that established previously (48), although this extension has not been experimentally tested. The primary sequence alignments that support the proposed structures shown in panel B are presented in the supplemental material (Fig. S1A). The regions highlighted in grey in the S. cerevisiae and K. lactis RNAs (B) indicate sequences capable of forming a potential pseudoknot (56).
An additional stem-loop structure immediately adjacent to the Est1-interacting hairpin has been previously identified as an element that is required for telomere replication in the Kluyveromyces budding yeasts (56). Sequence inspection of the six Saccharomyces TLC1 RNAs revealed a small block of conserved sequences in the equivalent region (see Fig. S1 in the supplemental material), which could be folded into a similar stem-loop (Fig. 2B). Covariation of base pairing within the stem region supports the conclusion that this is a phylogenetically conserved structure present in the budding yeast telomerase RNAs (see Fig. 7A, below).
Interaction of Est2 with TLC1 requires a stem-loop structure.
The stem-loop described in Fig. 2B was located within a region that has previously been shown to be required for the interaction of the Est2 telomerase protein with TLC1 (33). To test whether this structure was responsible for mediating an Est2-RNA interaction, a 62-nucleotide deletion (tlc1-59) that precisely removed the stem-loop was constructed (Fig. 3A). This mutation resulted in a complete telomere replication defect: both the senescence phenotype and telomere length of a tlc1-59 strain were indistinguishable from that displayed by a strain that lacked the entire TLC1 RNA (Fig. 3B and C).
The effect of this deletion on telomerase RNP assembly was examined by assessing the ability of this mutant RNA to associate with the Est1 and Est2 telomerase proteins. The mutant tlc1-59 gene was introduced into strains bearing tagged versions of Est1 or Est2 (HA3-Est1 and ProA-Est2) as well as an additional copy of the TLC1 gene (tlc1-Δ148-440). The tlc1-Δ148-440 derivative, which lacks 300 nucleotides, retains full association with both Est1 and Est2 (33) and can be easily distinguished on Northern blots by its altered size relative to the full-length TLC1 RNA. It therefore serves as an internal control for immunoprecipitation efficiency. With these strains, the association of Est1 and Est2 with tlc1-59 as well as with tlc1-47 RNA (deleted for the bulge in the Est1-interacting stem-loop) was monitored.
Interaction of Est2 with the tlc1-59 RNA was decreased by >200-fold, whereas association with Est1 was largely unaffected (Fig. 3D). Thus, the inability of Est2 to interact with a telomerase RNA deleted for this specific stem-loop was not due to overall impaired RNP assembly due to either reduced RNA levels or global misfolding of the tlc1-59 RNA (although we cannot rule out misfolding of individual domains of the RNA). We also examined the association of the Est1 and Est2 telomerase proteins with the mutant tlc1-47 RNA: Est1 association with tlc1-47 was lost, but interaction with Est2 was retained at roughly wild-type levels (Fig. 3D), as had been observed previously (48).
To test whether formation of the duplex stem was required for Est2 interaction, mutations that either disrupted or restored base pairing were introduced into the proposed stem region and examined for effects on telomere replication and Est2 association. Two sets of sequence changes (tlc1-29 and tlc1-30) were introduced into either strand of the duplex, both of which would be predicted to disrupt base pairing (Fig. 4A). When introduced into a tlc1-Δ strain, both mutations resulted in short telomeres (Fig. 4B) and a greatly reduced association with the Est2 protein (Fig. 4C). To ask if these defects were due to disrupted base pairing, a third derivative of TLC1 (tlc1-31), which contained both sets of sequence changes and should therefore restore base pairing (Fig. 4A), was also examined. In a tlc1-31 mutant strain, both telomere length and Est2 association were restored to wild-type levels (Fig. 4B and 4C).
Curiously, tlc1-29 and tlc1-30, which should each disrupt pairing of the same set of nucleotides, did not have identical phenotypes. Telomeres were only moderately short in the tlc1-29 strain compared to the more severe telomere shortening displayed by the tlc1-30 mutant strain (Fig. 4B). The severity of the telomere length defect also correlated with Est2 association, which was reduced by 30-fold and 100-fold for tlc1-29 and tlc1-30, respectively (Fig. 4C). These differences were not the consequence of altered RNA levels: the tlc1-29 and tlc1-30 mutant RNAs were both expressed at levels comparable to that of the tlc1-Δ148-440 control RNA (0.87 and 1.0, respectively; data not shown).
To explore this in more detail, a second set of mutations that would also be predicted to disrupt the duplex were constructed (tlc1-43 and tlc1-44), as well as the compensatory mutation (tlc1-45) that combined both sets of mutations. Strains bearing the tlc1-43 and tlc1-44 mutations both exhibited extremely short telomeres as well as a substantial decline in Est2 association (Fig. S2 in the supplemental material). The tlc1-45 mutation reversed these defects, although telomere length and Est2 association were not fully restored to wild-type levels (Fig. S2). Collectively, the results of these two sets of compensatory mutations establish that formation of this stem is required for association of the Est2 protein with the telomerase RNP.
Sequences adjacent to the stem-loop are required for Est2 association with TLC1.
Notably, the tlc1-29 and tlc1-30 mutations, as well as other mutations predicted to disrupt base pairing of this stem (such as tlc1-34; see Fig. 4A), did not result in a complete null phenotype; such mutations resulted in short telomeres, but the defect was not severe enough to confer a senescence phenotype (Fig. 4D and data not shown). Furthermore, overexpression of the Est2 protein was capable of partial suppression of the telomere replication defect of the tlc1-59 strain but not the tlc1-47 strain (Fig. 3B). This suggested that additional sequences and/or structural elements besides the Est2-interacting stem-loop contribute to association of Est2 with the telomerase RNA. To examine this in more detail, a series of deletions were introduced on either side of the Est2-interacting stem-loop (Fig. 5A) and examined for effects on telomere replication and Est2 association (Fig. 5B and 5C).
On the 3′ side of the Est2 interaction stem, three small successive deletions were examined. A 10-nucleotide deletion immediately adjacent to the stem (tlc1-74) resulted in a moderate reduction in telomere length (Fig. 5B). Est2 association with this mutant RNA was reduced 30-fold, whereas Est1 association was largely unaffected (Fig. 5C). One possible explanation for the phenotype of the tlc1-74 mutant could be destabilization of the base of the duplex stem. This possibility was ruled out by a mutation that prevented base pairing of the last two nucleotides of the stem and yet had no effect on telomere length (data not shown). Two additional deletions which successively removed 5 to 6 nucleotides of adjacent sequences (tlc1-124 and tlc1-125) reduced Est2 association by only two- to threefold, with correspondingly little effect on telomere length (Fig. 5B and C). These results, combined with prior work (33), argue that fewer than 10 nucleotides on the 3′ side of this stem contribute to the specific interaction between the Est2 protein and TLC1.
In a similar manner, a series of deletions were constructed on the 5′ side of the Est2-interacting stem-loop that encompassed the 80-nucleotide stretch between the Est1- and Est2-interacting stem-loops. As shown in Fig. 5C, this set of deletions exhibited a roughly reciprocal relationship with regard to the effects on Est1 and Est2 association: those deletions closest to the Est1-interacting structure had the greatest impact on Est1 association, whereas deletions closer to the Est2-interacting stem-loop had a greater effect on Est2 association. The tlc1-120 deletion, which removed 33 nucleotides immediately adjacent to the Est1-interacting stem-loop, as well as the tlc1-47 mutation were severely reduced for interaction with the Est1 protein but retained wild-type levels of association with Est2. Two centrally located deletion alleles (tlc1-126 and tlc1-127) had little or no impact on either Est1 or Est2 association. However, approximately half-way through the region, a reproducible two- to threefold reduction in Est2 association, accompanied by a modest but detectable effect on telomere length, was observed as the result of two adjacent deletions (tlc1-111 and tlc1-64). A more extensive deletion (tlc1-112), which extended to within 2 nucleotides of the base of the Est2-interacting stem-loop, had an even more pronounced effect. Telomere replication was noticeably impaired in the tlc1-112 mutant strain, as evidenced by a substantial effect on telomere length along with a fivefold reduction in Est2 association.
These experiments establish that additional sequences, and thus potentially additional structural elements, in addition to the Est2-interacting stem-loop described in Fig. 3 are important for Est2 interaction. Subsequent experiments, described in the next two sections, were directed at elucidating the potential contribution of sequences to the 5′ side of this stem-loop.
Chimeric TLC1 RNAs support an extended 5′ region required for Est2 association.
As an alternative approach towards analyzing additional sequence or structural requirements for Est2 association, we constructed a set of chimeric TLC1 molecules by replacing sequences in the S. cerevisiae TLC1 RNA with the corresponding sequences from the S. kluyveri RNA (Fig. 6A). The S. cerevisiae Est2-interacting stem-loop structure, with no adjacent flanking sequences, was replaced with the analogous S. kluyveri sequences, generating a chimeric RNA (tlc1-76) that exhibited intermediate function: a tlc1-Δ strain expressing this chimeric RNA maintained telomeres at a stable but short length (Fig. 6B).
To ask whether introduction of additional S. kluyveri flanking sequences could increase the level of function, a second chimeric TLC1 molecule (tlc1-91) that extended the boundary of the chimera by 45 nucleotides on the 5′ side was constructed (Fig. 6A). Strikingly, this chimera was now capable of maintaining telomeres at wild-type length (Fig. 6B) and exhibited wild-type levels of association with the Est2 telomerase subunit (Fig. 6C). Similar results were obtained with a comparable set of chimeras in which the same regions of the S. cerevisiae RNA were replaced with S. castellii TLC1 sequences (data not shown). These results, combined with the experiments shown in Fig. 5, argue that an additional determinant for Est2 interaction resides in the region 5′ to the stem-loop.
Two alternative models for an additional structural element required for Est2 interaction.
Sequence inspection of the 5′-flanking region adjacent to the stem-loop structure suggested that this region had the potential to form a second, smaller stem-loop. Inspection of the sequences of all six of the Saccharomyces RNAs as well as the six Kluyveromyces RNAs indicated that all 12 RNAs were capable of base pairing to form this second proposed stem-loop (Fig. 7A). Although the extent of base pairing in the duplex region of this second stem was small, this proposed structure was reasonably well supported by phylogenetic data, based on covariation of base pairing among both the Saccharomyces and Kluyveromyces RNAs in this region, as indicated in Fig. 7A. In fact, the covariation data supporting this second stem are stronger than the comparable phylogenetic data in favor of the larger stem, although the compensatory mutagenesis shown in Fig. 4 and supplemental Fig. S2 clearly establish that the larger stem forms.
However, while this work was in progress, Blackburn and colleagues proposed, on the basis of phylogenetic conservation among Kluyveromyces telomerase RNAs, that this region instead formed a pseudoknot that involved 5′-flanking sequences base-paired to the loop of the Est2-interacting structure (56). Although the S. cerevisiae RNA can also potentially form a pseudoknot (Fig. 7B), sequence inspection of the other Saccharomyces RNAs did not readily support the formation of a similar, phylogenetically conserved pseudoknot in these additional RNAs (Fig. 2B). Therefore, we performed several sets of experiments with the S. cerevisiae RNA designed to differentiate between these two predicted structures.
To ask whether nucleotides in the duplex region of the proposed smaller stem-loop shown in Fig. 7A were required for base pairing, a series of nucleotide substitution mutations were constructed in the S. cerevisiae RNA. Each of these mutations (tlc1-65, tlc1-66, tlc1-71, and tlc1-72, depicted in Fig. 7C) should disrupt base pairing of this proposed duplex. If this second stem-loop structure forms in vivo and makes a critical contribution to telomerase function, these mutations should confer a telomere replication defect. In contrast to this prediction, however, these mutant RNAs had very little effect on telomere replication. These four substitution mutations conferred telomere lengths that were either wild type or only slightly reduced relative to those of a TLC1 strain (Fig. 7D). Mutant RNAs in which base pairing of this predicted duplex should have been restored (tlc1-67 and tlc1-73) still exhibited the same modest decline in telomere length (Fig. 7D). Furthermore, deletion of one strand of this proposed duplex (tlc1-111; Fig. 7C) also caused only a mild telomere length defect (Fig. 5B). Therefore, despite the apparent phylogenetic conservation of this proposed stem-loop structure, mutational analysis of this region suggests that this second stem does not form in vivo, or if it does, its formation is not crucial for telomere replication under standard laboratory conditions.
In a second approach, we extended the chimera analysis presented in the previous section. A third chimeric RNA (tlc1-129) was constructed by deleting 21 nucleotides of S. kluyveri sequences from the 5′ boundary of the tlc1-91 chimeric RNA. This deletion was designed so that it removed one strand of the proposed second duplex depicted in Fig. 7A. If the flanking S. kluyveri sequences present in the tlc1-91 RNA conferred full function through the formation of a second stem-loop, deletion of a portion of this stem-loop should reduce function. Strikingly, however, the tlc1-129 RNA was still capable of promoting wild-type telomere replication (Fig. 6B). The results from this experiment parallel those shown in Fig. 7D and suggest that formation of a second stem-loop is not an essential determinant in the S. kluyveri telomerase RNA.
As a complement to the above analysis, we tested a structural requirement of the pseudoknot that has been proposed to form in the K. lactis telomerase RNA (56). In the S. cerevisiae RNA, a comparable pseudoknot would involve base pairing between two sets of sequences (nucleotides 735 to 740 and 768 to 773; depicted in Fig. 7B), one of which resides in the loop of the Est2-interacting hairpin. Therefore, we examined whether removal of the loop region in the S. cerevisiae TLC1 RNA would alter either telomere replication or Est2 association with the telomerase RNP.
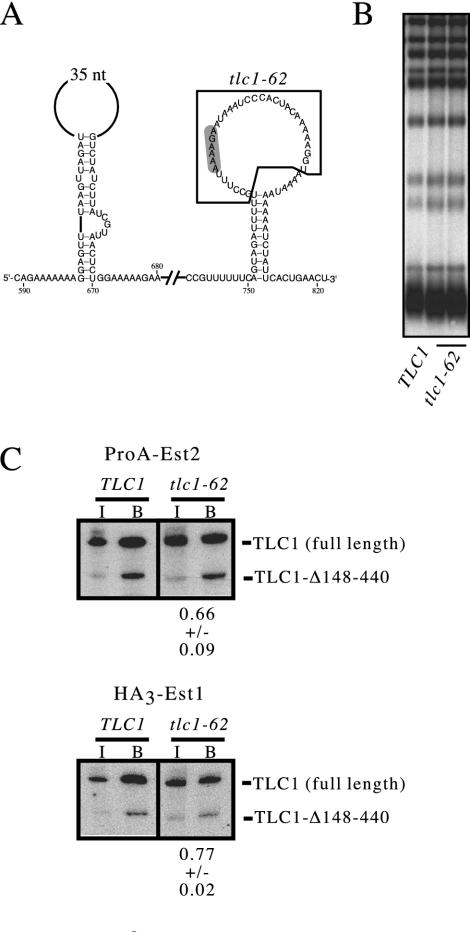
Strikingly, a deletion (tlc1-62) that removed all but 6 nucleotides of the loop region (Δ762-795) had no detectable effect on telomere replication. The telomere length of the tlc1-62 strain was indistinguishable from that of a TLC1 strain (Fig. 8B), and Est2 association with the tlc1-62 mutant RNA was reduced by less than twofold (Fig. 8C). Furthermore, the tlc1-64 deletion shown in Fig. 5, which deleted nucleotides 728 to 739 and thus removed the other strand of the proposed pseudoknot, also exhibited only minimal effects on telomere replication (Fig. 5B). Therefore, removal of sequences that should be essential to the formation of a pseudoknot in the S. cerevisiae RNA, analogous to that proposed for the K. lactis RNA (56), had essentially no effect on either telomere length maintenance or telomerase assembly.
FIG. 8.
The loop of the Est2-interacting stem-loop is not required for association of Est2 with the telomerase RNP. (A) Sequences deleted in the tlc1-62 mutant are indicated. (B) Telomere length of a tlc1-62 strain (lanes 2 and 3) compared to that of a TLC1 strain (lane 1). (C) Coimmunoprecipitation of ProA-Est2 (top) and HA3-Est1 (bottom) with either TLC1 or tlc1-62 in parallel with tlc1-Δ148-440.
A pseudoknot can replace the 95-nucleotide Est2 interaction domain.
Pseudoknot structures have been observed in telomerase RNAs from a diverse range of species, indicating that this is a phylogenetically conserved motif (8, 32, 52, 56). Furthermore, the ability of the closely related Kluyveromyces budding yeast telomerase RNAs to form a pseudoknot in this region is well supported by phylogenetic covariation among the Kluyveromyces telomerase RNAs (56). However, the experiment presented in Fig. 8 argued that sequence elements that would be required for the formation of a similar pseudoknot in the S. cerevisiae RNA were dispensable for normal laboratory growth of the strain.
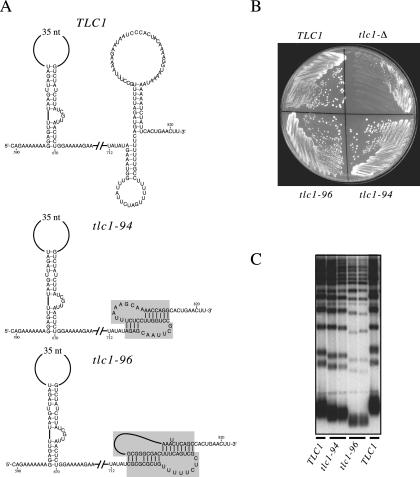
As an alternative approach towards addressing this topic, we constructed an additional set of chimeric RNAs in which the Est2-interacting region was replaced with well-characterized telomerase RNA pseudoknots from two phylogenetically distant species. Two chimeric RNAs were constructed in which a 95-nucleotide region of TLC1, encompassing both the large Est2-interacting stem-loop and the smaller proposed stem-loop depicted in Fig. 7A, was replaced with pseudoknots from the Oxytricha nova and human telomerase RNAs (tlc1-94 and tlc1-96, respectively; Fig. 9A). A tlc1 mutant strain deleted for this 95-nucleotide region (tlc1-122) displayed a full telomere replication defect (data not shown), comparable to that of the tlc1-59 deletion mutant (Fig. 3). Amazingly, the O. nova chimera, in which a 39-nucleotide O. nova sequence capable of forming a pseudoknot (32) was introduced in place of this 95-nucleotide stretch, retained almost full function: a tlc1-Δ strain expressing the tlc1-94 chimeric RNA showed no signs of senescence, and telomeres were maintained at a length only slightly below that of a wild-type strain (Fig. 9C). The human chimera (tlc1-96), which introduced a portion of the human pseudoknot in place of TLC1 sequences, was more reduced in its ability to replicate telomeres; nevertheless, this strain was also capable of long-term healthy growth, and telomeres were stably maintained, albeit at a relatively short length (Fig. 9B and C).
FIG. 9.
O. nova and human pseudoknots can replace Est2-interacting sequences of the S. cerevisiae RNA. (A) Diagram of two chimeric TLC1 molecules (tlc1-94 and tlc1-96), constructed by replacing nucleotides 717 to 812 in the S. cerevisiae TLC1 RNA with the pseudoknot from the O. nova telomerase RNA (nucleotides 59 to 98) or a minimal version of the pseudoknot from the human telomerase RNA (nucleotides 90 to 183), respectively. (B) Growth after ≈75 generations of a tlc1-Δ strain harboring plasmids expressing TLC1, tlc1-94, tlc1-96, or vector. (C) Telomere length of tlc1 chimeras shown in part A. Plasmids were transformed into a tlc1-Δ strain and grown for ≈60 generations prior to analysis.
Consistent with their ability to promote telomere replication, these chimeric RNAs were also capable of mediating association of the yeast Est2 protein with the telomerase RNP. The tlc1-94 RNA could be coimmunoprecipitated with the Est2 protein at levels that were reduced only 16.1-fold relative to controls. Association between the tlc1-96 human chimeric RNA and the Est2 protein was more severely reduced (50-fold relative to controls), consistent with the more impaired ability of this chimeric RNA to maintain telomeres. Since the introduced O. nova and human sequences did not exhibit any notable sequence similarity with the 95-nucleotide region of TLC1 that was replaced, this argues that the ability to rescue the telomere replication defect of the tlc1-122 strain was not due to introduction of a specific sequence element. Instead, these data indicate that introduction of a highly ordered structure, with no sequence identity to the region of TLC1 that is replaced, can restore Est2 interaction and the telomere replication defect of the tlc1-122 strain.
DISCUSSION
Determination of the secondary structures of ciliate and metazoan telomerase RNAs has provided the basis for numerous studies that have addressed contributions by the RNA subunit to enzyme function as well as characterization of specific protein-RNA interactions. The large size and the rapid sequence divergence of the yeast telomerase RNAs have not permitted simple extrapolation from the previously determined secondary structures for the ciliate and metazoan RNAs. However, recent work from several laboratories has begun to identify specific regions of the budding yeast telomerase RNAs that are required for either enzyme activity (47, 49, 55) or interaction with specific proteins (33, 45, 48).
These prior studies included deletion mapping of the TLC1 RNA, which defined two regions of the RNA that were required for association of the Est1 and Est2 telomerase subunits (33). The Est1-interacting domain has been shown to employ a bulged stem that tethers Est1 to the telomerase RNP (48). In this work, we used genetic and phylogenetic analyses to characterize structural elements that similarly mediate the association of Est2 with the telomerase RNA. The phylogenetic portion of our analysis relied on both genetic and molecular techniques to recover TLC1 homologs from other Saccharomyces species. Identification of telomerase RNA genes by in vivo complementation has a particular advantage in that functional screens do not necessarily rely on the substantial DNA sequence identity that is required when hybridization techniques are used to recover homologues. Indeed, the S. castellii TLC1 gene, which shows only 55% overall sequence identity to S. cerevisiae TLC1, was successfully recovered through in vivo complementation.
A phylogenetically conserved structure is required for interaction with Est2.
Despite the large degree of sequence divergence among the TLC1 RNAs that were analyzed, a conserved stem-loop that is required for interaction with the Est2 catalytic subunit was readily identifiable. This structure corresponds to an analogous hairpin that was reported in the Kluyveromyces telomerase RNAs (56) while this work was in progress. In S. cerevisiae, deletion of this stem-loop or disruption of base pairing of the duplex portion of the structure results in loss of the interaction between Est2 and TLC1 and a corresponding severe telomere replication defect, arguing that this stem makes a substantial contribution to the association of Est2 with the telomerase RNP. This hairpin consists of an 11-bp stem, with no unpaired nucleotides or bulges that disrupt this duplex. Furthermore, the loop of this hairpin is dispensable for Est2 interaction. Since specific RNA-protein interactions usually require either a single-stranded RNA sequence or a structural perturbation in a double-stranded RNA helix, such as a bulge or hairpin (14), this suggests that additional sequences and/or structures in the TLC1 RNA are required to mediate its interaction with Est2. Consistent with this, residues immediately surrounding the 62-nucleotide stem-loop (Fig. 5), as well as a small stretch of residues at the 5′ end of TLC1 (33), also contribute to the interaction between Est2 and TLC1.
Two not necessarily exclusive models for additional structures in this region.
One structural proposal for the more extended region required for Est2 interaction is that a second stem-loop forms adjacent to the larger Est2-interacting hairpin. All 12 Saccharomyces and Kluyveromyces telomerase RNAs exhibit the ability to form this smaller stem-loop. This second stem is moderately supported by the degree of covariation exhibited within the base-paired region in both the Saccharomyces and Kluyveromyces telomerase RNAs (Fig. 7A). In addition, mutations in this region reproducibly conferred, albeit modestly, a reduced Est2 interaction with the telomerase RNP (Fig. 5), arguing that this region does contribute to Est2 association. However, mutations predicted to disrupt base pairing in this region had an extremely minimal effect on telomere length, and mutations that should restore base pairing did not reverse this minimal defect (although, as Fig. S2 shows, not all compensatory mutations that are predicted to restore base pairing gave the expected in vivo restoration to a wild-type phenotype).
We also considered in this study whether this extended Est2-interacting region forms a pseudoknot structure (as depicted in Fig. 7B), similar to that proposed for other telomerase RNAs, including the relatively closely related Kluyveromyces RNAs (56). Formation of this potential pseudoknot would present a single-stranded stretch of uracil residues (Fig. 7B), which are highly conserved among all 12 budding yeast RNAs (supplemental Fig. S1), suggesting that the function of the pseudoknot may be, at least in part, to maintain this uracil-rich region as a single-stranded region available for protein interaction. However, two sets of observations, based on analysis of the loop region, which should be involved in base pairing to form the pseudoknot, are not easily reconciled with the proposal that this regions forms such a structure.
First, in contrast to the situation with the Kluyveromyces RNAs (56), phylogenetic analysis of the Saccharomyces RNAs failed to identify a conserved region within this loop that would support the proposed base-pairing (Fig. 2B and data not shown). The sequence within the loop of these six Saccharomyces RNAs is fairly diverged, however, which prevents easy alignment of the primary sequence in this region and thus identification of conserved structures based on covariation. Accumulation of additional Saccharomyces RNA sequences may help elucidate whether base pairing, which may be conserved at the structural rather than the sequence level, will contribute to the formation of a pseudoknot in this region of the RNA. We also cannot rule out noncanonical base pairing in this region; although noncanonical base pairing is rare, these types of interaction have been observed in some RNAs (19).
A second observation that is not apparently consistent with the formation of a pseudoknot is that deletion of either of the two sets of sequences that would be required for its formation did not confer a notable telomere replication defect or loss of Est2 association. These data argue that either a pseudoknot does not form or it is not critical for telomerase activity under standard laboratory conditions. However, we note that other studies have suggested that telomerase pseudoknots may dictate repeat addition processivity but not impair DNA synthesis per se (43). The TLC1 variant that was deleted for the loop region was only assayed in an otherwise wild-type background, where the requirements for telomerase processivity may not be very high. It is possible that monitoring a more sensitive parameter, with an assay such as that developed very recently by Lingner and colleagues (51), may reveal more subtle defects as a consequence of this mutation.
The above discussion weighs the evidence in favor of the formation of one or the other of these two proposed structures. One additional hypothesis worth considering in future studies is that there may be an equilibrium between the formation of the smaller stem-loop and the pseudoknot, potentially as a response to alterations in telomere length. For example, a switch between the pseudoknot shown in Fig. 7B and the stem-loop shown in Fig. 7A might contribute to the control of a long-range novel pseudoknot that has been proposed by Tzfati, Blackburn, and colleagues to modulate template usage (56).
Interaction of Est2 with the telomerase RNA may rely on a highly structured domain.
In an extension of our chimeric RNA studies, a chimeric TLC1 RNA was generated in which a 95-nucleotide region required for Est2 interaction was replaced with a 39-nucleotide pseudoknot from the distantly related O. nova telomerase RNA. The replaced region of TLC1 is essential for telomere replication and Est2 association, as a TLC1 RNA deleted for this 95-nucleotide region (tlc1-122) was completely defective for both aspects. Strikingly, this defect was substantially bypassed by the insertion of the O. nova pseudoknot, and a similarly constructed human pseudoknot chimera also retained partial function (Fig. 9). This suggests that this 95-nucleotide region of TLC1 may primarily contribute to Est2 association by providing a highly structured region, although we certainly cannot exclude the possibility that sequence-specific elements may also facilitate protein binding.
One such element may be a stretch of uracil residues which, as mentioned above, are highly conserved among all 12 budding yeast RNAs and could provide a single-stranded region for protein interaction. Notably, both the O. nova and the human chimeras also have a uracil-rich region that is presented in a single-stranded conformation available for potential protein binding. A second element that may mediate Est2-specific binding is a small stretch of residues at the 5′ end of TLC1 that was previously identified by Cech and colleagues (33). We were unable to ascertain whether the additional Saccharomyces RNAs analyzed in this study possessed this sequence motif, although this may simply be a consequence of the degree of sequence divergence among these six RNAs.
One proposal that could explain all of the data presented in this study, and by other laboratories, is that several small sequence-specific elements, combined with a sequence-independent structure, may provide the elements that dictate the specificity for Est2 binding. Thus, one simple explanation for the surprisingly high level of activity displayed by the O. nova and human chimeric RNAs is that Est2 association with the RNA is not dependent on the formation of a specific structure, such as a pseudoknot or the two stem-loops shown in Fig. 7A. Instead, the formation of any secondary structure in this region that is capable of properly presenting specific sequence elements that mediate Est2 binding, such as a uracil-rich region, may be sufficient. Further analysis, such as an extension of the phylogenetic studies presented here, as well as RNA-Est2 cross-linking studies, will be required in order to directly test this proposal.
Supplementary Material
Acknowledgments
We thank April Livengood, Anita Seto, Art Zaug, Tom Cech, Dan Gottschling, and Ted Young for generously providing plasmids and strains and Tom Cech, Deborah Wuttke, and members of the Lundblad laboratory for many helpful comments during the progress of this work. We also thank Raymund Wellinger, David Zappulla, and Tom Cech for sharing results prior to publication.
This work was supported by a U.S. Army Medical Research and Materiel Command Breast Cancer Research Predoctoral Fellowship DMAD17-00-0134 to A.S.C. and National Institutes of Health grant AG11728 grant to V.L.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22:3474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antal, M., E. Boros, F. Solymosy, and T. Kiss. 2002. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 30:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachand, F., and C. Autexier. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol. Cell. Biol. 21:1888-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 5.Cawthon, R. M., K. R. Smith, E. O'Brien, A. Sivatchenko, and R. A. Kerber. 2003. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361:393-395. [DOI] [PubMed] [Google Scholar]
- 6.Cech, T. R. 2004. Beginning to understand the end of the chromosome. Cell 116:273-279. [DOI] [PubMed] [Google Scholar]
- 7.Chappell, A. S. Molecular and phylogenetic analysis of the Saccharomyces cerevisiae EST3 and TLC1 telomerase subunits. Ph.D. dissertation. Baylor College of Medicine, Houston, Tex.
- 8.Chen, J. L., M. A. Blasco, and C. W. Greider. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100:503-514. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. L., K. K. Opperman, and C. W. Greider. 2002. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 30:592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Cohn, M., M. J. McEachern, and E. H. Blackburn. 1998. Telomeric sequence diversity within the genus Saccharomyces. Curr. Genet. 33:83-91. [DOI] [PubMed] [Google Scholar]
- 12.Comolli, L. R., I. Smirnov, L. Xu, E. H. Blackburn, and T. L. James. 2002. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. USA 99:16998-17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong, Y. S., W. E. Wright, and J. W. Shay. 2002. Hum. telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66:407-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Draper, D. E. 1999. Themes in RNA-protein recognition. J. Mol. Biol. 293:255-270. [DOI] [PubMed] [Google Scholar]
- 15.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 16.Evans, S. K., and V. Lundblad. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162:1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira, M. G., K. M. Miller, and J. P. Cooper. 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 13:7-18. [DOI] [PubMed] [Google Scholar]
- 19.Hermann, T., and E. Westhof. 1999. Non-Watson-Crick base pairs in RNA-protein recognition. Chem. Biol. 6:R335-343. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, T. R., S. K. Evans, R. G. Weilbaecher, and V. Lundblad. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10:809-812. [DOI] [PubMed] [Google Scholar]
- 22.Jacob, N. K., K. E. Kirk, and C. M. Price. 2003. Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell 11:1021-1032. [DOI] [PubMed] [Google Scholar]
- 23.Jia, X., T. Weinert, and D. Lydall. 2004. Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 166:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanoh, J., and F. Ishikawa. 2003. Composition and conservation of the telomeric complex. Cell. Mol. Life Sci. 60:2295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 26.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 28.Lai, C. K., J. R. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, S. Y., and S. J. Elledge. 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113:881-889. [DOI] [PubMed] [Google Scholar]
- 31.Lingner, J., J. P. Cooper, and T. R. Cech. 1995. Telomerase and DNA end replication: no longer a lagging strand problem? Science 269:1533-1534. [DOI] [PubMed] [Google Scholar]
- 32.Lingner, J., L. L. Hendrick, and T. R. Cech. 1994. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 8:1984-1998. [DOI] [PubMed] [Google Scholar]
- 32a.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 33.Livengood, A. J., A. J. Zaug, and T. R. Cech. 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 22:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loayza, D., and T. de Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 424:1013-1018. [DOI] [PubMed] [Google Scholar]
- 35.Lundblad, V. 2003. Telomere replication: an Est fest. Curr. Biol. 13:R439-R441. [DOI] [PubMed] [Google Scholar]
- 36.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 37.Lustig, A. J. 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4:916-923. [DOI] [PubMed] [Google Scholar]
- 38.Marcand, S., V. Brevet, and E. Gilson. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 40.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Rivera, L., and M. A. Blasco. 2001. Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J. Biol. Chem. 276:5856-5865. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 43.Moriarty, T. J., D. T. Marie-Egyptienne, and C. Autexier. 2004. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 24:3720-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 45.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger, C. O. Johnson, M. Tzoneva, and D. E. Gottschling. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27:64-67. [DOI] [PubMed] [Google Scholar]
- 46.Romero, D. P., and E. H. Blackburn. 1991. A conserved secondary structure for telomerase RNA. Cell 67:343-353. [DOI] [PubMed] [Google Scholar]
- 47.Roy, J., T. B. Fulton, and E. H. Blackburn. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 12:3286-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seto, A. G., A. J. Livengood, Y. Tzfati, E. H. Blackburn, and T. R. Cech. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 16:2800-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seto, A. G., K. Umansky, Y. Tzfati, A. J. Zaug, E. H. Blackburn, and T. R. Cech. 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9:1323-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stellwagen, A. E., Z. W. Haimberger, J. R. Veatch, and D. E. Gottschling. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17:2384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teixeira, M. T., M. Arneric, P. Sperisen, and J. Lingner. 2004. Telomere length homeostasis is achieved via a switch between telomerase extendible and non-extendible states. Cell 117:323-335. [DOI] [PubMed] [Google Scholar]
- 52.ten Dam, E., A. van Belkum, and K. Pleij. 1991. A conserved pseudoknot in telomerase RNA. Nucleic Acids Res. 19:6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tesmer, V. M., L. P. Ford, S. E. Holt, B. C. Frank, X. Yi, D. L. Aisner, M. Ouellette, J. W. Shay, and W. E. Wright. 1999. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol. 19:6207-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theimer, C. A., L. D. Finger, L. Trantirek, and J. Feigon. 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. USA 100:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288:863-867. [DOI] [PubMed] [Google Scholar]
- 56.Tzfati, Y., Z. Knight, J. Roy, and E. H. Blackburn. 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 17:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 58.Vulliamy, T., A. Marrone, I. Dokal, and P. J. Mason. 2002. Association between aplastic anaemia and mutations in telomerase DNA. Lancet 359:2168-2170. [DOI] [PubMed] [Google Scholar]
- 59.Vulliamy, T., A. Marrone, F. Goldman, A. Dearlove, M. Bessler, P. J. Mason, and I. Dokal. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413:432-435. [DOI] [PubMed] [Google Scholar]
- 60.Yu, G. L., J. D. Bradley, L. D. Attardi, and E. H. Blackburn. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344:126-132. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, J., K. Hidaka, and B. Futcher. 2000. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 20:1947-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12:1489-1498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.