Abstract
trans splicing of a spliced-leader RNA (SL RNA) to the 5′ ends of mRNAs has been shown to have a limited and sporadic distribution among eukaryotes. Within metazoans, only nematodes are known to process polycistronic pre-mRNAs, produced from operon units of transcription, into mature monocistronic mRNAs via an SL RNA trans-splicing mechanism. Here we demonstrate that a chordate with a highly compact genome, Oikopleura dioica, now joins Caenorhabditis elegans in coupling trans splicing with processing of polycistronic transcipts. We identified a single SL RNA which associates with Sm proteins and has a trimethyl guanosine cap structure reminiscent of spliceosomal snRNPs. The same SL RNA, estimated to be trans-spliced to at least 25% of O. dioica mRNAs, is used for the processing of both isolated or first cistrons and downstream cistrons in a polycistronic precursor. Remarkably, intercistronic regions in O. dioica are far more reduced than those in either nematodes or kinetoplastids, implying minimal cis-regulatory elements for coupling of 3′-end formation and trans splicing.
Spliced-leader RNA (SL RNA) trans splicing is a modified splicing reaction in which the leader portion of the SL RNA is covalently linked to the 5′ ends of mRNAs. SL RNA trans splicing has been reported in euglenoids, kinetoplastids, cnidarians, nematodes, and Platyhelminthes (1, 8, 16, 31, 36). Discovery of trans splicing in the urochordate Ciona intestinalis (39) extended the phylogenetic range of trans splicing to deuterostomes, but an important unresolved question is whether trans splicing is an ancestral or a derived trait. At present, either hypothesis would invoke multiple gain or loss events (36).
SL RNAs consist of a 5′ exon and a 3′ intron with a conserved consensus 5′ splice donor site at the exon-intron boundary. They are small (<150 nucleotides [nt]), but there is little conservation in length or nucleotide sequence across phyla. The exon-intron boundary forms part of a conserved stem-loop secondary structure (17). During trans splicing, the capped SL RNA exon moiety is covalently linked to the 5′ ends of mRNAs, forming a leader sequence ranging from 16 nt in C. intestinalis to 41 nt in trypanosomatids. SL RNAs of cnidarians and nematodes have a trimethyl guanosine (TMG) cap (1, 36), though in kinetoplastids, there is an m7G cap with additional modifications of the three downstream residues (cap 4) (2). SL RNA introns contain determinants for the trans-splicing reaction, including a putative Sm binding motif, and association with the Sm complex is essential for trans splicing in kinetoplastids and nematodes (8, 10, 15, 27, 37). These features are shared with snRNPs: snRNAs are TMG capped, and the Sm protein complex was originally characterized in spliceosomal snRNPs, where it is required for cis splicing (40). Parallels between SL RNPs and snRNPs are reinforced by the similar mechanics of cis and trans splicing. In cis splicing, intron boundaries are usually defined by a 5′ donor site and a 3′ acceptor site, often containing a polypyrimide tract in addition to the 3′ AG dinucleotide acceptor site (28). In trans splicing, analogue elements are shared between the SL RNA containing the 5′ donor site and the acceptor mRNA containing the 3′ acceptor site. The latter is defined by a polypyrimidine tract and the AG dinucleotide in kinetoplastids, cnidarians, and C. intestinalis (21, 30, 36, 39), whereas in nematodes, the trans-splice acceptor is contracted to UUUCAG, there is no polypyrimidine tract, and integrity of the 3′ trans-acceptor splice site is required for accurate trans splicing (6, 33). Finally, both trans and cis splicing depend on additional exonic enhancer sequences and on SR proteins (26, 33, 34).
To date, the trematode Schistosoma mansoni and the nematodes Caenorhabditis elegans, Caenorhabditis briggsae, Oscheius (formerly Dolichorabditis) dolichura, and Pristionchus pacificus are the only metazoans known to transcribe a portion of their genes as polycistronic pre-mRNAs (1, 9, 14, 25). Other nematodes that trans-splice have not been shown to have operons (8, 29). These polycistronic pre-mRNAs are processed to mature monocistronic mRNAs via SL RNA trans splicing and intron cis splicing. In C. elegans, two SL RNAs are involved: SL1 RNA trans-splices the first cistron, and SL2 RNA trans-splices all downstream cistrons. trans-spliced mRNAs are TMG capped and polyadenylated, and bona fide AAUAAA polyadenylation consensus signals occur within 30 nt upstream of the polyadenylation cleavage site (1). In SL2 intercistronic regions, a U-rich element is present, and both the U-rich element and the polyadenylation signal are required for correct polyadenylation and SL-2 trans splicing of the upstream and downstream cistrons, respectively (20, 22). An analogous mRNA expression pathway is observed in the protist kinetoplastids. In Leishmania and Trypanosoma spp., all protein-coding genes are expressed as polycistrons, with a single type of SL RNA trans-splicing every cistron. A noticeable difference is that kinetoplastid 3′ untranslated regions (3′ UTRs) do not contain any consensus polyadenylation signal sequence (4). Instead, the polyadenylation cleavage site appears to be determined at a fixed distance upstream of the trans-splice site (24, 30).
The above data led to the view that SL RNA trans splicing occurs sporadically in distinct subphyla, where somewhat different modes of utilization have evolved (31). Among deuterostomes, SL RNA trans splicing has been reported only for one urochordate, the ascidian C. intestinalis. Ascidians, appendicularians, and thalacians form three classes in the subphylum Urochordata, the sister group to all other chordates. Recent interest in this phylogenetic group is manifested by several genome-sequencing projects at or nearing completion in different classes, including the appendicularian Oikopleura dioica. Appendicularians are the most abundant zooplankton group after copepods, with a panglobal distribution. As a chordate, O. dioica is remarkable for its small (<70 Mb), compact (1 gene per 4 kb) genome (35) as well as for its extremely short life cycle (6 days at 15°C) (38). Here we show that O. dioica not only trans-splices SL RNAs to mRNAs, as does C. intestinalis, but also uses trans splicing in resolving polycistronic transcripts. This is the first demonstration of coupling of trans splicing with polycistronic transcription in the deuterosome, and more particularly, the chordate lineage. This finding brings new insight into the evolution of both mechanisms in eukaryotes.
MATERIALS AND METHODS
Animal culture and sampling.
O. dioica is maintained year-round in culture at 15°C in our laboratory (3, 38). Unfertilized oocytes and juvenile day 4 animals were concentrated in watch glasses and rinsed with sterile-filtered seawater. Samples were transferred to Eppendorf tubes and centrifuged for 2 min at 4°C and 5,000 × g. Excess seawater was removed, a volume of RNAlater (Sigma) sufficient to immerse the specimens was added, and samples were flash-frozen in liquid nitrogen and stored at −80°C.
In silico analyses.
Resources included a nonredundant genomic shotgun data set, prepared from purified O. dioica sperm DNA, covering ∼70% of the O. dioica genome; 1,155 nonredundant expressed sequence tags (ESTs); and 7 sequenced bacterial artificial chromosome (BAC) clones. Similarity searches were carried out with BLAST, and divergent genes were predicted by using GENSCAN (www.hgmp.mrc.ac.uk). Other alignments were performed using ClustalW. RNA secondary structures were calculated by using mfold, version 3.1 (44).
Cloning and sequencing.
Genomic sequences of the O. dioica cyclin D3 gene cluster were obtained by genome walking using primers designed from cDNA and genome contig sequences. Total RNAs from oocytes and day 4 juveniles were templates for rapid amplification of 5′ cDNA ends (5′ RACE) using a GeneRacer kit (Invitrogen) to produce reverse transcription products enriched in full-length cDNAs. A day 4 cDNA library served as a template for 3′ RACE via a vector-anchored strategy using pairs of gene-specific nested primers. For all genes of the cluster, the different pairs of specific nested primers used for 5′ RACE were designed downstream of the specific nested primers used for 3′ RACE, allowing reconstruction of full-length cDNAs after sequencing of the overlapping RACE products. For RNase A/T1 protection, the SL RNA was cloned by genomic PCR using primers SLUP (ACTCATCCCATTTTTGAGTCCG) and T7SLDW (TAATACGACTCACTATAGGGTATTTGTAAGAGGCGAGAGGGATAGG), corresponding to the 5′ and 3′ ends of the gene, respectively. U5 snRNA was amplified by using primers U5UP (TAGCTCTGGTCTCTCTTCAAAACG) and U5DW (TACGACTAGGTCGGAATTGAGG). Histone H4 cDNAs have been described previously (3). The pDD probe, encompassing the 3′ end of the dynein gene and the 5′ end of the delta-tubulin gene, and probe pMC, encompassing the 3′ end of the multiple bridging factor 1 (MBF) gene and the 5′ end of the cyclin D gene, were produced by PCR on genomic DNA. U5 snRNA was identified in shotgun genome sequences, whereas the trans-spliced RPL31 5′-end cDNA was cloned by 5′ RACE.
RNA extraction and Northern blotting.
RNA was extracted by a standard guanidium thiocyanate-acid-phenol method. Total RNA was treated with RQ1-DNase (Promega) and proteinase K prior to final phenol-chloroform extraction and ethanol precipitation. A dideoxy sequencing reaction, as well as a 32P- 5′-end-labeled size marker and 5 μg of total RNAs from unfertilized oocytes and day 4 animals, was run on 6% acrylamide-8 M urea or 1% agarose-MOPS (morpholinepropanesulfonic acid)-formaldehyde gels. Products were transferred to N+ membranes (Amersham) and UV cross-linked. Oligonucleotide probes for the SL RNA (TTAGACAATCGAAATCGGACTCAAA) and 5S rRNA (CGGTCACCCATGTAAGTACTAAC) were labeled by using T4 polynucleotide kinase and [γ-32P]ATP. Membranes were hybridized in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-5× Denhardt's solution-0.5% sodium dodecyl sulfate (SDS)-100 μg of yeast tRNA (Sigma)/ml with 32P 5′-end-labeled probes at 50°C. The final stringency washing was done with 0.1× SSC at 25°C. Membranes were reprobed after boiling in 0.1% SDS.
RNase A/T1 protection.
Labeled antisense riboprobes specific for the SL RNA coding region, the 5′ end of the RPL31 gene, U5 snRNA, histone H4, pDD, and pMC were in vitro transcribed from T7, T3, or SP6 promoters after linearization, with a ribonucleotide mix containing [α-32P]UTP. Protocols for in vitro transcription and RNase protection were performed as described previously (3). Samples were run on 6% polyacrylamide-8 M urea gels. Autoradiographs were analyzed by using a phosphorimager (Fuji).
RT-PCR.
Animals were left on ice for 15 min prior to RNA extraction, in an attempt to slow down metabolic processes. Total RNAs from unfertilized oocytes and day-4 animals were subjected to a second RQ1-DNase treatment (1 U per μg of total RNA) in the presence of an RNase inhibitor. A single batch of 5 μg of total RNA from each stage was subjected to reverse transcription as follows. RNA was denatured at 90°C for 5 min and chilled on ice. Moloney murine leukemia virus (Invitrogen) reverse transcriptase, or H2O as a reverse transcriptase-negative control, was added to the reaction mixture on ice along with 100 pmol of random hexamers. The reaction mixture was incubated for 10 min at room temperature, followed by 1 h at 37°C. Tris-EDTA was added to a final volume of 50 μl, and reverse transcriptase was inactivated by incubation at 95°C for 5 min. PCRs were carried out with Dynazyme Taq (Finnzymes) under the following conditions: 94°C for 2 min; 30 cycles of 94°C for 20 s, 55°C for 45 s, and 72°C for 20 s; and a final elongation at 72°C for 3 min. Primer sequences are given item S3 in the supplemental data. For detection of reverse-transcribed mature mRNA (i.e., reaction products from primer pairs B-Ae and L-Ae), the equivalent of 100 ng of total RNA (1 μl of the reverse transcription product per 100 μl of PCR mix) was used as a template. For detection of reverse-transcribed pre-mRNA, the equivalent of 400 ng of total RNA was used as a template. Genomic DNA and a poly(A)+ phage cDNA library were used as controls. PCR products were run on agarose gels, purified, cloned, and sequenced. DNA contamination of the RNA samples was assessed by semiquantitative reverse transcription-PCR (RT-PCR) with a Roche LightCycler (unpublished).
Immunoprecipitation.
For anti-Sm immunoprecipitation, protein G-Sepharose (Amersham) was incubated for 1 h at room temperature in NET-2 (50 mM Tris [pH 7.4], 150 mM NaCl, 0.05% Nonidet P-40) with the anti-Sm antibody Y12 (Neomarkers). Unfertilized oocytes (1,000) were rinsed once in 40 mM Tris (pH 7.4)-400 mM NaCl and then crushed on ice in NET-2 supplemented with a protease inhibitor cocktail (Sigma), RNase-out (Promega), and protein G-Sepharose. After 15 min, the extract was centrifuged for 5 min at 4°C and 13,500 × g, and the supernatant was recovered. The oocyte crude extract was supplemented with a Y12-bound protein G-Sepharose preparation and incubated for 1 h at 4°C, then washed three times in NET-2. The supernatant and pellet of the last washing step were the unbound and bound fractions, respectively. A portion of the initial precleared extract (2.5% of total volume) was analyzed as the input sample. Input, bound, and unbound samples were treated with proteinase K and SDS before phenol-chloroform extraction and ethanol precipitation of RNA. Each RNA fraction was divided into four tubes and subjected to RNase protection. For anti-TMG immunoprecipitation, 15 μg of total RNA was incubated in NET-2 with an agarose-conjugated anti-TMG antibody (NA02A; Oncogene Research Products) for 1 h at 4°C. Beads were washed three times in NET-2. The supernatant and pellet from the last washing step were subjected to RNA extraction and RNase protection as described above.
Nucleotide sequence accession numbers.
BMBL accession numbers for O. dioica sequences used in this study are as follows: AJ628164 (U5 snRNA gene), AJ628165 (5S rRNA gene), AJ628166 (SL RNA gene), AJ628167 (ribosomal protein L31 [RPL31] partial cDNA), and AJ628168 (cycD3 gene cluster).
RESULTS
A subset of O. dioica mRNAs contain a 5′ leader SL RNA.
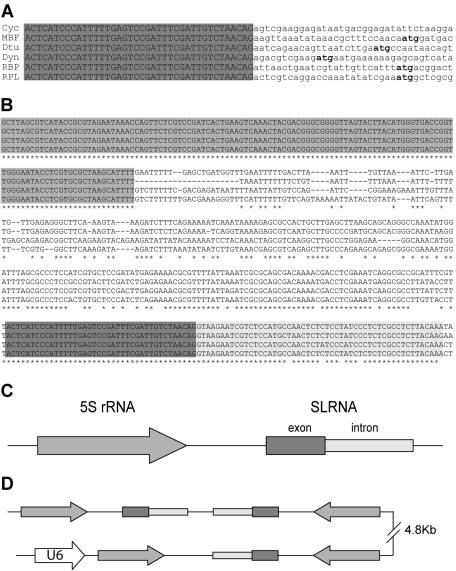
In a survey of O. dioica genes, we observed that the 5′ ends of some of these cDNAs began with an identical 40-nt leader sequence (Fig. 1A). The presence of the leader at different developmental stages was confirmed by 5′ RACE on oocytes and day 4 animals and by leader-anchored PCR on tadpole and day 2 cDNA libraries (data not shown). Alignment of the cDNAs with corresponding genomic sequences revealed that the common leader was never encoded upstream of the first exon. Instead, distinct genes were predicted in the immediate vicinity, and in each case, sequences at the beginning of the first exon contained a consensus for a 3′ acceptor splice site. This suggested the use of a trans-spliced leader RNA. Alternatively, considering the compact genome of O. dioica, a mechanism by which a promoter leader sequence would be alternatively cis spliced at a distance could not be ruled out.
FIG. 1.
O. dioica SL RNA genes. (A) Alignment of 5′ ends of cDNAs for the O. dioica cyclin D3-like homologue (Cyc), MBF, delta-tubulin (Dtu), dynein light chain (Dyn), RBP, and RPL31 (RPL). The common 40-nt leader sequence is capitalized and highlighted in grey, and the deduced methionine initiation codon is boldfaced. The deduced initiation codon for the cyclin cDNA is further downstream. (B) Alignment of SL RNA genomic loci. The 4 most divergent sequences out of 19 independent loci are shown. The 5′ region highlighted in grey is the 5S rRNA coding sequence. Dark and light grey backgrounds in the 3′ region indicate SL RNA exon and intron sequences, respectively. Asterisks indicate conserved nucleotides. (C) Schematic representation of the 5S rRNA-SL RNA locus assembled from contigs in the genomic shotgun database. (D) 5S rRNA-SL RNA head-to-tail repeats in a BAC sequence. Elements are represented as in panel C. The U6 snRNA gene is upstream of one of the 5S rRNA genes.
The SL RNA coding region is a genomic repeat element associated with the 5S rRNA.
BLASTN with the leader sequence against a nonredundant partial O. dioica genomic database revealed at least 40 occurrences in the genome. Alignments of these loci showed conservation of the element a further 52 bp downstream of the common 40-nt leader sequence identified above (Fig. 1B). Since the second part of the element began with a consensus 5′ splice donor site (CAG-GTAAGAA) and was not detected in mature cDNA leaders, it was likely to be the intron moiety of a putative SL RNA. In all cases, at an average distance of 220 bp upstream of the putative SL RNA, the 5S rRNA gene was detected in the same transcriptional orientation (Fig. 1B and C). Northern blotting confirmed expression of the 124-nt 5S rRNA (Fig. 2A). BLASTN of the 5S rRNA sequence against the O. dioica genomic database revealed that the 5S rRNA was associated with the putative SL RNA at a two-thirds frequency. The remaining 5S rRNA loci showed no association with any recognizable repeat element. The 5S-SL RNA locus was also identified in sequenced BAC clones, where arrangements of head-to-tail repeats were observed. In one case, another polymerase III transcribed gene, U6 snRNA, was also present (Fig. 1D).
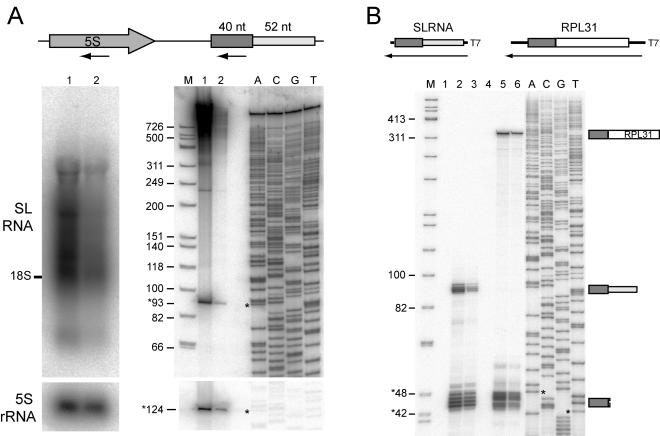
FIG. 2.
SL RNA expression. (A) Northern blot, with arrows in the schema at the top indicating regions used as probes. A 1% denaturing agarose gel (left panels) and a 6% polyacrylamide denaturing gel (right panels) were probed with oligonucleotides specific to the SL RNA exon (upper gels) and 5S rRNA (lower gels). Lanes: 1, oocyte RNA; 2, day 4 RNA; M, molecular size marker (with sizes given in thousands on the left); ACGT, dideoxy sequencing reaction. Sizes of bands indicated by asterisks are given. Intense smearing in the upper portion of lane 1 in the right panel may be partially due to nonspecific binding of polysaccharides in addition to the specific detection of trans-spliced RNAs seen in lane 2 or the left panel. (B) RNase protection, with schema at the top showing the antisense probes used for full-length SL RNA (lanes 1, 2, and 3) and the 5′ end of RPL31 RNA (lanes 4, 5, and 6). Lanes: 1 and 4, yeast tRNA control; 2 and 5, oocyte total RNA; 3 and 6, day 4 total RNA; M and ACGT, as explained for panel A. Diagrams to the right of the autoradiograph indicate protected fragments corresponding to visualized bands. Minor variation in band lengths observed for full-length SL RNA likely results from slight sequence differences in the 3′-terminal region, yielding alteration over a few base pairs in the length of the protected fragments.
The SL RNA component of an snRNP is trans spliced to the 5′ end of a subset of mRNAs.
Northern blotting and RNase A/T1 protection demonstrated expression of a single 93-nt SL RNA in oocytes and day 4 juveniles. Northern blotting with an SL RNA exon probe confirmed the expression of a single 93-nt RNA, in agreement with a 40- plus 52-nt RNA containing a 5′ cap structure. Higher-molecular-weight RNAs, probably reflecting the presence of the leader sequence at the 5′ end of mRNAs, were also detected. RNase A/T1 protection with a probe corresponding to the coding sequence of the SL RNA also protected RNA of around 93 nt and low-molecular-weight fragments (42 to 48 nt). These fragments most likely represented 5′ leaders, as supported by control experiments using probes for RPL31 cDNA, which contains the leader sequence (Fig. 2B). The slight size difference between RNase-protected 42- to 48-nt fragments and the 40-nt leader sequence can be explained by sequence identity between the beginning of the SL RNA intron sequence and the acceptor exons. We conclude that the exon moiety from the stably expressed 93-nt SL RNA was trans spliced onto the 5′ end of a subset of mRNAs.
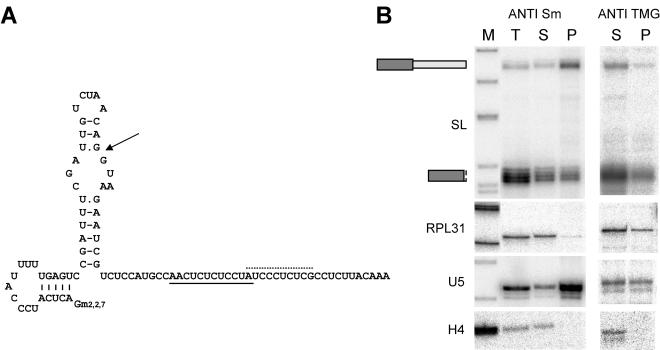
Trypanosome and nematode SL RNAs have a secondary structure of three stem-loops, with the Sm binding site between stem-loops II and III (8, 27). SL RNAs of C. elegans and cnidarians have a 2,2,7-TMG cap. In ascidians, the proposed secondary structure is simpler, with a single 5′ stem-loop and a 3′ single-stranded region containing a putative Sm binding motif (39). The O. dioica SL RNA is predicted to fold into two stem-loops, with both the exon and the 5′ trans-splice site in the 5′ portion of the molecule (Fig. 3). In all SL RNAs modeled, the conserved stem-loop I contains the part of the exon engaged in base pairing with the exon-intron junction, implying that this stem-loop is no longer present on the leader after mRNA trans splicing. In the O. dioica SL RNA, the analogous structure would be formed by the second stem-loop. Therefore, the first stem-loop would remain present on the leader sequence and represents a novel SL RNA secondary structure. The presence of this hairpin structure on a subset of mRNAs raises questions as to potential regulatory functions, especially in light of recently reported regulatory functions for the leader in mRNA translation (29, 36, 42). No 3′ stem-loop was predicted, an absence reminiscent of the secondary structure of the C. intestinalis SL RNA intron. Two possible Sm binding motifs were identified in the SL RNA intron, and anti-Sm immunoprecipitation on oocyte extracts confirmed Sm association. As expected, Sm-associated U5 snRNA was enriched in the Sm-bound fraction, whereas histone H4 mRNA was detected only in the unbound fraction. As predicted, only full-length SL RNA was enriched in the Sm-bound fraction, as opposed to the 42- to 48-nt ladder or the RPL31 mRNA, indicating that Sm association is mediated through the intron moiety. Anti-TMG cap immunoprecipitation enriched the SL RNA, trans-spliced RPL31 RNA, and TMG-capped U5 snRNA, but not H4 RNA, in the precipitate. We conclude that the SL RNA has a TMG cap, as do cognate snRNAs, and this cap is brought via the leader to trans-spliced mRNAs. Thus, the SL RNA of O. dioica has characteristics of a bona fide snRNP, like the SL RNAs of lower eukaryotes.
FIG. 3.
O. dioica SL-RNP. (A) Model of SL RNA secondary structure. Arrow points to the exon-intron boundary; dotted and solid lines, possible Sm binding sites within the intron. Gm2,2,7, TMG cap. (B) Immunoprecipitation with anti-Sm (left) and anti-TMG (right) antibodies evaluated by RNase protection with probes specific for SL RNA, RPL31 mRNA, U5 snRNA, and histone H4 mRNA. Lanes: M, molecular size marker; T, total input RNA; S, unbound RNA fraction; P, antigen-bound RNA fraction.
To identify genes affected by trans splicing, we searched for the splice leader sequence in 1,155 nonredundant ESTs from a nonnormalized, directional day-4 cDNA library (Table 1). Identities at the start of 143 sequences (12.4%) aligned with the leader. No other trans-spliced leader was detected. To estimate the proportion of ESTs that lacked the leader due to insufficient cDNA elongation, BLASTX of conserved sequences (e < 10−20) revealed a putative start codon in 158 of 375 cases (42%), with 90 sequences (24%) containing the leader. The average distance between the leader and the predicted start codon was 22 nt, but this distance was 5 nt or less for 20% of the genes, with the start codon immediately following the leader in some instances. Most leader-containing ESTs had corresponding genomic contigs in the O. dioica shotgun data set. Alignment of the ESTs and genomic contigs confirmed that the leader sequence was spliced in trans. In comparing 52 distinct trans-splice acceptor sites to 605 cis-splice acceptor sites, the same consensus sequence, TTT(C/T/A)AG, was observed at both intron and intercistronic region 3′ ends. A notable difference from cis splicing was that most exons trans spliced to the leader (133 of 145) started with an adenosine (Fig. 4), whereas the start of cis-spliced exons did not show any nucleotide preference. BLASTX of 145 trans-spliced ESTs (see item S1 in the supplemental data) detected 43 distinct ribosomal proteins, accounting for half of the trans-spliced ESTs, presumably due to high expression levels. Other sequences included housekeeping proteins such as translation initiation or elongation factors or heat shock proteins, but also a variety of translation products, including 20 showing little or no similarity with databases. Overall, this suggests that a significant proportion of genes are trans spliced by a single SL RNA.
TABLE 1.
trans-Spliced ESTs
| Feature | No. (%) |
|---|---|
| Nonredundant ESTs | 1,155 (100) |
| Containing SL RNA leader | 145 (12.6) |
| Containing another leader | 0 (0) |
| Conserved sequencesa | 375 (100) |
| Containing putative start codon | 158 (42) |
| Containing SL RNA leader | 90 (24) |
BLASTX e value, <10−20. See the text for details.
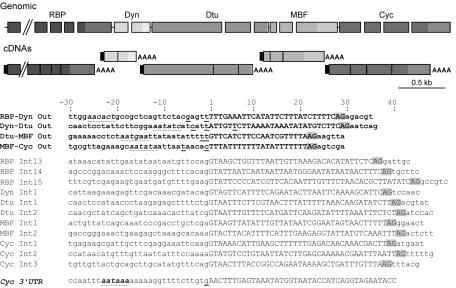
FIG. 4.
Intergenic sequences in a cluster of trans-spliced genes are very short and lack a consensus polyadenylation signal. (Top) Genomic organization of the cluster and its processed mRNAs, with gene nomenclature as explained for Fig. 1. Rectangles, gaps, and lines represent exons, introns, and intercistronic regions, respectively. Rectangles of the same greyscale represent exons of the same gene, with 3′ UTRs in a lighter shade. On mature mRNAs, black rectangles indicate the trans-spliced leader and AAAA indicates the poly(A) tail. Schema is drawn to scale except for the 4,446-bp RBP16 gene, which has been truncated (parallel diagonal lines). A putative exon is present immediately upstream of the RBP16 gene, suggesting that the compact gene cluster continues further upstream. (Bottom) Sequence alignments of intercistronic regions (boldfaced) and introns. Flanking residues 30 nt upstream and 6 nt downstream of the splice sites are included. A portion of the sequence of the 3′ UTR of the cyclin gene is also shown. Capital letters, intronic and intergenic sequences. AG (highlighted in grey), 3′ cis or trans acceptor splice site. For each gene, sequencing of several cDNAs revealed alternative poly(A) cleavage sites. The various mapped cleavage sites are underlined, with the statistically most frequent occurring at the exon-intercistronic region boundary. Position +1 corresponds to the first nucleotide of the intercistronic region or intron. The consensus polyadenylation signal of the cyclin gene is boldfaced with a dotted underline, and sequences most closely approximating a polyadenylation signal in the 3′ regions of the upstream genes are indicated with a dotted underline. Polyadenylation cleavage sites were assigned as the first C, G, or U upstream of the sequenced poly(A) tail.
Resolving polycistronic transcripts via SL RNA trans splicing.
The compact genome of O. dioica raises the question of whether trans-spliced mRNAs may also arise from operon units of transcription, as in rhabditid nematodes. Indeed, presumptive exons of other genes were closely juxtaposed upstream of some trans-spliced genes. Among candidate operons, we focused on one containing a cyclin D homologue (Fig. 4) (unpublished). No putative exon was detected within the 667 bp downstream of the cyclin D gene polyadenylation cleavage site. On the other hand, the Ran binding protein 16 (RBP), dynein light chain 1 (Dyn), delta-tubulin (Dtu), and MBF genes were detected immediately upstream. A further putative exon similar to a human protein of unknown function was detected upstream of the RBP gene. All corresponding full-length cDNAs were polyadenylated and contained the 40-nt leader sequence. Alignment of the cDNAs and genomic sequences mapped the reading frame boundaries. As for many genes in O. dioica, introns were short, about 40 nt. Surprisingly, intergenic distances were even shorter, ranging from 23 to 30 nt (see below).
To determine if mRNAs of these genes resulted from processing of a probably short-lived, polycistronic pre-mRNA, RT-PCR analyses were conducted on total RNAs from unfertilized oocytes and day 4 animals. Lack of DNA contamination was assessed through several controls. First, amplification between the histone H2A and H2B genes was detected only with a genomic DNA template (Fig. 5, bottom panel) and not on reverse-transcribed RNA preparations. Second, omission of reverse transcriptase failed to generate any RT-PCR product. Finally, semiquantitative LightCycler PCR analysis showed no amplification at all from the samples lacking reverse transcriptase after 47 cycles, whereas samples with reverse transcriptase amplified the expected product after 41 cycles (see item S3 in the supplemental data).
FIG. 5.
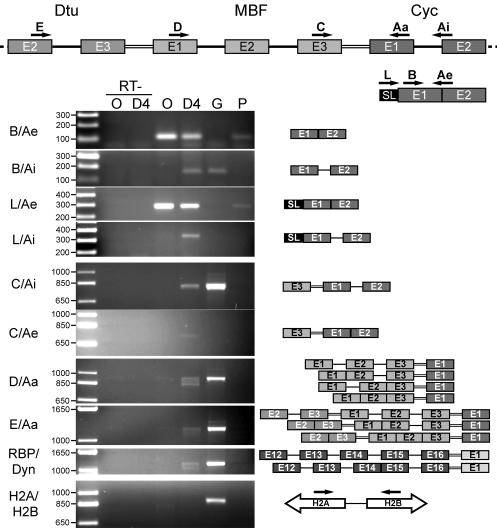
The gene cluster containing the cyclin D3-like gene is transcribed as a polycistronic transcript. Schema at the top shows the positions and orientations of primers used for RT-PCR analysis of RNA products from portions of the cyclin D3-like gene cluster. Rectangles, exons; single lines, introns; double lines, intercistronic regions. Primers were exon specific except for Ai and Ae. Ai amplifies cDNAs produced from mRNAs containing the first intron of the cyclin D gene, and Ae amplifies the spliced product between exons 1 and 2 of the same gene. Gels show PCR products resulting from use of the primer pairs given at the left. Leftmost lanes, molecular weight ladders, with sizes (in thousands) to the left. Template DNAs: O, oocyte cDNA; D4, cDNA from day 4 animals; G, genomic DNA; P, phage cDNA library from day 4 animals. RT− lanes, controls run in the absence of reverse transcriptase. The bottom two panels show RT-PCR products amplified between exon 12 of the RBP gene and exon 1 of the Dyn gene (RBP/Dyn) and the divergently transcribed histone H2A and H2B genes (H2A/H2B), respectively. Schema to the right of the gels depicts results from sequencing of RT-PCR products, showing various cis- and trans-spliced RNA intermediates. Schema for H2A and H2B shows genomic organization of the histone genes targeted for amplification.
The polycistronic pre-mRNA was characterized by using exon (Aa, B, C, D, and E)-, exon-intron (Ai)-, exon-exon boundary (Ae)-, and SL RNA exon (L)-specific primers (Fig. 5). Identification of the RT-PCR products was confirmed by cloning and sequencing of the amplified band and by Southern blotting of the gels with appropriate probes (Fig. 5) (see item S3 in the supplemental data). The cycD3 spliced form (primer pair Ae-B or Ae-L) was amplified from oocytes and day 4 cDNA, as well as from a control phage poly(A)+ cDNA library. The cycD3 pre-mRNA-specific Ai-B primer pair revealed precursor only in day 4 reverse-transcribed RNA and genomic DNA. No pre-mRNA was detected from oocyte RNA, and this was true for all primer pairs tested. Remarkably, RT-PCR with primer pair L-Ai detected a trans-spliced cycD3 pre-mRNA that had not been cis spliced. In assaying for a polycistronic pre-mRNA, primer pair C-Ai detected the cDNA copy of a bicistronic MBF-cycD3 pre-mRNA. Polycistronic pre-mRNA was also revealed by using primer D (MBF exon 1) or E (Dtu exon 2) with Aa (cycD3 exon 1), as well as using primers amplifying between the RBP and Dyn genes. Lower-molecular-weight products were also amplified. Sequencing revealed these to be alternate cis-splicing intermediates of the polycistron, eliminating the possibility that they represented genomic copies (Fig. 5). This also indicated that cis splicing could occur prior to trans splicing. Interestingly, cis-spliced but not trans-spliced maturation intermediates have also been observed in S. mansoni (8).
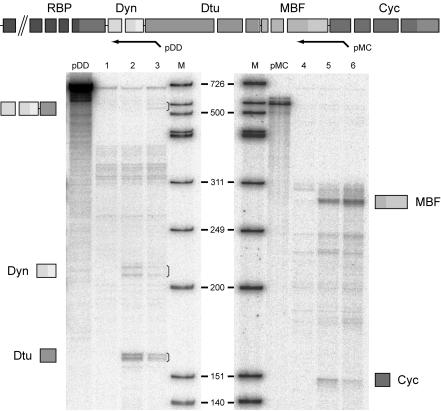
Further evidence of polycistronic pre-mRNA was obtained by RNase protection (Fig. 6). Probe pDD protected the dynein-delta-tubulin bicistronic pre-mRNA product, specific for day 4, in addition to the individual mature mRNA exons detected at the oocyte and day 4 stages. Taken together, these results show that the cycD3-like homologue is transcribed on the same pre-mRNA as (at least) the four genes immediately upstream and that this polycistronic pre-mRNA is processed into monocistronic mRNAs. At present, we lack the capability to confirm the processing of polycistronic transcripts by an independent transformation approach in O. dioica. However, the very small size of the intercistronic regions (<30 nt) argues against individual promoters and makes alternative explanations for the tight gene clustering and the existence of polycistronic pre-mRNAs improbable. This is, to our knowledge, the first demonstration of such an mRNA expression mechanism in the chordate lineage.
FIG. 6.
Expression of genes in the cyclin D3-like gene cluster. A schematic representation of probes used for the RNase protection assay is shown at the top. pDD encompasses exon 1 (partially), intron 1, and exon 2 of the dynein gene, the intergenic region between the dynein and delta-tubulin genes, and part of exon 1 of the delta-tubulin gene. pMC encompasses part of exon 3 of the MBF gene, the intergenic region, and part of exon 1 of the cyclin gene. Drawings beside the autoradiographs represent identities of protected fragments corresponding to visualized bands (indicated by brackets). Lanes: 1 and 4, yeast tRNA control; 2 and 5, oocyte total RNA; 3 and 6, day 4 total RNA; pDD and pMC, 1/1,000 dilution of the probe alone; M, molecular size marker.
Absence of bona fide polyadenylation signals in unusually short intercistronic regions.
One surprising feature of O. dioica intercistronic regions is their size, as short as 23 nt (Fig. 4), compared to the 100- to 400-nt intercistronic regions characteristic of trypanosomes and nematodes. In the four cases mapped here, the longest intercistronic region was smaller than the shortest intron. Though similar at their 3′ ends, introns and intercistronic regions differ at their 5′ ends, where intercistronic regions lack a consensus 5′ donor splice site. Several cDNAs (n > 5) were sequenced for each gene for 3′-end determination. Polyadenylation cleavage sites were imprecise for the RBP, Dyn, Dtu, and MBF mRNAs, with two to three alternate poly(A) starts, in contrast to the unique site for the cycD mRNA. When the polyadenylation cleavage site occurs after an adenosine residue, 3′ RACE experiments cannot differentiate whether initial A residues in the sequenced poly(A) tail were encoded in the transcribed mRNA or added during polyadenylation. With this proviso, all but one cleavage site was followed by the consensus triplet NUU, including the cycD gene, which appears to be the last cistron in the gene cluster. Inspection of the sequence upstream of the polyadenylation cleavage site revealed that only the cycD gene had the consensus AAUAAA signal, 17 nt upstream of the poly(A) tail. No convincing consensus polyadenylation signal, with respect to sequence or position, was detected (Fig. 4) (see item S2 in the supplemental data). Therefore, though only one gene cluster has been characterized here, posttranscriptional polycistronic 3′-end formation in O. dioica appears to employ cis-regulatory elements divergent from those of nematodes and kinetoplastids.
Posttranscriptional regulation of polycistronic pre-mRNAs.
In higher vertebrates, cyclin D genes are regulated transcriptionally both within the cell cycle and through development (7). In O. dioica, cyclin D3-like mRNA levels are regulated at least at the developmental level (P. Ganot et al. unpublished data), though information within individual cell cycles is lacking. To assess the possibility that the Dyn, Dtu, MBF, and Cyc genes are functionally clustered to permit coordinately regulated gene expression, we monitored mRNA levels in oocytes and day 4 juveniles (Fig. 6). Though genes arranged in the cyclin D cluster have common transcription, individual steady-state mRNA levels were not strictly coordinated. Evaluation of expression levels of the four cyclin D cluster genes revealed that the cyclin, dynein, and delta-tubulin mRNAs were expressed at low levels, which decreased from the oocyte stage to day 4. This was in direct contrast to MBF mRNA expression levels, which were higher and increased between these two developmental stages.
trans splicing and its relation to gene spacing.
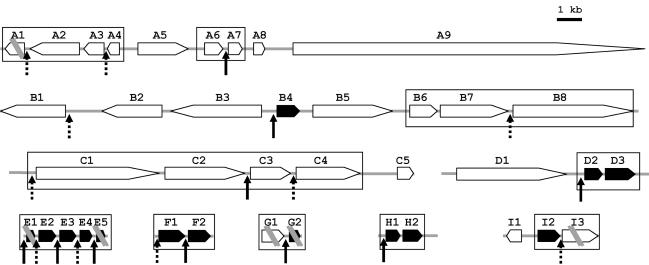
To determine whether trans splicing specifically or preferentially affected genes transcribed as polycistronic messages, we examined gene proximity around genes that matched sequenced ESTs. We chose ESTs of conserved genes (e < 10−20) and annotated the shotgun contigs with which they aligned. Genomic environments upstream of start codons and downstream of stop codons were inspected for neighboring genes. In most cases, adjacent genes were in the same orientation. Between genes whose mRNAs were subject to trans splicing and their immediate upstream and downstream neighbors, intergenic distances were shorter than average. Genome fragments containing genes that are trans spliced, or potentially trans spliced, revealed variability in the environment of such genes but included high gene density regions suggestive of additional cases of trans-spliced polycistronic messages (see, e.g., E1 to E5, a compact cluster of ribosomal protein genes, in Fig. 7). One notable feature was that the first gene in a given orientation (e.g., B4, encoding ribosomal protein S2) contained the same leader sequence as the interior trans-spliced genes in the cyclin D3-like cluster. Thus, unlike the specialized SL1 and SL2 RNAs of C. elegans, O. dioica uses the same SL RNA for the first and downstream cistrons in a gene cluster. This agrees with the EST analysis (Table 1), indicating that this is the major SL RNA in O. dioica, if not the only one. Overall, the data are compatible with a preferential but not systematic association of trans splicing with candidate operon-like organizations.
FIG. 7.
Putative operons in the O. dioica genome. Shown is the genomic organization of nine regions containing genes that are trans spliced (acceptor site [arrows]) or possibly trans spliced (candidate acceptor site localized next to the start codon [dotted arrows]). Solid arrows, ribosomal protein genes. Candidate operons (distance between translated regions, <300 bp) are boxed. Double parallel diagonal bars indicate regions of discontinuity in sequence information. A1, phospholipid-transporting ATPase VA; A2, cisplatin resistance-related protein CRR9P; A3, LMP7-like protein; A4, CG9166 protein; A5, speckle-type POZ protein; A6, prefoldin subunit 2; A7, dUTP nucleotidohydrolase; A8, prediction; A9, basement membrane-specific heparan sulfate proteoglycan core protein precursor; B1, myeloblast KIAA0230; B2, prediction; B3, CG14213 protein; B4, ribosomal protein S2; B5, prediction; B6, prediction; B7, adenosine deaminase; B8, autoantigen; C1, mediator subunit SUR2; C2, Trp4-associated protein TAP1; C3, retinoblastoma binding proteins 4 and 7; C4, protein phosphatase 2, regulatory subunit A (PR 65); C5, similar to SMAD; D1, nucleoporin 155; D2, ribosomal protein L17; D3, ribosomal protein L8; E1, ribosomal protein S20; E2, ribosomal protein L26; E3, ribosomal protein L11; E4, ribosomal protein S13; E5, ribosomal protein S5; F1, ribosomal protein S16; F2, ribosomal protein L6; G1, related to guanine nucleotide-binding protein; G2, ribosomal protein L10; H1, ribosomal protein L23; H2, ubiquitin A 52-residue ribosomal protein fusion product 1 (UBA52) (includes ribosomal protein L40); I1, related to thioredoxin; I2, ribosomal protein L24; I3, DNA replication licensing factor mcm2.
DISCUSSION
The central finding here is that the chordate O. dioica uses a single or at least a predominant SL RNA species to trans-splice a significant fraction of mRNAs, including those expressed as polycistronic pre-mRNAs. The novelty of trans splicing of polycistronic pre-mRNAs in the chordate lineage is accompanied by the observation that O. dioica carries out this process in a context where cis-regulatory elements in the intercistronic region are far more compact or reduced than those in either nematodes or kinetoplastids. Like other organisms for which investigation of trans splicing has been carried out in sufficient detail, the O. dioica SL RNA has a TMG cap and associates with Sm proteins through its intron moiety. Extension of Sm association and the TMG cap to chordate SL RNAs emphasizes the conserved nature of the SL-RNP and its participation in the splicing reaction, as both characteristics are features of the spliceosomal snRNPs.
The SL RNA loci.
SL RNA genes are usually multiple copy and can be clustered (like the SL1 RNA genes of C. elegans [15]) or dispersed. One recurring evolutionary trait of SL RNA genes is their tandem arrangement and association with the 5S rRNA gene, though they are also found outside the 5S rRNA cluster (12, 13, 36). However, the observation that linkage of the 5S rDNA and SL RNA genes is maintained in urochordates, though these linked genes are dispersed throughout the genome, suggests some selective pressure for this arrangement. In kinetoplastids and nematodes, promoter elements have been characterized both upstream and within the coding sequence of the SL RNA. The intergenic stretch between the 5S rRNA and the putative SL RNA had more limited sequence conservation, and Northern blot analysis failed to detect any RNA product from this region (data not shown), consistent with a possible function as a promoter.
trans splicing of polycistrons in O. dioica.
Often, where trans splicing has been identified, a single SL RNA is involved. In kinetoplastids, the same SL RNA is responsible for trans splicing of the first and downstream cistrons of the operon. Thus far, only in cnidarians and in some nematodes have two distinct SL RNA subtypes been shown to coexist (1, 36). Although we cannot rule out the existence of other, minor SL RNA types in O. dioica, our EST analysis indicates that the SL RNA characterized here is the principal one, since no other leader is detected. Also, though the same leader sequence could in theory be present on more than one type of SL RNA, genome database mining with the SL RNA sequence failed to detect more variation within the different gene copies than appears in the alignment presented in Fig. 1. Importantly, the same SL RNA is used in the processing of both isolated or first cistrons and downstream cistrons in a polycistronic message.
Polycistronic processing into monocistrons involves both trans splicing of the downstream cistron and the cleavage and polyadenylation of the upstream cistron, processes that depend on specific recognition of intergenic cis elements. The cyclin D3-like gene cluster in O. dioica revealed several unexpected features regarding the requisite cis elements. Intercistronic regions were very short, 23 to 30 nt, in marked contrast to the polycistrons of other eukaryotes, suggesting that O. dioica contains minimal cis elements for intercistronic region definition. The 3′ acceptor site has a strong UUU(C/U/A)AG consensus instead of a polypyrimidine tract distinctly separated from the AG dinucleotide. The same contracted 3′ splice site occurs in C. elegans intercistronic regions, in which alteration of the UUUCAG consensus leads to abnormal trans splicing (6). Polypyrimidine tracts or U-rich elements, present in kinetoplastid or C. elegans intercistronic regions, respectively, were not observed in O. dioica intercistronic regions, and no lariat branch point consensus was detected. The absence of the branch point consensus is also observed in trypanosomatids and nematodes (6, 18, 21, 33, 43). Thus, it seems that the trans-splice acceptor site is defined only by a strong 3′ splice site. Such a simple sequence likely occurs at a number of positions along pre-mRNAs, suggesting a requirement for additional cis elements beyond intercistronic region boundaries to enable specific recognition. Indeed, 5′ exonic sequences downstream of the splice site are required for correct trans splicing in nematodes and kinetoplastids (26, 33). In our survey, a noticeable signature was that trans-spliced exons usually began with an A residue, though other elements are likely present.
The link between trans splicing and 3′-end formation.
In nematodes and kinetoplastids, trans splicing and 3′-end formation within polycistronic pre-mRNAs appear to be coupled, and polyadenylation is partly defined by the SL RNP trans-splicing machinery. General cis requirements for 3′-end formation of eukaryotic mRNAs, the consensus AAUAAA polyadenylation signal and the GU-rich polyadenylation regulatory element, are recognized by the cleavage and polyadenylation specificity factor (CPSF) and the cleavage and stimulation factor (CstF), respectively. Binding and interaction of these complexes triggers synthesis of the poly(A) tail (5). Nuclear magnetic resonance studies have revealed that human CstF-64 recognizes with strong affinity a UU dinucleotide in the GU-rich element (32). In nematodes, 3′-end formation of individual mRNAs from polycistronic pre-mRNAs requires the AAUAAA consensus. Mutation of the AAUAAA sequence in a bicistron abolishes 3′-end formation of the upstream gene and reduces expression of the downstream gene. A further effect of such mutations is that SL2 trans splicing is partially replaced by SL1 trans splicing (20, 22). There is a minor class of naturally occurring SL1 trans-spliced operons in C. elegans in which the intercistronic region is essentially nonexistent, with the AAUAAA located only a few base pairs upstream of the trans-splice site. In these operons, however, production of upstream and downstream mature mRNAs appears to be mutually exclusive (41). Also, CstF-64 and the SL2 snRNP, but not the SL-1 snRNP, interact in vivo (13). One bicistron has been characterized in the trematode, where only the downstream cistron is trans spliced. In this particular case, the intercistronic region is very small (54 nt), with a clear AAUAAA consensus and a short polypyrimidine tract upstream of the poly(A) site and the AG 3′ acceptor, respectively (9). In kinetoplastids, where all mRNAs seem to be produced from polycistronic pre-mRNAs, the trans-splice acceptor site is selected as the first AG downstream of the conserved polypyrimidine tract (4, 21). No polyadenylation signal has been reported, and 3′-end cleavage is imprecise over a few nucleotides (24, 26). Destruction of the 3′ UTR of a given gene does not alter the 3′-end cleavage site, which occurs at a fixed distance upstream of the trans-splice signal (24, 30). Conversely, mutation of the trans-splice acceptor site (the first AG downstream of the conserved polypyrimidine tract) causes deficient 3′-end formation in addition to deficient SL RNA trans splicing (21, 30). Interestingly, when the trypanosome CSPF-30 factor is depleted, 3′-end formation is impaired, as is trans splicing (19).
Inside the O. dioica cyclin D gene cluster, the presence of polyadenylation signals within intercistronic regions appears unlikely, unless their sequence and positioning are highly divergent. Consistent with this, 3′ cleavage sites in this polycistron are inaccurate but tend to follow the rule of preceding a NUU triplet, the only potential motif for CstF-64 homologue recognition. Of note, the AAUAAA consensus is present, and probably required, for many other genes we have mapped in O. dioica (data not shown). The lack of obvious cis polyadenylation signals in the interior cistrons of the O. dioica cyclin D gene cluster argues against a scenario of 3′-end recognition by the polyadenylation machinery alone; rather, it suggests that 3′-end formation is partly defined by the SL RNP trans-splicing machinery. This similarity to mechanisms in C. elegans or kinetoplastids is only partial, given the very few conserved elements within the small intercistronic regions of O. dioica.
Coevolution of trans splicing and polycistronic transcription in metazoans?
The revelation of trans splicing in a growing number of organisms across eukaryotic phyla supports an ancestral origin for this mechanism. Our results strengthen the view that its features have been strongly adapted during evolution, since divergent pre-mRNA cis element requirements and SL RNA sequences are employed in distinct taxa, including taxa within the same phylum. Some components may be conserved while others, such as SL RNA genes, are recruited independently in distinct lineages, when recourse to trans splicing has been strongly augmented. Two SL RNP specific proteins, of 30 and 175 kDa, have been characterized in the nematode Ascaris lumbricoides (11), but their degree of conservation with homologues in C. elegans is limited. A search for similar proteins in the O. dioica genomic database failed to detect any translation product with significant similarity (best e value, 0.15). Definition of the functional protein components of the SL RNP in O. dioica and their evolutionary conservation will be important in solving this puzzle.
Our finding of polycistronic transcription in O. dioica complements information on this process in another family of metazoans, the rhabditid nematodes, where it is a recurrent theme. Increasing instances of eukaryotic polycistronic transcripts have been described, including those of Drosophila melanogaster and vertebrates (23). As for trans splicing, it is unclear whether polycistronic transcription is ancestral or has appeared several times during evolution. trans splicing may have allowed, in part, the processing of polycistronic transcripts. Alternatively, operon transcription may have adopted different processing pathways, including trans splicing. In either scenario, it is intriguing to consider the possible coevolution of the utilization of trans splicing in a given phylum and transcription of polycistronic precursors in a taxon within this phylum. One force that may have an influence on either mechanism, or their coevolution, could be genome compaction. Significant changes in genome size through expansion and compaction have occurred numerous times during evolution, with occasional extremes. High genome compaction, such as that observed in C. elegans and O. dioica, brings genes close to each other, permits cotranscription, and may necessitate the dissociation of polycistronic RNAs into single units. In this respect, O. dioica has a miniature genome, less than half the size of that of the urochordate C. intestinalis, and the rhabditid C. elegans has a fourfold-more-compact genome than the ascarid A. lumbricoides (www.genomesize.com). In both the urochordate and nematode examples, the species with the more compact genome trans-splices both individual and polycistronic messages, whereas that with the larger genome is only known to trans-splice monocistronic messages. Under such a driving force, trans splicing and polycistronic transcription could have reappeared or been strongly reamplified from background levels in a few lineages. On the other hand, genome expansion could dissociate operons, with trans splicing possibly preserved at variable levels to play a role in the regulation of posttranscriptional events.
Supplementary Material
Acknowledgments
We thank members of the Sars Oikopleura culture facility for a continuous supply of animals.
This work was supported by grants from the Norwegian Research Council and the Ministry of Education and by NFR Biotechnology grant 146653/431.
Footnotes
Supplemental data for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Blumenthal, T. 1995. trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, D. A., N. R. Sturm, and M. C. Yu. 2000. Transcription of the kinetoplastid spliced leader RNA gene. Parasitol. Today 16:78-82. [DOI] [PubMed] [Google Scholar]
- 3.Chioda, M., R. Eskeland, and E. M. Thompson. 2002. Histone gene complement, variant expression, and mRNA processing in a urochordate Oikopleura dioica that undergoes extensive polyploidization. Mol. Biol. Evol. 19:2247-2260. [DOI] [PubMed] [Google Scholar]
- 4.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 6.Conrad, R., R. F. Liou, and T. Blumenthal. 1993. Functional analysis of a C. elegans trans-splice acceptor. Nucleic Acids Res. 21:913-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coqueret, O. 2002. Linking cyclins to transcriptional control. Gene 299:35-55. [DOI] [PubMed] [Google Scholar]
- 8.Davis, R. E. 1997. Surprising diversity and distribution of spliced leader RNAs in flatworms. Mol. Biochem. Parasitol. 87:29-48. [DOI] [PubMed] [Google Scholar]
- 9.Davis, R. E., and S. Hodgson. 1997. Gene linkage and steady state RNAs suggest trans-splicing may be associated with a polycistronic transcript in Schistosoma mansoni. Mol. Biochem. Parasitol. 89:25-39. [DOI] [PubMed] [Google Scholar]
- 10.Denker, J. A., P. A. Maroney, Y. T. Yu, R. A. Kanost, and T. W. Nilsen. 1996. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA 2:746-755. [PMC free article] [PubMed] [Google Scholar]
- 11.Denker, J. A., D. M. Zuckerman, P. A. Maroney, and T. W. Nilsen. 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417:667-670. [DOI] [PubMed] [Google Scholar]
- 12.Drouin, G., and M. M. de Sa. 1995. The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol. Biol. Evol. 12:481-493. [DOI] [PubMed] [Google Scholar]
- 13.Evans, D., I. Perez, M. MacMorris, D. Leake, C. J. Wilusz, and T. Blumenthal. 2001. A complex containing CstF-64 and the SL2 snRNP connects mRNA 3′ end formation and trans-splicing in C. elegans operons. Genes Dev. 15:2562-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, D., D. Zorio, M. MacMorris, C. E. Winter, K. Lea, and T. Blumenthal. 1997. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc. Natl. Acad. Sci. USA 94:9751-9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson, K. C., P. J. Heid, and J. H. Rothman. 1996. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev. 10:1543-1556. [DOI] [PubMed] [Google Scholar]
- 16.Frantz, C., C. Ebel, F. Paulus, and P. Imbault. 2000. Characterization of trans-splicing in Euglenoids. Curr. Genet. 37:349-355. [DOI] [PubMed] [Google Scholar]
- 17.Greenbaum, N. L., I. Radhakrishnan, D. J. Patel, and D. Hirsh. 1996. Solution structure of the donor site of a trans-splicing RNA. Structure 4:725-733. [DOI] [PubMed] [Google Scholar]
- 18.Hannon, G. J., P. A. Maroney, J. A. Denker, and T. W. Nilsen. 1990. trans-splicing of nematode pre-messenger RNA in vitro. Cell 61:1247-1255. [DOI] [PubMed] [Google Scholar]
- 19.Hendriks, E. F., A. Abdul-Razak, and K. R. Matthews. 2003. tbCPSF30 depletion by RNA interference disrupts polycistronic RNA processing in Trypanosoma brucei. J. Biol. Chem. 278:26870-26878. [DOI] [PubMed] [Google Scholar]
- 20.Huang, T., S. Kuersten, A. M. Deshpande, J. Spieth, M. MacMorris, and T. Blumenthal. 2001. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol. Cell. Biol. 21:1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummel, H. S., R. D. Gillespie, and J. Swindle. 2000. Mutational analysis of 3′ splice site selection during trans-splicing. J. Biol. Chem. 275:35522-35531. [DOI] [PubMed] [Google Scholar]
- 22.Kuersten, S., K. Lea, M. MacMorris, J. Spieth, and T. Blumenthal. 1997. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA 3:269-278. [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence, J. G. 2002. Shared strategies in gene organization among prokaryotes and eukaryotes. Cell 110:407-413. [DOI] [PubMed] [Google Scholar]
- 24.LeBowitz, J. H., H. Q. Smith, L. Rusche, and S. M. Beverley. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7:996-1007. [DOI] [PubMed] [Google Scholar]
- 25.Lee, K. Z., and R. J. Sommer. 2003. Operon structure and trans-splicing in the nematode Pristionchus pacificus. Mol. Biol. Evol. 20:2097-2103. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Estrano, C., C. Tschudi, and E. Ullu. 1998. Exonic sequences in the 5′ untranslated region of α-tubulin mRNA modulate trans-splicing in Trypanosoma brucei. Mol. Cell. Biol. 18:4620-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelboim, M., C. L. Estrano, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 277:35210-35218. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 29.Maroney, P. A., J. A. Denker, E. Darzynkiewicz, R. Laneve, and T. W. Nilsen. 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA 1:714-723. [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews, K. R., C. Tschudi, and E. Ullu. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8:491-501. [DOI] [PubMed] [Google Scholar]
- 31.Nilsen, T. W. 2001. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends Genet. 17:678-680. [DOI] [PubMed] [Google Scholar]
- 32.Perez Canadillas, J. M., and G. Varani. 2003. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 22:2821-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romfo, C. M., P. A. Maroney, S. Wu, and T. W. Nilsen. 2001. 3′ splice site recognition in nematode trans-splicing involves enhancer-dependent recruitment of U2 snRNP. RNA 7:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanford, J. R., and J. P. Bruzik. 1999. SR proteins are required for nematode trans-splicing in vitro. RNA 5:918-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo, H. C., M. Kube, R. B. Edvardsen, M. F. Jensen, A. Beck, E. Spriet, G. Gorsky, E. M. Thompson, H. Lehrach, R. Reinhardt, and D. Chourrout. 2001. Miniature genome in the marine chordate Oikopleura dioica. Science 294:2506. [DOI] [PubMed] [Google Scholar]
- 36.Stover, N. A., and R. E. Steele. 2001. trans-spliced leader addition to mRNAs in a cnidarian. Proc. Natl. Acad. Sci. USA 98:5693-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troedsson, C., J.-M. Bouquet, D. L. Aksnes, and E. M. Thompson. 2002. Resource allocation between somatic growth and reproductive output in the pelagic chordate Oikopleura dioica allows opportunistic response to nutritional variation. Mar. Ecol. Prog. Ser. 243:83-91. [Google Scholar]
- 39.Vandenberghe, A. E., T. H. Meedel, and K. E. Hastings. 2001. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 15:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 41.Williams, C., L. Xu, and T. Blumenthal. 1999. SL1 trans-splicing and 3′-end formation in a novel class of Caenorhabditis elegans operon. Mol. Cell. Biol. 19:376-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J. Biol. Chem. 278:38269-38275. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, H., and T. Blumenthal. 1996. Functional analysis of an intron 3′ splice site in Caenorhabditis elegans. RNA 2:380-388. [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.