Abstract
Mammalian target of rapamycin (mTOR) is a key regulator in various cellular processes, including cell growth, gene expression, and synaptic functions. Autism spectrum disorder (ASD) is frequently accompanied by monogenic disorders, such as tuberous sclerosis complex, phosphatase and tensin homolog tumor hamartoma syndrome, neurofibromatosis 1, and fragile X syndrome, in which mTOR is hyperactive. Mutations in the genes involved in the mTOR-mediated signaling pathway have been identified in some cases of syndromic ASD. Evidences indicate a pathogenic role for hyperactive mTOR-mediated signaling in ASD associated with these monogenic disorders, and mTOR inhibitors are a potential pharmacotherapy for ASD. Abnormal synaptic transmission through metabotropic glutamate receptor 5 may underlie in a part of ASD associated with hyperactive mTOR-mediated signaling. In this review, the relationship between mTOR and ASD is discussed.
Keywords: Autophagy, fragile X syndrome, mammalian target of rapamycin, metabotropic glutamate receptor 5, neurofibromatosis, phosphatase and tensin homolog, tuberous sclerosis complex
Introduction
Mammalian target of rapamycin (mTOR) is a serine/ threonine kinase that critically regulates important cellular physiology, such as protein synthesis and mRNA translation. This regulation occurs in response to a diverse range of stimuli from outside the cell, such as growth factors, cytokines, energy starvation, and hypoxia, and affects cell growth, cell differentiation, and metabolism. The relevance of mTOR in human diseases is increasingly appreciated in the field of oncology [1], immunology [2], metabolism [3], and neurology [4], including autism spectrum disorder (ASD).
ASD is a neurodevelopmental disorder of which the core symptoms include impairment in reciprocal social interaction and restrictive, repetitive behaviors and interests [5]. Its incidence has been reported to be increasing, and the current prevalence is estimated to be approximately 1% of the general population. Twin studies revealed that the monozygotic concordance reaches as high as 90%, whereas the concordance is substantially lower in dizygotic twins, implying that genetics have significant impact on an individuals’ susceptibility to ASD [6]. Thus, individuals with heritable disorders, such as tuberous sclerosis complex (TSC) and fragile X syndrome (FXS), demonstrate ASD symptoms at a particularly high frequency. Recent advances in the genetic research of ASD have revealed that many genetic disruptions are found in individuals with ASD, including single gene mutations and copy number variations, in different chromosomal regions [7]. Understanding the molecular pathophysiology of ASD-associated disorders, such as TSC and FXS, has also progressed. An accumulating body of evidence highlights the causal role of dysregulated mTOR signaling pathway in a subset of individuals with ASD and provides us with an insight to understand the molecular pathophysiology of ASD and potential pharmacological therapies.
Mammalian target of rapamycin
mTOR associates with several protein components to form two distinct complexes. mTOR complex 1 (mTORC1) is characterized by a critical component called raptor [8, 9] and regulates protein synthesis, energy metabolism, cell growth and proliferation, and cap-dependent mRNA translation [3]. mTOR inhibitors, such as rapamycin and everolimus, block mTOR-raptor coupling and inactivate mTORC1 [8, 9]. The other mTOR is mTOR complex 2 (mTORC2), which has a rictor instead of a raptor. The absence of a raptor makes mTORC2 insensitive to mTOR inhibitors [10, 11]. mTORC2 controls cellular functions, such as cytoskeletal rearrangement and cell survival [10, 11].
Upstream of mTORC1, a number of stimuli, such as growth factors, cytokines, energy starvation, and hypoxia, are integrated by TSC 1/2 complex. The TSC1 product hamartin and the TSC2 product tuberin form a heterodimer complex [12, 13] (Fig. 1). TSC2 has GTPase-activating protein (GAP) activity that is regarded as the most important of TSC1/2 complex [14, 15]. The central role of TSC1 is to stabilize the complex as TSC1 couples with TSC2 and prevents TSC2 from ubiquitin-mediated degradation [16]. In addition, TSC1 may have mTORC1-independent functions. Tsc1-deficient cells showed deficient rearrangement of the actin cytoskeleton that did not respond to rapamycin, and this phenotype was rescued by inhibiting the Rho-associated protein kinase (ROCK), a downstream effector of the small GTPase RhoA [17]. Its significance is, however, less clear particularly with respect to human disorders. The TSC1/2 complex decreases the GTP-bound active form of the small G-protein Ras homolog enriched in brain (Rheb) and results in constitutive activation of mTORC1 [18].
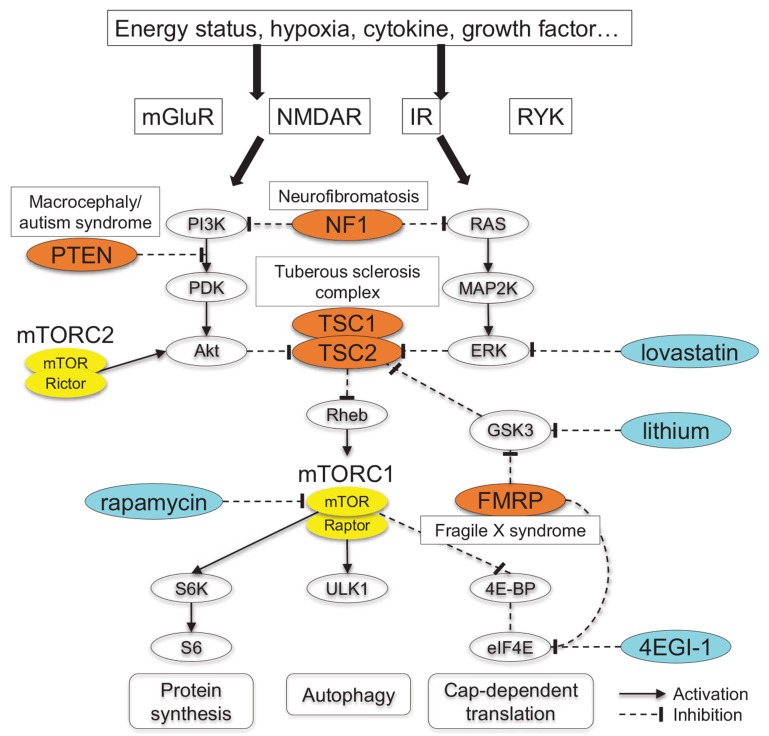
Fig. (1).
Signaling pathway involving mTORC1. Various stimuli converge on mTORC1 pathway via a number of routes. mTORC1 activation impacts on different cellular physiology, such as protein synthesis, cap-dependent mRNA translation, and autophagy. Here mutations in TSC1/2, PTEN, FMR1, and NF1 cause constitutive mTORC1 activation and syndromic ASD. Akt, protein kinase B; eIF4E, eukaryotic initiation factor 4E; ERK, extracellular signal-regulated kinase; 4E-BP, 4E-binding protein; FMRP, fragile X mental retardation protein; GSK3, glycogen synthase kinase 3; IR, insulin receptor; MAP2K, mitogen-activated protein kinase kinase; mGluR, metabotropic glutamate receptor; mTOR, mammalian target of rapamycin; NF1, neurofibromin; NMDAR, NMDA receptor; PDK, phosphoinositide-dependent kinase; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog; RAS, rat sarcoma; Rheb, Ras homolog enriched in brain; RYK, receptor-like tyrosine kinase; S6K, p70 ribosomal S6 kinase; TSC, tuberous sclerosis complex; ULK1, unc51-like kinase 1.
mTORC1 exerts its kinase activity on target proteins ribosomal protein S6 kinases (S6Ks) and eukaryotic translation initiation factor-4E (eIF4E)-binding proteins (4E-BPs). Two S6K proteins S6K1 and S6K2 exist in mammalian cells. mTORC1 phosphorylates both S6K1 and S6K2, whereas other regulators phosphorylate either S6K1 or S6K2. For example, Neurabin acts on S6K1 and ERK on S6K2. Most of the known regulators activate S6Ks while S6K1 dephosphorylation by PP2A inhibits S6K1. The active form of S6Ks then phosphorylates ribosomal protein S6 and increases general protein synthesis. S6K1 has several isoforms, and p70-S6K1 is most extensively studied [19]. There are three paralogues of 4E-BPs (4E-BP1, 4E-BP2, and 4E-BP3) [20], and 4E-BP2 is the major form in the mammalian brains. 4E-BP1 and its phosphorylated form are used as markers of mTORC1 activity. 4E-BPs bind to the cap-dependent transcription factor eIF4E to repress cap-dependent mRNA translation. mTORC1 phosphorylates 4E-BPs and make them lose their binding activity to eIF4E. This leads to the deregulation of eIF4E and enhances the subsequent initiation of cap-dependent mRNA translation [21-23]. In this manner, the two genes critically control mTORC1-mediated protein synthesis. The third major substrate of mTORC1 is Unk-51-like kinase 1 (ULK1) that acts as a repressor of autophagy [24, 25], a process that removes damaged organelles and generates energy. mTORC1 activation suppresses ULK1 and enhances autophagy.
Syndromic ASD associated with MTORC1 hyperactivation
As noted above, the mTORC1 signaling pathway has a central role in cell growth and proliferation. A number of human disorders that are caused by mutations of the genes involved in this pathway, such as TSC and neurofibromatosis type 1 (NF1), are characterized by the high susceptibility to tumor development (Fig. 1). The associated tumors are basically benign in TSC [26] and NF1 [27], whereas those seen in Cowden syndrome caused by mutations in the phosphatase and tensin homolog (PTEN) gene are at a high risk of malignancies [28]. Tumor susceptibility in these disorders can be explained by the germline loss of heterozygosity and additional mutations in the other allele. In contrast, FXS individuals are not at an increased risk of tumor development [29], and enlarged testes are a common physiological feature in the male patients.
ASD and intellectual disability are common neurological features in TSC, FXS, NF1, and a set of disorders caused by PTEN mutations. Considering that ASD manifests from infancy when other neurological and neoplastic symptoms do not yet appear, it is speculated that germline haploinsufficiency and consequent mTORC1 activation are sufficient for the development ASD in these disorders. In the following context, the above ASD-related monogenic disorders are discussed with respect to dysregulated mTORC1-mediated signaling.
There are several of genes in this signaling pathway that are associated with ASD [7]. Of note, four human monogenic disorders, TSC, FXS, macrocephaly/autism syndrome caused by a mutation in PTEN, and NF1, are caused by mutations in the genes upstream of mTORC1. These findings imply that mTORC1 overactivation is one of the common pathological events underlying ASD.
Tuberous Sclerosis Complex
TSC (MIM#191100, #613254) is an autosomal dominant disorder presenting with disease manifestations in different organs, such as skin, brain, and kidney. Long before the discovery of the causative genes, the disorder was recognized in individuals presenting with the classical “triad” of a facial skin lesion (angiofibroma), epilepsy, and intellectual disability [30]. Later TSC1 [31] and TSC2 [32] were identified as the causative genes. TSC is estimated to occur in 1 in 6,000 live births [33], according to the former diagnostic criteria [34]. The recently revised criteria include the results of genetic testing in addition to the original clinical hallmarks including skin features (facial angiofibroma, hypopigmented macules, shagreen patch), neurological features (cortical tuber, subependymal nodule), and hamartomatous lesions in different organs (renal and retinal angiomyolipoma, cardiac rhabdomyoma, lung lymphangiomyomatosis) [26]. TSC1 and TSC2 mutations basically cause similar features although individuals with TSC2 mutations are more severely affected in certain aspects: more severe skin and kidney involvement [35, 36], more severe intellectual disability and heavier seizure burden [36, 37], and ASD [37, 38].
TSC is characterized by various CNS pathological complications, many of which are also specific including cortical tubers and subependymal giant cell astrocytoma [26]. Neuropsychiatric manifestations of TSC are also diverse: epilepsy that is often intractable [39], intellectual disability, ASD, attention-deficit hyperactivity disorder, and anxiety [40]. These manifestations are not specific, but they are highly prevalent and make daily life difficult for individuals with TSC. The concept of TSC-associated neuropsychiatric disorders is thus introduced in the latest guideline [41, 42]. As for ASD, approximately half of the individuals with TSC have ASD [40, 43]. TSC accounts for 1%−4% of all ASD cases and is a typical monogenic ASD [44].
As described above, TSC1 and TSC2 coordinate to inhibit downstream Rheb and mTORC1 (Fig. 1). Somatic mutations of TSC1 and TSC2, mainly the loss of heterozygosity, abolish the inhibition of mTORC1 [35, 36]. The consequent constitutive overactivation of mTORC1, particularly the enhanced phosphorylation of S6Ks and 4E-BPs leading to increased global protein synthesis and cap-dependent mRNA translation, is considered to constitute the central molecular pathology of TSC.
Two groups of rodent models of TSC are currently available, each resulting from different genetic manipulations (Table 1 ). The first group includes germ-line haploinsufficient models, such as Tsc1+/− [45], Tsc2+/− mice [46], and Eker rats [47]. Conditional knockout (KO) mice that lack Tsc1 or Tsc2 in glial fibrillary acidic protein (GFAP)-positive cells [48, 49] or in Purkinje cells [50, 51] have also been analyzed in detail. Haploinsufficient models present with impaired social interaction, the core behavioral feature of ASD [52-54], and learning deficits in the domains of spatial and working memory [55]. Of note, these animals basically have no epileptic seizures and less severe morphological changes in the brain compared with human TSC individuals [45, 46, 52]. Mice lacking Tsc1 or Tsc2 in Purkinje cells also show no evidence of epileptic seizures, but more autistic-like behavioral alterations, such as social impairment, restrictive behavior, and abnormal ultrasound vocalizations, are observed [50, 51]. The neurological phenotype is much more severe in mice with astrocyte-specific deletion of Tsc genes. The affected animals frequently suffer from premature death. The animals that survive develop spontaneous seizures and progressive macrocephaly and die before 5 months of age. Intriguingly and in accordance to clinical observation, Tsc2 KO mice experience more frequent seizures and more premature death with higher levels of mTORC1 activity compared to Tsc1 KO mice [56]. However, the precise mechanism relating the higher mTORC1 activity with the Tsc2 deletion is yet to be determined.
Table 1. mTOR-related disorders and their animal models discussed in this review.
| Genes | Human Disorders | Animal Models |
|---|---|---|
|
TSC1 TSC2 |
Tuberous sclerosis complex TSC1 or TSC2 haploinsufficiency Skin (e.g. facial angiofibroma) Brain (e.g. cortical tuber) Hamartomas (e.g. renal angiomyolipoma) Often presents with epilepsy, intellectual disability. ASD in approximately half of the patients. |
Haploinsufficient mice No brain lesions, no epilepsy ASD-like social behavior Learning deficits Eker rats (Tsc2 haploinsufficiency) Brain lesions in rare cases, no epilepsy Reduced social interaction Purkinje cell-specific KO mice Progressive loss of PCs ASD-like social behavior GFAP-positive cell-specific KO mice Seizures, progressive brain enlargement Early mortality |
| PTEN | Macrocephaly/autism syndrome PTEN haploinsufficiency Marked macrocephaly Found in 7%-17% of ASD with macrocephaly. Note: also the cause of Bannayan-Riley-Ruvalcaba syndrome Lhermitte-Duclos syndrome Cowden syndrome |
Haploinsufficient mice ASD-like social behavior Increased brain weight NSE-positive cell specific KO mice Reduced social interaction Seizures, increased anxiety, increased lomotor activity Early mortality |
| FMR1 | Fragile X syndrome > 200 CGG repeats in FMR1 X-linked inheritance Intellectual disability, macrocephaly, macroorchidism in males ASD more often in males (30%) |
Fmr1 KO mice (only males studied) ASD-like social behavior Increased anxiety, increased locomotor activity, cognitive deficits |
| NF1 | Neurofibromatosis type 1 NF1 haploinsufficiency Skin (e.g. café-au-lait spots) Neurofibromas ASD in 15%-30% of the patients |
Haploinsufficient mice Learning deficits, attention deficits Impaired social discrimination |
NSE, Neuron Specific Enolase.
At the neuronal level, the TSC1/2 complex regulates neuronal morphology and synaptic function in the brain. Complete deletion of Tsc1 and Tsc2 in the Purkinje cells leads to progressive Purkinje cell loss and abnormal dendritic morphology including increased spine density, and heterozygous deletion is sufficient to reduce excitability of Purkinje cells [50, 51]. GFAP-positive cell-specific loss of the genes causes unregulated astrocyte proliferation, neuronal death independent of seizures, and neuronal disorganization in the brain [48, 56]. Altered structure, an increased number of dendritic spines, and impaired axon guidance are also associated with TSC [57, 58]. An interesting key to link synaptic deficits and ASD is deficient mTORC1-mediated autophagy. Distorted synaptic morphology was observed in the brains of Tsc2+/− mice and human sporadic ASD, and impaired autophagy was detected in both conditions [59]. The mechanism of autophagy controlled by the mTORC1/ULK1 pathway may be complicated as autophagy was unexpectedly enhanced in Tsc1/2 deleted dividing cells [60].
The TSC/mTORC1 pathway also controls synaptic plasticity and long-term memory via metabotropic glutamate receptor (mGluR)-mediated long-term potentiation (LTP) [55, 61] and long-term depression (LTD) [62, 63]. Impaired LTP in the hippocampus of Tsc1 KO mice is due to elevated levels of extracellular glutamate [61]. Deregulation of mTORC1 perturbs neural excitatory/inhibitory balance at the network level by exaggerating excitability and reducing inhibitory synaptic transmission [64]. The above synaptic and cellular alterations may ultimately converge on the development of neurocognitive impairments in TSC including ASD.
The therapeutic implication of TSC-associated neurological problems is raised by the observation that the mTOR inhibitor rapamycin has effects for reversing behavioral and molecular abnormalities. In haploinsufficient models, transient treatment with rapamycin is sufficient to effectively suppress mTORC1 [53, 55], rescue normal LTP [55] and LTD [62], and correct learning deficits [55] and autistic-like behavioral deficits [53]. Rapamycin shows similar benefits in conditional KO model; prevention of brain enlargement, epileptic seizure, and early mortality [56]; autistic-like deficient social approach [50]; and Purkinje cell loss and abnormal morphology [50, 51]. Moreover, the clinical use of mTOR inhibitor improves cognitive function and relieves behavioral problems, as will be discussed later in more detail.
Macrocephaly/Autism Syndrome Caused by PTEN Mutation
Increased head size is an occasional physical feature of individuals with ASD, particularly in early life [65]. Mutations in the PTEN gene were identified in those with ASD accompanied by extreme macrocephaly, and this condition was named thereafter “macrocephaly/autism syndrome” (MIM#605309) [66-68]. With respect to human diseases, the PTEN gene was originally found as the cause of Bannayan–Riley–Ruvalcaba syndrome (MIM#153480) (manifestation: macrocephaly, intellectual disability, multiple intestinal hamartoma, etc.), Lhermitte–Duclos syndrome (manifestation: cerebellar ataxia, seizure, etc.), and Cowden syndrome (MIM#158350) (manifestation: macrocephaly, mucocutaneous lesions, intestinal polyps, increased risk of malignancies) [69, 70]. As these disorders and macrocephaly/autism syndrome share the PTEN gene mutations, they are now recognized as the spectrum called PTEN hamartoma tumor syndrome [28]. From a neurological aspect, macrocephaly and developmental problems, including ASD, are common features along the spectrum, whereas hamartomatous and other features are characteristic in each disorder [28]. PTEN mutations can be detected in 7%-17% of individuals with ASD with extreme macrocephaly [62, 66, 67], and they are estimated to be found in 1% or more of all individuals with ASD.
The PTEN gene product negatively regulates the phosphoinositol 3-kinase (PI3K), which decreases the levels of phosphatidylinositol (3,4,5)-triphosphate (PIP3; see figure). This then suppresses PDK and Akt, activates the TSC1/2 complex, and keeps mTORC1 repressed, constituting the PI3K/Akt/TSC/mTORC1 signaling pathway [71]. Mutations in the PTEN gene abolish inhibition of PI3K that results in Akt activation, TSC1/2 complex inhibition, and constitutive mTORC1 activation (Fig. 1).
The neurological issues accompanied by PTEN mutations have been extensively studied using haploinsufficient mice [72, 73] and conditional KO mice [74, 75] (Table 1). Similar to the mouse models of TSC, Pten +/− mice show a mild neurological phenotype: increased brain weight, impaired prepulse inhibition, and altered social behavior reminiscent of ASD [76-78]. Most of neuron-specific enolase-positive cell-specific Pten conditional KO mice die in early postnatal weeks. The mice who survive exhibit macrocephaly that is progressive and more severe than that observed in Pten+/− mice impaired prepulse inhibition and prominent behavioral abnormalities, such as diminished reciprocal social interaction, increased locomotor activity, more anxiety, memory deficit, and spontaneous seizures [74, 75].
Neurons from Pten KO mice display increased phosphorylation of Akt and S6, indicating the overactivation of the PI3K/Akt/TSC/mTORC1 signaling pathway [74, 75]. Neuronal alteration includes increased soma size and axonal growth, hypertrophic and ectopic axonal projections, and abnormal synapses with increased presynaptic varicosities. Dendrites are also hypertrophic and accompanied by increased spine density [74]. According to Zhou, et al. [75], chronic administration of rapamycin from an early postnatal period onward prevented brain enlargement and neuronal soma hypertrophy and alleviated axonal and dendritic hypertrophy. This treatment kept the mutant mice in a generally healthy condition as long as the treatment continued, and it prevented phenotypic abnormalities observed in the nontreated mutants, such as impaired social interaction, elevated anxiety, and the development and worsening seizures [75]. Older mice also displayed improvements in neuronal abnormalities and seizures although the behavioral effect was difficult to assess because of handling-related death. It is therefore suggested that the abnormal features of Pten conditional KO mice are due to elevated mTORC1 activity and preventable by keeping mTORC1 inhibited.
Fragile X Syndrome
FXS (MIM#300624) is the most common cause of inherited intellectual disability [79]. It is an X-linked disorder with intellectual disability, ASD, increased anxiety, and physical features including macrocephaly and macroorchidism in males [80]. Unlike other disorders mentioned in this review, the genetic defect in most FXS cases is abnormal elongation of CGG repeats in the 5′-untranslated region of fragile X mental retardation 1 (FMR1) gene [81]. Typical FXS individuals have a CGG repeat size more than 200, called as “full mutation” [82]. An FMR1 repeat size between 55 and 200 (premutation) is shorter than that in typical FXS individuals but longer than that in the normal population that has a repeat size <55. Those with such a repeat size do not show typical FXS manifestation, whereas some are affected with the characteristic movement disorder named fragile X-associated tremor/ataxia syndrome (FXTAS) in later life [83]. An individual with a premutation with or without FXTAS is at risk of neuropsychiatric disorders, such as a mood disorder or a panic disorder [84]. FXS as diagnosed by detection of the full mutation is found in 1 in 7,100 males and 1 in 11,000 females [85]. Twenty-two percent of individuals have ASD, and the prevalence rises to 30% in males [43]. Approximately 1%-5% of cases of ASD are accompanied by FXS, and it is yet one of the most common syndromic ASD [44].
FMR1 encodes the protein fragile X mental retardation protein (FMRP), an RNA-binding protein involved in gene translation and enhancing global protein synthesis. Its binding targets are very diverse and include postsynaptic density-95, the GluR1 and GluR2 subunits of the AMPA glutamate receptor, and several other synaptic proteins important for neurotransmission and structure [86, 87]. Although the precise mechanism is less clear, deficiencies in FMRP are associated with increased mTORC1 signaling [88]. CGG repeat elongation silences the FMR1 gene via hypermethylation of CpG island in its promoter region. Point mutations in the FMR1 gene (for example, nonsense mutation, missense mutation, and frameshift mutation) are rarely found in FXS. Both FXS-causing mutations, CGG repeat elongation and point mutation, commonly lead to the development of FXS by abolishing the production of FMRP.
Regarding animal models of FXS, Fmr1 KO mice have been extensively analyzed [89] (Table 1). Several kinds of independently generated Fmr1 KO mice exhibit prominent behavioral abnormalities (see reviews [90, 91]). With regard to autistic-like behavioral deficits, a number of studies using the three-chambered social approach test report normal behavior, whereas impaired sociability, the absence of a preference to explore the novel mouse over the novel inanimate object, has been observed in other research [92, 93]. Detailed analysis reveals that Fmr1 KO mice display fewer affiliative behaviors during social interactions with female mice [94]. Repetitive behaviors, the other major behavioral alteration in ASD, are also observed Fmr1 KO mice. For example, the mutant animals exhibit higher levels of self-grooming behavior [94]. Fmr1 KO mice show diverse abnormal features, such as increased anxiety, increased locomotor activity, cognitive deficits in different behavioral tasks, and deficient prepulse inhibition (see reviews [90, 91]).
At the neuronal level, the brain of young Fmr1 KO mice shows increased spine density and length, and increased immature, thin spines are observed in the brain of adult mice [95]. This parallels the findings from postmortem analyses of the brain from individuals with FXS [96]. FMRP depletion also affects synaptic function, such as LTP and LTD, involving mGluR. An analysis of synaptic plasticity in Fmr1 KO mice revealed that LTD triggered by mGluR was enhanced, whereas NMDA-dependent LTD was normal [97], leading to the proposal of “mGluR theory of FXS” [98]. In line with this theory, Fmr1 KO mice exhibited persistent of mGluR-mediated LTD [99] and deficient LTP [100]. The causal role of mGluR5 in the FXS phenotype was suggested by the observation that the mGluR5 negative allosteric modulator MPEP inhibited audiogenic seizures in the mutant [101]. An analysis of Fmr1 KO mice crossed with Gmr5+/− mice, mutants that express half the normal level of mGluR5, demonstrated the critical role of enhanced mGluR5 activity in a wide range of neurological symptoms of FXS [102]. Increased dendritic spine density and elevated levels of protein synthesis in the hippocampus of Fmr1 KO mice was rescued by reducing mGluR5 expression. These findings were accompanied by the reversal of exaggerated memory extinction and susceptibility to audiogenic seizures [102]. Thus “mGluR theory of FXS” is now widely accepted to explain the diverse neurological phenotype in FXS [103]. Moreover, inhibitors of mGluR5 are being investigated as candidate agents to improve cognitive deficits in human FXS [104].
Recently, attention is being paid to the link between the FMRP and mTORC1 pathway as another pathomechanism in neurocognitive deficits in FXS. Increased levels of phosphorylated S6K1, indicating hyperactivation of mTORC1, were found in the hippocampus of Fmr1 KO mice [88]. In line with this, evidence of mTORC1 activation was also identified in fibroblasts, lymphocytes, and postmortem brain tissue of human individuals with FXS [105, 106]. Accumulating evidence suggests a connection between the deletion of FMRP and deregulation of mTROC1 in FXS. Upon FMRP depletion, exaggerated activity of PI3K and Akt was found in Fmr1 KO mice, suggesting a pathological role of excess signaling through the PI3K/Akt pathway to enhance mTORC1 activity [107]. Increased expression of the FMRP binding protein CYFIP1 was another finding in Fmr1 KO mice to link FXS to excess protein synthesis via eIF4E [108]. To test whether mTORC1 activation contributes to the development of ASD-like phenotype in FXS, S6K1 KO mice were crossed to Fmr1 KO mice to genetically suppress mTORC1-mediated protein synthesis. The Fmr1-S6K1 double mutants displayed a reversal in excessive protein synthesis and autistic-like behavioral deficits in Fmr1 KO mice [109]. Fmr1 KO mice exhibited increased glycogen synthase kinase 3 (GSK3) activity that lowers TSC1/2 complex activity, which likely results in mTORC1 activation [110, 111]. The pathogenic role of GSK3 activation in autistic-like deficient social recognition of Fmr1 KO mice was demonstrated by pharmacological and genetic intervention. The abnormal behavior was improved by lithium, which acts as a GSK3 inhibitor, and also corrected by knocking-in the Gsk3b gene with an FMRP-resistant inactive mutation [110, 111]. Another example for discussing the relationship between mGluR5 and mTORC1 is the BTBR T + tf/J (BTBR) inbred strain of mice. This strain of mouse has autistic-like behavioral abnormalities that were rescued by an mGluR5 antagonist, as seen in Fmr1 KO mice [112]. Strikingly, rapamycin treatment also alleviated impaired social behaviors of BTBR mice [113]. The preclinical evidence suggests the therapeutic possibility of mTOR inhibitors to correct autistic-like behavioral features in FXS.
Neurofibromatosis Type 1
NF1 (MIN#162200) is an autosomal-dominant and common neurocutaneous disorder (a group of diseases whose features primarily include the brain and skin) that occurs in 1 in 2,500–3,000 live births [114]. A clinical diagnosis of NF1 is made depending on the criteria that mainly require various skin manifestations [27]. Detailed information about neurocognitive features is not required for the diagnosis although the entire phenotype is much more diverse than described in the criteria [114]. With respect to neurological symptoms, NF1 individuals frequently show signs of visuospatial and visuomotor deficits, attention deficit/hyperactivity disorder, and ASD. According to recent epidemiological studies, the frequency of ASD in NF1 is estimated to be as high as 15%–30% [43] [115-117]. Based on these observations, NF1 is now one of the most prevalent causes of monogenic ASD.
With respect to signaling cascades, NF1 is a RASopathy, a set of disorders associated with deregulation of the rat sarcoma (RAS)/MAPK pathway. Other RASopathies include Noonan syndrome and Costello syndrome [118]. This pathway begins with the activation of receptor tyrosine kinase and downstream RAS proteins (H-RAS, K-RAS, N-RAS). This then stimulates mitogen-activated protein kinase kinase and extracellular signal-regulated kinase (ERK). The TSC1/2 complex, which is located downstream of ERK, receives inhibitory inputs from ERK. In this pathway, NF1 works as a negative regulator of RAS proteins. The heterozygous loss of the NF1 gene abolishes the inhibition of the RAS signaling pathway and results in constitutive activation of ERK1/2 and mTORC1 [120, 121], which are considered as the central mechanism underlying NF1 (Fig. 1). With respect to mTORC1, NF1 has another major function as a repressor of PI3K/Akt pathway. Analysis of NF1-deleted primary cells revealed that the NF1 loss was associated with enhanced activity of Akt and S6K1. The PI3K inhibitor wortmannin inhibited both Akt and S6K1, whereas rapamycin suppressed S6K1 but not Akt [119], suggesting that NF1 deletion affects different signaling pathways that converge on the common effector, mTORC1 (Fig. 1).
Mice with a heterozygous loss in the Nf1 gene show neurocognitive deficits compatible with those of human NF1 individuals (Table 1): spatial learning deficits [121], attention deficits [120], and impaired discrimination of social stimuli relevant to ASD [122]. These behavioral changes in mice are associated with the activation of ERK and Akt, both of which enhance signal transmission mediated by mTORC1 [119, 120]. With this regard, Nf1+/− mice have decreased hippocampal LTP, both in the early and late phases [121, 123]. Studies using several lines of Nf1 conditional KO mice suggest that neurocognitive features of NF1 are centrally attributable to neuronal loss of the gene and consequent increases in GABA release [124]. Lovastatin, a farnesylation inhibitor that negatively acts on RAS and ERK, reverses the behavioral deficits seen in Nf1+/− mice [120, 121], and GABA receptor antagonists also restored hippocampal LTP in the mutants [121]. This body of evidence led to clinical trials evaluating the neurocognitive effects of the drugs. In spite of clear preclinical findings, individuals with NF1 showed minimal beneficial changes after treatment with simvastatin, whereas the participants with intellectual disabilities responded substantially [125]. To effectively prevent abnormal signaling in NF1, a molecular target that is even more downstream in the signaling pathway may need to be identified and studied. Considering that both the RAS/MAPK and PI3K/Akt pathways finally converge on mTORC1, the evaluation of mTOR inhibitors for neurocognitive deficits will be anticipated.
mTORC1 TARGET PROTEINS THAT INCREASE AUTISTIC-LIKE PHENOTYPES
There is a large body of evidence showing that the heterozygous loss of different genes, TSC 1/2, PTEN, FMR1, and NF1, is linked to ASD through unregulated mTORC1-mediated signal transduction. The next question is what actually causes ASD when mTORC1 is unregulated. One possible explanation is that mTORC1 activation suppresses 4E-BPs and subsequent cap-dependent mRNA translation is elevated (Fig. 1). 4E-BPs normally inhibit translation initiation by repressing eIF4F complex formation [126]. The complex includes eIF4E (the cap-binding protein), eIF4A (the RNA helicase), and eIF4G (the scaffolding protein bridging RNA to ribosome) [127]. eIF4E preferentially promotes a set of mRNAs with extensive secondary structure [124, 128]. The causal role of eIF4E in ASD was first pointed out by the discovery of individuals with ASD whose eIF4E gene had an activating mutation in its promoter region [129]. Further studies revealed that the 4E-BP2 KO and eIF4E overexpression generated an autistic-like behavioral manifestation, impaired social approach, and repetitive behaviors in mice [130, 131]. The ASD-like behavioral alteration was mediated by enhanced cap-dependent translation and protein synthesis, resulting in an imbalance in excitatory/inhibitory transmission and mGluR-LTD [130, 131]. Furthermore, pharmacological inhibition of this cap-dependent translation using eIF4E/eIF4G interaction inhibitor 4EGI-1 rescued the behavioral, translational, and synaptic deficits [119, 120], emphasizing the pathogenic role of exaggerated cap-dependent mRNA translation in ASD.
The development of ASD via 4EBP/eIF4E-mediated gene translation may be attributable to the expression of specific genes. The deletion of the 4E-BP2 gene preferentially enhanced mRNA translation of synaptic proteins neuroligins (Nlgns), and its significance was demonstrated by the observation that Nlgn1 knockdown alleviated autistic-like behaviors in 4E-BP2 KO mice [130]. The causal role of altered 4E-BP/eIF4E-mediated translational control in ASD is further supported by the loss of FMRP/CYFIP1-mediated translational repression that results in eIF4E-dependent translation in FXS [93]. Another example is the matrix metallopeptidase 9 (MMP9) gene. Fmr1 KO mice exhibit overexpression of the Mmp9 gene dependent on FMRP and eIF4E and pharmacological suppression of Mmp9 translation rescued FXS-like phenotypes [121].
Altered mGluR5-LTD is another clue to connect mTORC1 activation to ASD. The idea derives from the observation that the mouse FXS phenotype was rescued by two ways: genetic suppression of mGluR5 expression [102] and genetic deletion of S6K1 mimicking mTORC1 inhibition [109]. The evidence in Fmr1 KO mice led to the speculation that mTORC1 activation causes ASD via excess mGluR5-LTD. However, it is controversial whether mTORC1 activation exaggerates or attenuates mGluR5-LTD. In one study, Tsc2+/− mice exhibited reduced mGluR5-LTD and impaired memory. This deficit was rescued pharmacologically by mTORC1 inhibition and mGluR5 activation and furthermore genetically by crossing with an Fmr1 KO mice [62]. In contrast, in another report in which Tsc2+/− mice were examined, the deficient mGluR5-LTD was observed in the juvenile period, but it was restored in adulthood. Moreover, the learning deficit and perseverative behavior of the mice improved in response to mGluR5 inhibition [132, 133]. Further research is mandatory to overcome these conflicting findings and to better understand the pathophysiology of TSC with respect to mGluR5-LTD.
Pharmacological interventions for MTORC1-related ASD
Based on the above discussion emphasizing the pathogenic role of mTORC1 dysregulation, mTOR inhibitors are expected to ameliorate ASD or cognitive impairment in selected sets of individuals. This issue is addressed most intensively in TSC. In the clinical trial of sirolimus for renal angiomyolipomas accompanied by TSC and lymphangiomyomatosis with or without TSC, immediate recall memory and executive function of the participants showed a substantial improvement after sirolimus treatment [134]. Everolimus reduced seizure frequency in individuals with TSC with or without subependymal giant cell astrocytoma. Moreover, quality-of-life scores, particularly on items related to ASD symptoms, significantly increased after the treatment [135-137]. Based on these promising results, several clinical trials are under way testing mTOR inhibitors for neurocognitive deficits in TSC [104]. Considering the large body of preclinical evidence reviewed above, other disorders that are associated with ASD and increased mTORC1 activity may benefit from mTOR inhibitors, such as FXS, NF1, and macrocephaly/autism syndrome with the PTEN mutation. Clinical investigation is anticipated to examine the therapeutic efficiency of mTOR inhibitors for mTORC1-related ASD in these disorders.
Conclusion
Constitutive mTORC1 activation observed in single-gene, ASD-associated disorders TSC, PHTS, FXS, and NF1 offers insight in understanding the as of yet unclear molecular pathophysiology of ASD. Rodent models of these disorders display autism-related behavioral phenotype and morphological and electrophysiological abnormalities. Evidence is accumulating that mTOR inhibitors mostly rescue these neuronal and behavioral alterations, suggesting the possibility of pharmacological intervention of mTORC1-associated ASD using mTOR inhibitors. On the basis of this preclinical evidence, new translational research will be expected to elucidate the therapeutic efficacy of mTOR inhibitors for ASD in the near future.
ACKNOWLEDGEMENTS
The authors greatly appreciate Masashi Mizuguchi (The University of Tokyo) for critical advice to this article. The authors would like to thank Enago (www.enago.jp) for the English language review. This work was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI Grant Number 26860836 and by the Practical Research Project for Rare/Intractable Diseases Program from Japan Agency for Medical Research and Development [H27-Itaku(Nan)-Ippan-015].
List of Abbreviations
- ASD
Autism Spectrum Disorder
- eIF4E
Eukaryotic Initiation Factor 4E
- 4E-BP
Eukaryotic Initiation Factor 4E-Binding Protein
- FMRP
Fragile X Mental Retardation Protein
- FXS
Fragile X Syndrome
- KO
Knockout
- LTD
Long Term Depression
- LTP
Long Term Potentiation
- mGluR
Metabotropic Glutamate Receptor
- mTOR
Mammalian Target of Rapamycin
- mTORC1
Mammalian Target of Rapamycin Complex 1
- NF1
Neurofibromatosis Type 1
- NSE
Neuron Specific Enolase
- PTEN
Phosphatase and Tensin Homolog
- Rheb
Ras Homolog Enriched in Brain
- S6K
Ribosomal Protein S6 Kinase
- TSC
Tuberous Sclerosis Complex
CONFLICT OF INTEREST
The author confirms that this article content has no conflict of interest.
REFERENCES
- 1.Moschetta M., Reale A., Marasco C., Vacca A., Carratù M.R. Therapeutic targeting of the mTOR-signalling pathway in cancer: benefits and limitations. Br. J. Pharmacol. 2014;171:3801–3813. doi: 10.1111/bph.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell J.D., Pollizzi K.N., Heikamp E.B., Horton M.R. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong Z.Z., Shang Y.C., Wang S., Maiese K. A Critical Kinase Cascade in Neurological Disorders: PI 3-K, Akt, and mTOR. Future Neurol. 2012;7:733–748. doi: 10.2217/fnl.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 5th ed. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- 6.Perou R., Bitsko R.H., Blumberg S.J., et al. Mental health surveillance among children--United States, 2005-2011. MMWR Surveill. Summ. 2013;62:1–35. [PubMed] [Google Scholar]
- 7.Liu X., Takumi T. Genomic and genetic aspects of autism spectrum disorder. Biochem. Biophys. Res. Commun. 2014;452:244–253. doi: 10.1016/j.bbrc.2014.08.108. [DOI] [PubMed] [Google Scholar]
- 8.Kim D.H., Sarbassov D.D., Ali S.M., et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 9.Hara K., Maruki Y., Long X., et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 10.Jacinto E., Loewith R., Schmidt A., et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 11.Sarbassov D.D., Ali S.M., Kim D.H., et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 12.van Slegtenhorst M., Nellist M., Nagelkerken B., et al. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum. Mol. Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- 13.Plank T.L., Yeung R.S., Henske E.P. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998;58:4766–4770. [PubMed] [Google Scholar]
- 14.Wienecke R., König A., DeClue J.E. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. J. Biol. Chem. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- 15.Xiao G.H., Shoarinejad F., Jin F., Golemis E.A., Yeung R.S. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J. Biol. Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- 16.Benvenuto G., Li S., Brown S.J., et al. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 2000;19:6306–6316. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- 17.Ohsawa M., Kobayashi T., Okura H., Igarashi T., Mizuguchi M., Hino O. TSC1 controls distribution of actin fibers through its effect on function of Rho family of small GTPases and regulates cell migration and polarity. PLoS One. 2013;8:e54503. doi: 10.1371/journal.pone.0054503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 19.Tavares M.R., Pavan I.C., Amaral C.L., Meneguello L., Luchessi A.D., Simabuco F.M. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Poulin F., Gingras A.C., Olsen H., Chevalier S., Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 21.Sund R., Pukkala E., Pause A., et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 22.Gingras A.C., Raught B., Gygi S.P., et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moerke N.J., Aktas H., Chen H., et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 24.Chang Y.Y., Neufeld T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganley I.G. Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Northrup H., Krueger D.A., International Tuberous Sclerosis Complex Consensus Group Tuberous sclerosis complex diagnostic criteria update: recommendations of the 2012 Iinternational Tuberous Sclerosis Complex Consensus Conference. Pediatr. Neurol. 2013;49:243–254. doi: 10.1016/j.pediatrneurol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Institutes of Health Consensus Development Conference Statement: neurofibromatosis—Bethesda, MD, USA, July 13–15, 1987. Neurofibromatosis. 1988;1:172–178. [PubMed] [Google Scholar]
- 28.Blumenthal G.M., Dennis P.A. PTEN hamartoma tumor syndromes. Eur. J. Hum. Genet. 2008;16:1289–1300. doi: 10.1038/ejhg.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patja K. Cancer incidence among persons with fragile X syndrome in Finland: a population-based study. J. Intellect. Disabil. Res. 2009;53:85–90. doi: 10.1111/j.1365-2788.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 30.Bournevell D.M. Sclerose tubereuse des circonvolutions cerebrales: idiotie et epilepsie hemiplegique. Arch. Neurol. 1880;1:81–91. [Google Scholar]
- 31.van Slegtenhorst M., de Hoogt R., Hermans C., et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 32.European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 33.Curatolo P., Bombardieri R., Jozwiak S. Tuberous sclerosis. Lancet. 2008;372:657–668. doi: 10.1016/S0140-6736(08)61279-9. [DOI] [PubMed] [Google Scholar]
- 34.Roach E.S., Gomez M.R., Northrup H. Tuberous sclerosis complex consensus conference: revised clinical diagnostic criteria. J. Child Neurol. 1998;13:624–628. doi: 10.1177/088307389801301206. [DOI] [PubMed] [Google Scholar]
- 35.Jones A.C., Daniells C.E., Snell R.G., et al. Molecular genetic and phenotypic analysis reveals differences between TSC1 and TSC2 associated familial and sporadic tuberous sclerosis. Hum. Mol. Genet. 1997;6:2155–2161. doi: 10.1093/hmg/6.12.2155. [DOI] [PubMed] [Google Scholar]
- 36.Dabora S.L., Jozwiak S., Franz D.N., et al. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am. J. Hum. Genet. 2001;68:64–80. doi: 10.1086/316951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis J.C., Thomas H.V., Murphy K.C., Sampson J.R. Genotype and psychological phenotype in tuberous sclerosis. J. Med. Genet. 2004;41:203–207. doi: 10.1136/jmg.2003.012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Numis A.L., Major P., Montenegro M.A., Muzykewicz D.A., Pulsifer M.B., Thiele E.A. Identification of risk factors for autism spectrum disorders in tuberous sclerosis complex. Neurology. 2011;76:981–987. doi: 10.1212/WNL.0b013e3182104347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu-Shore C.J., Major P., Camposano S., Muzykewicz D., Thiele E.A. The natural history of epilepsy in tuberous sclerosis complex. Epilepsia. 2010;51:1236–1241. doi: 10.1111/j.1528-1167.2009.02474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries P.J., Hunt A., Bolton P.F. The psychopathologies of children and adolescents with tuberous sclerosis complex (TSC): a postal survey of UK families. Eur. Child Adolesc. Psychiatry. 2007;16:16–24. doi: 10.1007/s00787-006-0570-3. [DOI] [PubMed] [Google Scholar]
- 41.Krueger D.A., Northrup H., International Tuberous Sclerosis Complex Consensus Group Tuberous sclerosis complex surveillance and management: recommendations of the 2012 International Tuberous Sclerosis Complex Consensus Conference. Pediatr. Neurol. 2013;49:255–265. doi: 10.1016/j.pediatrneurol.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries P.J., Whittemore V.H., Leclezio L., et al. Tuberous sclerosis associated neuropsychiatric disorders (TAND) and the TAND Checklist. Pediatr. Neurol. 2015;52:25–35. doi: 10.1016/j.pediatrneurol.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards C., Jones C., Groves L., Moss J., Oliver C. Prevalence of autism spectrum disorder phenomenology in genetic disorders: a systematic review and meta-analysis. Lancet Psychiatry. 2015;2(10):909–916. doi: 10.1016/S2215-0366(15)00376-4. [DOI] [PubMed] [Google Scholar]
- 44.Caglayan A.O. Genetic causes of syndromic and non-syndromic autism. Dev. Med. Child Neurol. 2010;52:130–138. doi: 10.1111/j.1469-8749.2009.03523.x. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi T., Minowa O., Sugitani Y., et al. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc. Natl. Acad. Sci. USA. 2001;98:8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi T., Minowa O., Kuno J., Mitani H., Hino O., Noda T. Renal carcinogenesis, hepatic hemangiomatosis, and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 1999;59:1206–1211. [PubMed] [Google Scholar]
- 47.Kobayashi T., Hirayama Y., Kobayashi E., Kubo Y., Hino O. A germline insertion in the tuberous sclerosis (Tsc2) gene gives rise to the Eker rat model of dominantly inherited cancer. Nat. Genet. 1995;9:70–74. doi: 10.1038/ng0195-70. [DOI] [PubMed] [Google Scholar]
- 48.Uhlmann E.J., Wong M., Baldwin R.L., et al. Astrocyte-specific TSC1 conditional knockout mice exhibit abnormal neuronal organization and seizures. Ann. Neurol. 2002;52:285–296. doi: 10.1002/ana.10283. [DOI] [PubMed] [Google Scholar]
- 49.Way S.W., McKenna J., III, Mietzsch U., Reith R.M., Wu H.C., Gambello M.J. Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse. Hum. Mol. Genet. 2009;18:1252–1265. doi: 10.1093/hmg/ddp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai P.T., Hull C., Chu Y., et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reith R.M., McKenna J., Wu H., et al. Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol. Dis. 2013;51:93–103. doi: 10.1016/j.nbd.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 52.Goorden S.M., van Woerden G.M., van der Weerd L., Cheadle J.P., Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann. Neurol. 2007;62:648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- 53.Sato A., Kasai S., Kobayashi T., et al. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat. Commun. 2012;3:1292. doi: 10.1038/ncomms2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waltereit R., Japs B., Schneider M., de Vries P.J., Bartsch D. Epilepsy and Tsc2 haploinsufficiency lead to autistic-like social deficit behaviors in rats. Behav. Genet. 2011;41:364–372. doi: 10.1007/s10519-010-9399-0. [DOI] [PubMed] [Google Scholar]
- 55.Ehninger D., Han S., Shilyansky C., et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat. Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng L.H., Rensing N.R., Zhang B., Gutmann D.H., Gambello M.J., Wong M. Tsc2 gene inactivation causes a more severe epilepsy phenotype than Tsc1 inactivation in a mouse model of tuberous sclerosis complex. Hum. Mol. Genet. 2011;20:445–454. doi: 10.1093/hmg/ddq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavazoie S.F., Alvarez V.A., Ridenour D.A., Kwiatkowski D.J., Sabatini B.L. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat. Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- 58.Nie D., Di Nardo A., Han J.M., et al. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat. Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang G., Gudsnuk K., Kuo S.H., et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Nardo A., Wertz M.H., Kwiatkowski E., et al. Neuronal Tsc1/2 complex controls autophagy through AMPK-dependent regulation of ULK1. Hum. Mol. Genet. 2014;23:3865–3874. doi: 10.1093/hmg/ddu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng L.H., Ouyang Y., Gazit V., et al. Abnormal glutamate homeostasis and impaired synaptic plasticity and learning in a mouse model of tuberous sclerosis complex. Neurobiol. Dis. 2007;28:184–196. doi: 10.1016/j.nbd.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auerbach B.D., Osterweil E.K., Bear M.F. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bateup H.S., Takasaki K.T., Saulnier J.L., Denefrio C.L., Sabatini B.L. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J. Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bateup H.S., Johnson C.A., Denefrio C.L., Saulnier J.L., Kornacker K., Sabatini B.L. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78:510–522. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Courchesne E., Karns C.M., Davis H.R., et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57:245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- 66.Butler M.G., Dasouki M.J., Zhou X.P., et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varga E.A., Pastore M., Prior T., Herman G.E., McBride K.L. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet. Med. 2009;11:d11–d17. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 68.McBride K.L., Varga E.A., Pastore M.T., et al. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3:137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- 69.Marsh D.J., Kum J.B., Lunetta K.L., et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum. Mol. Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 70.Zhou X.P., Marsh D.J., Morrison C.D., et al. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. Am. J. Hum. Genet. 2003;73:1191–1198. doi: 10.1086/379382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sulis M.L., Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 72.Suzuki A., de la Pompa J.L., Stambolic V., et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 73.Podsypanina K., Ellenson L.H., Nemes A., et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwon C.H., Luikart B.W., Powell C.M., et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou J., Blundell J., Ogawa S., et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J. Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Page D.T., Kuti O.J., Prestia C., Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc. Natl. Acad. Sci. USA. 2009;106:1989–1994. doi: 10.1073/pnas.0804428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Napoli E., Ross-Inta C., Wong S., et al. Mitochondrial dysfunction in Pten haplo-insufficient mice with social deficits and repetitive behavior: interplay between Pten and p53. PLoS One. 2012;7:e42504. doi: 10.1371/journal.pone.0042504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clipperton-Allen A.E., Page D.T. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum. Mol. Genet. 2014;23:3490–3505. doi: 10.1093/hmg/ddu057. [DOI] [PubMed] [Google Scholar]
- 79.Turner G., Webb T., Wake S., Robinson H. Prevalence of fragile X syndrome. Am. J. Med. Genet. 1996;64:196–197. doi: 10.1002/(SICI)1096-8628(19960712)64:1<196::AID-AJMG35>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 80.Kidd S.A., Lachiewicz A., Barbouth D., et al. Fragile X syndrome: a review of associated medical problems. Pediatrics. 2014;134:995–1005. doi: 10.1542/peds.2013-4301. [DOI] [PubMed] [Google Scholar]
- 81.Verkerk A.J., Pieretti M., Sutcliffe J.S., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 82.Naumann A., Hochstein N., Weber S., Fanning E., Doerfler W. A distinct DNA-methylation boundary in the 5'- upstream sequence of the FMR1 promoter binds nuclear proteins and is lost in fragile X syndrome. Am. J. Hum. Genet. 2009;85:606–616. doi: 10.1016/j.ajhg.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brouwer J.R., Willemsen R., Oostra B.A. The FMR1 gene and fragile X-associated tremor/ataxia syndrome. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2009;150B:782–798. doi: 10.1002/ajmg.b.30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bourgeois J.A., Seritan A.L., Casillas E.M., et al. Lifetime prevalence of mood and anxiety disorders in fragile X premutation carriers. J. Clin. Psychiatry. 2011;72:175–182. doi: 10.4088/JCP.09m05407blu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hunter J., Rivero-Arias O., Angelov A., Kim E., Fotheringham I., Leal J. Epidemiology of fragile X syndrome: a systematic review and meta-analysis. Am. J. Med. Genet. A. 2014;164A:1648–1658. doi: 10.1002/ajmg.a.36511. [DOI] [PubMed] [Google Scholar]
- 86.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 87.Darnell J.C., Mostovetsky O., Darnell R.B. FMRP RNA targets: Identification and validation. Genes Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 88.Sharma A., Hoeffer C.A., Takayasu Y., et al. Dysregulation of mTOR signaling in fragile X syndrome. J. Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bakker C.E., Verheij C., Willemsen R., et al. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 90.Bernardet M., Crusio W.E. Fmr1 KO mice as a possible model of autistic features. ScientificWorldJournal. 2006;6:1164–1176. doi: 10.1100/tsw.2006.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kazdoba T.M., Leach P.T., Silverman J.L., Crawley J.N. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis. Res. 2014;3:118–133. doi: 10.5582/irdr.2014.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dahlhaus R., El-Husseini A. Altered neuroligin expression is involved in social deficits in a mouse model of the fragile X syndrome. Behav. Brain Res. 2010;208:96–105. doi: 10.1016/j.bbr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 93.McNaughton C.H., Moon J., Strawderman M.S., Maclean K.N., Evans J., Strupp B.J. Evidence for social anxiety and impaired social cognition in a mouse model of fragile X syndrome. Behav. Neurosci. 2008;122:293–300. doi: 10.1037/0735-7044.122.2.293. [DOI] [PubMed] [Google Scholar]
- 94.Pietropaolo S., Guilleminot A., Martin B., D'Amato F.R., Crusio W.E. Genetic-background modulation of core and variable autistic-like symptoms in Fmr1 knock-out mice. PLoS One. 2011;6:e17073. doi: 10.1371/journal.pone.0017073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grossman A.W., Elisseou N.M., McKinney B.C., Greenough W.T. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res. 2006;1084:158–164. doi: 10.1016/j.brainres.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 96.Rudelli R.D., Brown W.T., Wisniewski K., et al. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol. 1985;67:289–295. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- 97.Huber K.M., Gallagher S.M., Warren S.T., Bear M.F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bear M.F., Huber K.M., Warren S.T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 99.Nosyreva E.D., Huber K.M. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J. Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- 100.Seese R.R., Babayan A.H., Katz A.M., et al. LTP induction translocates cortactin at distant synapses in wild-type but not Fmr1 knock-out mice. J. Neurosci. 2012;32:7403–7413. doi: 10.1523/JNEUROSCI.0968-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yan Q.J., Rammal M., Tranfaglia M., Bauchwitz R.P. Suppression of two major Fragile X Syndrome mouse model phenotypes by the mGluR5 antagonist MPEP. Neuropharmacology. 2005;49:1053–1066. doi: 10.1016/j.neuropharm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 102.Dölen G., Osterweil E., Rao B.S., et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bhakar A.L., Dölen G., Bear M.F. The pathophysiology of fragile X (and what it teaches us about synapses). Annu. Rev. Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lipton J.O., Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoeffer C.A., Sanchez E., Hagerman R.J., et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 2012;11:332–341. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumari D., Bhattacharya A., Nadel J., et al. Identification of fragile X syndrome specific molecular markers in human fibroblasts: a useful model to test the efficacy of therapeutic drugs. Hum. Mutat. 2014;35:1485–1494. doi: 10.1002/humu.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gross C., Nakamoto M., Yao X., et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J. Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Napoli I., Mercaldo V., Boyl P.P., et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 109.Bhattacharya A., Kaphzan H., Alvarez-Dieppa A.C., Murphy J.P., Pierre P., Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Min W.W., Yuskaitis C.J., Yan Q., et al. Elevated glycogen synthase kinase-3 activity in Fragile X mice: key metabolic regulator with evidence for treatment potential. Neuropharmacology. 2009;56:463–472. doi: 10.1016/j.neuropharm.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mines M.A., Yuskaitis C.J., King M.K., Beurel E., Jope R.S. GSK3 influences social preference and anxiety-related behaviors during social interaction in a mouse model of fragile X syndrome and autism. PLoS One. 2010;5:e9706. doi: 10.1371/journal.pone.0009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silverman J.L., Smith D.G., Rizzo S.J., et al. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci. Transl. Med. 2012;4:131ra51. doi: 10.1126/scitranslmed.3003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burket J.A., Benson A.D., Tang A.H., Deutsch S.I. Rapamycin improves sociability in the BTBR T(+)Itpr3(tf)/J mouse model of autism spectrum disorders. Brain Res. Bull. 2014;100:70–75. doi: 10.1016/j.brainresbull.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hirbe A.C., Gutmann D.H. Neurofibromatosis type 1: a multidisciplinary approach to care. Lancet Neurol. 2014;13:834–843. doi: 10.1016/S1474-4422(14)70063-8. [DOI] [PubMed] [Google Scholar]
- 115.Acosta M.T., Gioia G.A., Silva A.J. Neurofi bromatosis type 1: new insights into neurocognitive issues. Curr. Neurol. Neurosci. Rep. 2006;6:136–143. doi: 10.1007/s11910-996-0036-5. [DOI] [PubMed] [Google Scholar]
- 116.Walsh K.S., Vélez J.I., Kardel P.G., et al. Symptomatology of autism spectrum disorder in a population with neurofibromatosis type 1. Dev. Med. Child Neurol. 2013;55:131–138. doi: 10.1111/dmcn.12038. [DOI] [PubMed] [Google Scholar]
- 117.Garg S., Lehtonen A., Huson S.M., et al. Autism and other psychiatric comorbidity in neurofibromatosis type 1: evidence from a population-based study. Dev. Med. Child Neurol. 2013;55:139–145. doi: 10.1111/dmcn.12043. [DOI] [PubMed] [Google Scholar]
- 118.Abramowicz A., Gos M. Neurofibromin in neurofibromatosis type 1 - mutations in NF1 gene as a cause of disease. Dev Period Med. 2014;18:297–306. [PubMed] [Google Scholar]
- 119.Johannessen C.M., Reczek E.E., James M.F., Brems H., Legius E., Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. USA. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li W., Cui Y., Kushner S.A., et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr. Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 121.Costa R.M., Federov N.B., Kogan J.H., et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 122.Molosh A.I., Johnson P.L., Spence J.P., et al. Social learning and amygdala disruptions in Nf1 mice are rescued by blocking p21-activated kinase. Nat. Neurosci. 2014;17:1583–1590. doi: 10.1038/nn.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guilding C., McNair K., Stone T.W., Morris B.J. Restored plasticity in a mouse model of neurofibromatosis type 1 via inhibition of hyperactive ERK and CREB. Eur. J. Neurosci. 2007;25:99–105. doi: 10.1111/j.1460-9568.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- 124.Cui Y., Costa R.M., Murphy G.G., et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krab L.C., de Goede-Bolder A., Aarsen F.K., et al. Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: a randomized controlled trial. JAMA. 2008;300:287–294. doi: 10.1001/jama.300.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pause A., Belsham G.J., Gingras A.C., et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 127.Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 128.Koromilas A.E., Lazaris-Karatzas A., Sonenberg N. mRNAs containing extensive secondary structure in their 59 non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:41538. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Neves-Pereira M., Müller B., Massie D., et al. Deregulation of EIF4E: a novel mechanism for autism. J. Med. Genet. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- 130.Gkogkas C.G., Khoutorsky A., Ran I., et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Santini E., Huynh T.N., MacAskill A.F., et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gkogkas C.G., Khoutorsky A., Cao R., et al. Pharmacogenetic inhibition of eIF4E-dependent Mmp9 mRNA translation reverses fragile X syndrome-like phenotypes. Cell Reports. 2014;9:1742–1755. doi: 10.1016/j.celrep.2014.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Potter W.B., Basu T., O'Riordan K.J., et al. Reduced juvenile long-term depression in tuberous sclerosis complex is mitigated in adults by compensatory recruitment of mGluR5 and Erk signaling. PLoS Biol. 2013;11:e1001627. doi: 10.1371/journal.pbio.1001627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Davies D.M., de Vries P.J., Johnson S.R., et al. Sirolimus therapy for angiomyolipoma in tuberous sclerosis and sporadic lymphangioleiomyomatosis: a phase 2 trial. Clin. Cancer Res. 2011;17:4071–4081. doi: 10.1158/1078-0432.CCR-11-0445. [DOI] [PubMed] [Google Scholar]
- 135.Krueger D.A., Care M.M., Holland K., et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N. Engl. J. Med. 2010;363:1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]
- 136.Krueger D.A., Wilfong A.A., Holland-Bouley K., et al. Everolimus treatment of refractory epilepsy in tuberous sclerosis complex. Ann. Neurol. 2013;74:679–687. doi: 10.1002/ana.23960. [DOI] [PubMed] [Google Scholar]
- 137.Franz D.N., Agricola K., Mays M., et al. Everolimus for subependymal giant cell astrocytoma: 5-year final analysis. Ann. Neurol. 2015;78(6):929–938. doi: 10.1002/ana.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]