Abstract
Background
Retrotransposed genes are different to other types of genes as they originate from a processed mRNA and are then inserted back into the genome. For a long time, the contribution of this mechanism to the origin of new genes, and hence to the evolutionary process, has been questioned as retrogenes usually lose their regulatory sequences upon insertion and generally decay into pseudogenes. In recent years, there is growing evidence, notably in mammals, that retrotransposition is an important process driving the origin of new genes, but the evidence in insects remains largely restricted to a few model species.
Findings
By sequencing the messenger RNA of three developmental stages (first and fifth instar larvae and adults) of the pest Helicoverpa armigera, we identified a second, intronless, long-wavelength sensitive opsin (that we called LWS2). We then amplified the partial CDS of LWS2 retrogenes from another six noctuid moths, and investigate the phylogenetic distribution of LWS2 in 15 complete Lepidoptera and 1 Trichoptera genomes. Our results suggests that LWS2 evolved within the noctuid. Furthermore, we found that all the LWS2 opsins have an intact ORF, and have an ω-value (ω = 0.08202) relatively higher compared to their paralog LWS1 (ω = 0.02536), suggesting that LWS2 opsins were under relaxed purifying selection. Finally, the LWS2 shows temporal compartmentalization of expression. LWS2 in H. armigera in adult is expressed at a significantly lower level compared to all other opsins in adults; while in the in 1st instar stage larvae, it is expressed at a significantly higher level compared to other opsins.
Conclusions
Together the results of our evolutionary sequence analyses and gene expression data suggest that LWS2 is a functional gene, however, the relatively low level of expression in adults suggests that LWS2 is most likely not involved in mediating the visual process.
Electronic supplementary material
The online version of this article (doi:10.1186/s13100-016-0074-8) contains supplementary material, which is available to authorized users.
Keywords: Gene duplication, Expression, Evolution, Noctuidae
Findings
Background
Gene duplication is a fundamental process in genome evolution generating new biological functions and promoting adaptation to the changing environment [1–6]. The classical model predicts that after gene duplication, one of the two genes usually degenerates in a few million years, or, in rare cases, one of the duplicates might evolve new gene functions [7]. There are four well established mechanisms by which the DNA duplicates. These are: 1) unequal crossing-over, 2) duplicative (DNA) transposition, 3) polyploidization, and 4) retrotransposition [8]. The contribution of these mechanisms, (with the exception of retrotransposition) to the origin of new genes is well-established. Retrotransposition is different to the other three mechanisms as it includes an intermediate RNA step. That is, a mature mRNA is reverse transcribed into a complementary DNA copy without introns and then inserted back into the genome randomly [9]. The location of insertion is presumed to be random, thereby implying that the copy usually lacks the regulatory apparatus responsible of driving the correct gene expression. This is one of the main reasons why, until recently, retrotransposed genes were not considered to be functional [10–12]. However, recent work has challenged this view suggesting that several retrotransposons have been important for the evolution of novel phenotypes [9]. Interestingly, Marques et al. proposed that between 0.5 and two retrogenes are fixed every million years in flies and mammals, respectively [13–15]. The integration of statistical and molecular methods can be employed to assess the retrotransposons functionality. As reviewed by Kaessman et al. [16] a ratio between non-synonymous and synonymous mutation (ω value) of less than 1 suggests that the gene is under selection and hence still functional. Additionally, other features such as the presence of an open reading frame (ORF), conserved sequence to other species and expression (e.g. qPCR or RNA-seq) are indicative of their functionality.
Opsins are a subfamily of G-protein coupled receptors crucial for the visual process in all the metazoans [17]. In insects, this process is mediated by four paralogs: Long wave sensitive (LWS), Ultraviolet sensitive (UV), Blue sensitive (B) and probably Rh7 [18, 19]. Additionally, it is believed c-opsin might be involved in circadian rhythms [20]. Mechanistically, photoreceptors expressing opsin of different wavelengths signal to the brain, which then perceive colours [21]. The type of wavelength and the separation between maximum peak of absorbance defines the visual capability [18, 19, 22]. Opsin retrotransposons have been identified in various species, including the diurnal moth Callimorpha dominula (superfamily Noctuidea) [19], the jellyfish Tripedalia cystophora [23], in cephalopods [24], and in teleost fish [25].
In this work, we investigated the evolutionary history of an opsin retrogene in 7 species of nocturnal moth (Noctuidea). Firstly, we sequenced the whole transcriptome for 3 different developmental stages and adults of the pest Helicoverpa armigera, including first instar larvae, fifth instar larvae and adults. We identified that in addition to the traditional opsin repertoire (LWS, B and UV), H. armigera possesses a second intronless LWS gene (that we call LWS2). Subsequently, we investigated the phylogenetic distribution of this gene in six other Noctuidea species, one Crambidae, 15 Lepidoptera and 1 Trichoptera species. Our results suggest that 1) LWS2 evolved within Noctuidea and 2) the ω-value is less than 1, suggesting a relaxed purifying selection acting on LWS2. Finally, expression levels of LWS2 in H. armigera strongly differ between developmental stages, suggesting that most likely this gene is not involved in the visual process.
Material and methods
Transcriptome, annotation and opsin identification
Using RNA-Seq, the transcriptome of the whole bodies of first instar larvae (four groups and n = 30 for each gourp), fifth instar larvae (four groups and n = 10 for each group), and adults (four gourps and n = 10 for each group) of the nocturnal moth H. armigera was sequenced with paired-end and 100-nt read length on the channels of an Illumina HiSeq™ instrument. Assembled contigs were annotated using BLASTx to align with the database of NR, String, Swissprot and KEGG (see Additional file 1 for details). The RNA-Seq data were submitted to the NCBI GEO database (accession number: GSE86914). Primers were designed and PCR undertaken to (Additional file 1: Table S4) amplify the full-length cDNA of LWS2. To understand whether this duplication was specific to H. armigera or species from Noctuidae, we amplified LWS2 in six other species of Noctuidae moth: Agrotis ypsilon, Agrotis segetum, Mamestra brassicae, Mythimna separata, Spodoptera exigua, Spodoptera litura and one species of Crambidae: Ostrinia nubilalis. Additionally, we employed BLASTn algorithm to investigated the presence of LWS2 in other 15 complete Lepidoptera and 1 Trichoptera complete genomes from LepBase [26], spanning at total of 12 insect families (see Additional file 1: Table S5). Sequences were identified as follow: LWS1 and 2 were used as seed in a BLASTn search. Each sequences with an e-value <10−20 was retained a putative good opsin. To discriminate between opsin and other GPCRs after the translation in protein using TranslatorX [27] and using InterProScan we identified all the sequences with the retinal binding domain.
Alignment, phylogenetic and evolutionary analysis
The data set including the newly identified LWS2 from Noctuidea, Callimorpha domicula opsins, Ostrinia nubilalis opsins and putative LWS2 from LepBase (81 sequences in total available in Additional file 2) was aligned using the codon model as implemented in PRANK [28]. The resulting alignment was manually curated to remove gap-rich regions. GTR-G was identified as the best-fitting substitution-model accordingly to the AIC as implemented in Modelgenerator [29]. The phylogenetic reconstruction was performed using Maximum likelihood (ML) using Iqtree [30] and confirmed using Bayesian analysis (BA) under site heterogeneous model CAT-G model as implemented Phylobayes 3.3e [31]. In ML reconstruction the nodal support was evaluated using UltraFast bootstrap (BS) and abayes (aBS) [32] while in BA using the Bayesian posterior probability (PP). In the BA the convergence among chains was estimated using bpcomp and chains were considered converged when the maxdiff value was < 0.3. The phylogenetic analyses were performed using a rooted tree i.e. the LWS tree was rooted using Rh7, UV and Blue opsin. Finally, in order to account for the possible misleading effect of using distant related outgroup, we repeated the analyses without outgroup.
In order to estimate whether LWS2 is under evolutionary constraint we estimated the ratio between non-synonymous and synonymous substitutions (ω-value) using a maximum likelihood approach [33] as implemented in CODEML [34]. If the ω < 1, this is indicative of purifying selection However, if the retrogenes were under relaxed purifying selection, we expect an elevated ω-value relative to its paralog LWS1, which suggests that it may be neo-functionality or sub-functionalization. We evaluated five hypotheses: (1) one ω-ratio for all branches (one-ratio model; assuming that all branches have been evolving at the same rate); (2) ω-ratio = 1 for all branches (neutral model; neutral evolution for all branches); (3) moth LWS2 lineage and LWS1 lineage have different ω-ratio (ω2 and ω1; two ratio model; allowing foreground branch to evolve under a different rate); (4) neutral evolution for moth LWS2 lineage (ω2 = 1); and (5) the free-ratio model with free ω-ratio for each branch. In addition, we used branch-site models: moth LWS2 lineage was defined as the foreground, rest lineages were defined as the background branch, and these were then specified in the tree file by using branch labels. Likelihood-ratio test (LRT) was employed to determine if the alternative model, indicating positive selection, was superior to the null model. We also performed CODEML test on H. armigera LWS2 lineage to see if nature selection acted on any of the LWS2 branches.
Quantitative expression of opsin
To test the expression of the opsin in H. armigera, we investigated the relative level of expression in the 3 different developmental stages using fragments per kilobase of exon per million fragments mapped (FPKM) [35]. Differential gene expression between paralogs at different developmental stages was evaluated using STATA v.9.0 and ANOVA. Bonferroni multiple comparisons were used to determine the level of significance between the relative levels of mRNA expression.
Results and discussion
We obtained about 4 gigabase (Gb) of sequence each sample from the RNA-Seq (total 63 Gb for 12 samples), and a total of 99,711 contigs (See Additional file 1 for details; with a total of 73, 709 unigenes). Using functional annotation, we were able to identify that in addition to the traditional insects opsin genes of LWS, UV and Blue [19, 36]. H. armigera possesses an additional opsin gene, named LWS2. Using PCR we confirmed the presence of this gene in H. armigera and in 6 other Noctuidea species. The phylogenetic analysis displayed in Fig. 1 supports the paralog relationship between the newly identified opsin and LWS2 from other species and LWS1 (PP = 0.76, BS = 1, aBS = 100, see Fig. 1). Additionally, our phylogenetic trees suggests that LWS2 orthologs are present only in Noctuidea. The analysis of the intron content suggests that while LWS1s noctuid species have seven introns, no introns are present in the LWS2s (Additional file 1: Figures S2a and S2b), this finding together with the monophyly of LWS1 and 2 suggests that the later (i.e. LWS2) originated as a retrocopy from LWS1 [8, 16]. Furthermore, our results strongly support the monophyly of intronless opsin in Noctuidea (PP = 0.9, BS = 100, aBS = 1) (Fig. 1). The tree topologies are invariant in respect to the sequences used for rooting the tree (compared to Additional file 1: Figures S3, S4, S5 and S6).
Fig. 1.
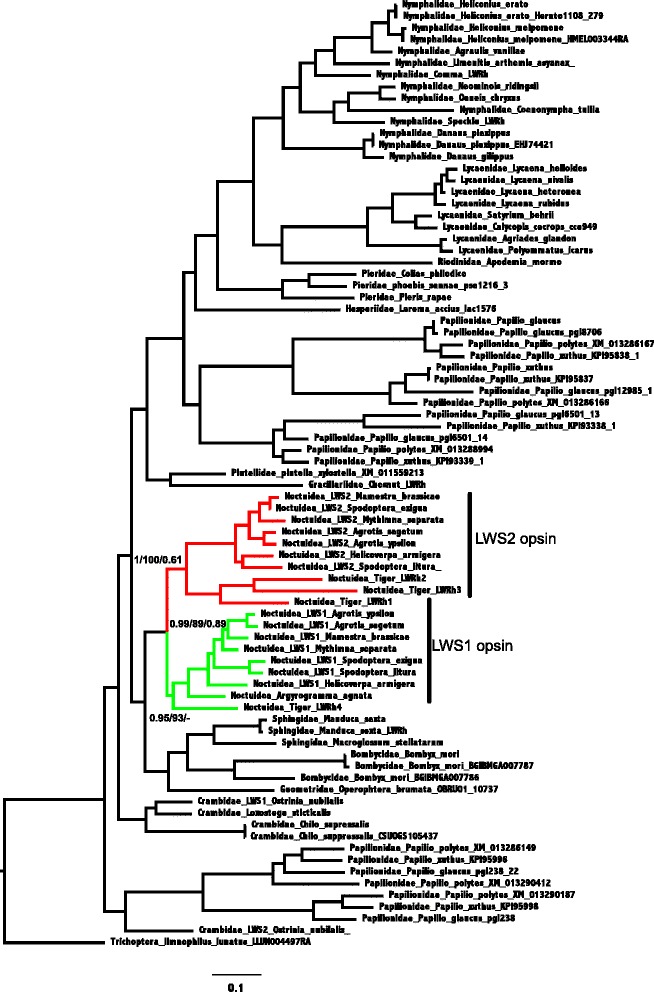
Maximum likelihood under GTR-G. Support values at nodes are from left to right aBayes, Ultrafast bootstrap and Bayesian Posterior Probability. LWS2 opsins of noctuid species denoted in red color. Noctuidae LWS1 opsins denoted in green color
In the next section, we investigated whether these LWS2 are functional. First we observed that all the LWS2 genes identified in this study have intact ORFs (see Additional file 1: Figure S2) arguing in favor of LWS2 functionality. Furthermore, our results suggest that LWS2 have an ω-value <1 (ω = 0.08202) indicating that is evolving under purifying selection as expected in functional genes. Finally we did not detect signal of positive selection acting on LWS2 (Table 1). These findings together indicate that LWS2 it is a functional genes.
Table 1.
Selective patterns for LWS opsins
| Model | npa | Ln Lb | Estimates of ω | Models compared | LRTc | P Values |
|---|---|---|---|---|---|---|
| Branch model | ||||||
| A: one ratio | 175 | −41372.52 | ω = 0.06527 | |||
| B: one ratio ω = 1 | 174 | −47529.36 | ω = 1 | B vs. A | 12313.68 | 0.0 |
| C: the LWS2 branch has ω2, the LWS1 branch has ω1 | 177 | −41347.92 | ω2 = 0.08202 | A vs. C | 49.2 | 0.0 |
| ω1 = 0.02536 | ||||||
| D: the LWS2 branch has ω2 = 1 | 176 | −41783.48 | ω2 = 1 | D vs. C | 871.12 | 0.0 |
| ω1 = 0.02515 | ||||||
| E: each branch has its own ω | 347 | −41045.37 | Variable ω by branch | A vs. E | 654.3 | 0.0 |
| Branch-site models | ||||||
| G: the LWS2 branch | 178 | −41152.48 | ||||
| H: the LWS2 branch has ω = 1 | 177 | −41152.48 | H vs. G | 0.0 | 1.0 | |
aNumber of parameters
bThe natural logarithm of the likelihood value
cTwice the log likelihood difference between the two models
Subsequently, we investigated whether the LWS2 might contribute to the evolution of visual capability in Noctuidea. The data from RNA-seq expression level in H. armigera suggested that LWS1, and more generally UV and Blue opsins in H. armigera, were significantly higher expressed in adults than LWS2 (Fig. 2a). The result suggested that LWS2 migth be not involved in the visual system of the adult. However, surprisingly, LWS2 in 1st-instar larvae has an higher relative level of expression compared to the other three opsins (Fig. 2b). The reasons for higher level of expression of LWS2 at the 1st-instar larvae are unclear. This finding is conceivable with a function of LWS2 other than vision [37, 38], another alternative is that observed level of expression represents transcriptional noise. The retrotransposed opsins are expressed as results of transcriptional activity in the new genomic location. However, additional experiments would be necessary to clarify between the competing hypothesis.
Fig. 2.
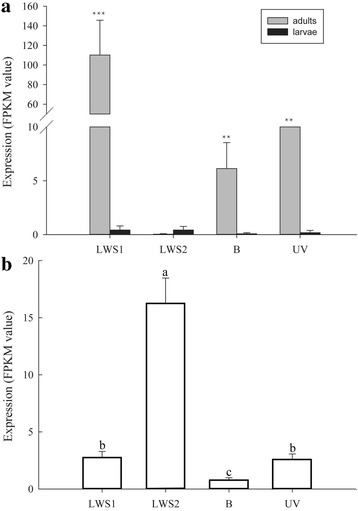
Quantitative expression levels for opsin genes in a larvae at 5th instar stage and adults and b larvae at 1st instar stage. a The “**” denote statistical significance of the expression levels at P < 0.01. The “***” denote statistical significance of the expression levels at P < 0.001. b Significant difference is shown using different letters (P < 0.0001, d.f. = 3, 12, F = 203.83). Mean ± SD. n = 4
In conclusion we report the existence of LWS2, originating as retrocopies from LWS1, in seven moths from the superfamily Noctuoidea. Furthermore, the intact ORF, the ω < 1, the phylogenetic conservation and expression independently suggests that LWS2 opsins are functional.
Acknowledgements
We thanks Dr. Tiantao Zhang, from Institute of Plant Protection, Chinese Academy of Agricultural Sciences, for providing the reference sequence of LWS1 and LWS2 in O. nubilalis.
Funding
This research was supported by Science Fund for Creative Research Groups of the National Science Foundation of China (No. 31321004), the National Natural Science Foundation of China (Grant No. 31401752).
Availability of data and material
The RNA-seq datasets generated during the current study are available in the NCBI GEO database (accession number: GSE86914), (https://www.ncbi.nlm.nih.gov/geo/).
Authors’ contributions
KW, PX, RF, BL and HX conceived the study. PX, BL and HX performed the experiments. KW, PX, RF, BL, HX and RIG wrote the manuscript. All of the authors critically reviewed and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The reared or wild-captured moths used in this study were serious pests in China, therefore, no permits were required for the described insect collection and experimentation.
Abbreviations
- aBS
a Bayes support
- B
Blue sensitive opsin
- BI
Bayesian inference
- BS
Bootstrap support
- FPKM
Fragments per kilobase of exon per million fragments mapped
- LRT
Likelihood-ratio test
- LWS
Long-wavelength sensitive opsin
- ORF
Open reading frame
- PP
Posterior probability
- UV
Ultraviolet sensitive opsin
Additional files
Materials and methods Detailed description of materials and methods used in RNA-seq and PCR amplification. Results Description of the results in RNA-seq and PCR amplification. Table S1. Summary of the sequence assembly after Illumina sequencing. Table S2. The opsin genes from H. armigera by RNA-seq. Table S3. The FKPM values of opsin genes in H. armgiera. Table S4. Primers used in this study. Table S5. The species with complete genome sequence used in this study. Figure S1. The discription of RNA-seq. (a) The distribution of sequences length. (b) The E-value distribution of the top matches in the nr database. (c) The species distribution of the matches in the nr database. (d) The sequence similarity distribution. Figure S2. The genomic sequence of LWS opsins in seven noctuid species. (a) LWS1 opsins showed seven introns. (b) The fragments od LWS2 opsins from seven noctuid species showed no introns in the region. The red letters showed the homology region of primers for amplifying partial sequence of LWS2 using DNA as templete. The introns are shaded. “.” = identical nucleotides; “-” = absence of nucleotides. AS = Agrotis segetum, AY = Agrotis ypsilon, HA = Helicoverpa armigera, MB = Mamestra brassicae, MS = Mythimna separata, SELWS1 = Spodoptera exigua, SL = Spodoptera litura. (c) The genomic sequence of LWS1 in O. nubilalis. (d) The genomic sequence of LWS2 in O. nubilalis. The exons were showed using black letters and the introns were showed using red letters. Figure S3. Maximum Likelihood tree with outgroup. Figure S4. Bayesian tree with outgroup. Figure S5. Maximum Likelihood tree with outgroup. Figure S6. Bayesian tree with outgroup. (DOC 2650 kb)
Alignment of sequences used in this study. (PHY 100 kb)
Contributor Information
Pengjun Xu, Email: xupengjun@163.com.
Roberto Feuda, Email: rfeuda@caltech.edu.
Bin Lu, Email: lvbin-3@163.com.
Haijun Xiao, Email: hjxiao@jxau.edu.cn.
Robert I. Graham, Email: rgraham@harper-adams.ac.uk
Kongming Wu, Email: kmwu@ippcaas.cn.
References
- 1.Ohno S. Evolution by gene duplication. London: George Allen and Unwin; 1970. p. 160. [Google Scholar]
- 2.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:R0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang P, Gu Z, Li W. Different evolutionary patternsbetween young duplicate genes in the human genome. Genome Biol. 2003;4:R56. doi: 10.1186/gb-2003-4-9-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Innan H, Kondrashov F. The evolution of gene duplications:classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 5.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B. 2012;279:5048–57. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian W, Zhang J. Genomic evidence for adaptation by gene duplication. Genome Res. 2014;24:1356–62. doi: 10.1101/gr.172098.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J. Evolution by gene duplicatin: an update. Trends Ecol Evol. 2003;18:292–8. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 8.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–67. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaessman H. Origins, evolution, and phylogenetic impact of new genes. Genome Res. 2010;20:1313–26. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffs P, Ashburner M. Processed pseudogenes in Drosophila. Proc Biol Sci. 1991;244:151–9. doi: 10.1098/rspb.1991.0064. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–58. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–7. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Casola C, Feschotte C, Betran E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zemojtel T, Duchniewicz M, Zhang Z, Paluch T, Luz H, Penzkofer T, Scheele JS, Zwartkruis FJ. Retrotransposition and mutation events yield Rap1 GTPases with differential signalling capacity. BMC Evol Biol. 2010;10:55. doi: 10.1186/1471-2148-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaessman H, Vinckenbosch N, Long M. RNS-based gene duplication mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feuda R, Hamilton SC, McInerney JO, Pisani D. Metazoan opsin evolution reveals a simple route to animal vision. Proc Natl Acad Sci U S A. 2012;109:18868–72. doi: 10.1073/pnas.1204609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briscoe AD, Chittka L. The evolution of color vision in insects. Annu Rev Entomol. 2001;46:471–510. doi: 10.1146/annurev.ento.46.1.471. [DOI] [PubMed] [Google Scholar]
- 19.Feuda R, Marletaz F, Bentley M, Holland PW. Conservation, duplication and divergence of five opsin genes in insect evolution. Genome Biol Evol. 2016;8:579–87. doi: 10.1093/gbe/evw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velarde RA, Sauer CD, Walden KK, Fahrbach SE, Robertson HM. Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem Mol Biol. 2005;35:1367–77. doi: 10.1016/j.ibmb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Solomon SG, Lennie P. The machinery of colour vision. Nat Rev Neurosci. 2007;8:276–86. doi: 10.1038/nrn2094. [DOI] [PubMed] [Google Scholar]
- 22.Neitz J, Neitz M. The genetics of normal and defective color vision. Vision Res. 2011;51:633–51. doi: 10.1016/j.visres.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liegertova M, Pergner J, Kozmikova I, Fabian P, Pombinho AR, Strnad H, Paces J, Vlcek C, Bartunek P, Kozmik Z. Cubozoan genome illuminates functional diversification of opsins and photoreceptor evolution. Sci Rep. 2015;5:11885. doi: 10.1038/srep11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris A, Bowmaker JK, Hunt D. The molecular basis of a spectral shift in the rhodopsin of two species of squid from different photic enviroments. Proc R Soc B Biol. 1993;254:233–40. doi: 10.1098/rspb.1993.0151. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM. The rhodopsin-encoding gene of bony fish lacks introns. Gene. 1995;164:273–7. doi: 10.1016/0378-1119(95)00458-I. [DOI] [PubMed] [Google Scholar]
- 26.Challis RJ, Kumar S, Dasmahapatra KKK, Jiggins CD, Blaxter M. Lepbase: the Lepidopteran genome database. bioRxiv. 2016; http://dx.doi.org/10.1101/056994.
- 27.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 2005;102:10557–62. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 32.Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–95. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–36. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 35.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P, Lu B, Xiao H, Fu X, Murphy RW, Wu K. The evolution and expression of the moth visual opsin family. PLoS One. 2013;8:e78140. doi: 10.1371/journal.pone.0078140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakane Y, Ikegami K, Ono H, Yamamoto N, Yoshida S, Hirunagi K, Ebihara S, Kubo Y, Yoshimura T. A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc Natl Acad Sci U S A. 2010;107:15264–8. doi: 10.1073/pnas.1006393107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriksson BJ, Fredman D, Steiner G, Schmid A. Characterisation and localisation of the opsin protein repertoire in the brain and retinas of a spider and an onychophoran. BMC Evol Biol. 2013;13:186. doi: 10.1186/1471-2148-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]


