Abstract
The fluctuation theorem (FT), which is a recent achievement in non-equilibrium statistical mechanics, has been suggested to be useful for measuring the driving forces of motor proteins. As an example of this application, we performed single-molecule experiments on F1-ATPase, which is a rotary motor protein, in which we measured its rotary torque by taking advantage of FT. Because fluctuation is inherent nature in biological small systems and because FT is a non-destructive force measurement method using fluctuation, it will be applied to a wide range of biological small systems in future.
Keywords: motor protein, fluctuation, non-equilibrium statistical mechanics
Fluctuation Theorem (FT)
A macroscopic motor generates heat, Q, when energy to move the motor is injected. Defining an entropy production, σ, generated during a time interval, Δt, by σ = Q/kBT where kB is the Boltzmann constant and T is the temperature of the environment, we find that σ > 0. This empirical rule for a macroscopic system is called the second law of thermodynamics. However is σ > 0 always observed for small systems such as motor proteins?
Before examining motor proteins, for simplicity, we considered a charged colloidal particle in water driven by an electric field (Fig. 1a). While motor proteins use energy from chemical reactions to move, the particle is driven by the force, F, exerted by the electric field. The position of the particle is denoted as X(t). Heat, Q, generated during a time interval, Δt, is described by Q=FΔX, where ΔX=X(t+Δt)−X(t). Because X(t) fluctuates in time due to thermal noise, Q has a diffierent value for each measurement. Q>0 when the particle moves in the same direction as that of the force while Q<0 when the particle moves against the force by being pushed by water molecules (which can happen if Δt is small) (Fig. 1b). In the latter case σ < 0. This apparent violation of the second law of thermodynamics is expressed by the fluctuation theorem (FT):
| (1) |
where P(σ) is the probability distribution of σ. Because Eq. (1) implies that the probability of σ > 0 exponentially increases when the mean value of σ becomes large, which is the case in a macroscopic system, Eq. (1) coincides with the second law of thermodynamics. FT was first advocated by Evans et al. in 19931, and it was experimentally verified in a colloidal particle system in 20022. Theoretically, many mathematical expressions of FT can be derived depending on system conditions3. Among them, the Crooks fluctuation theorem was experimentally verified in an RNA hairpin system4.
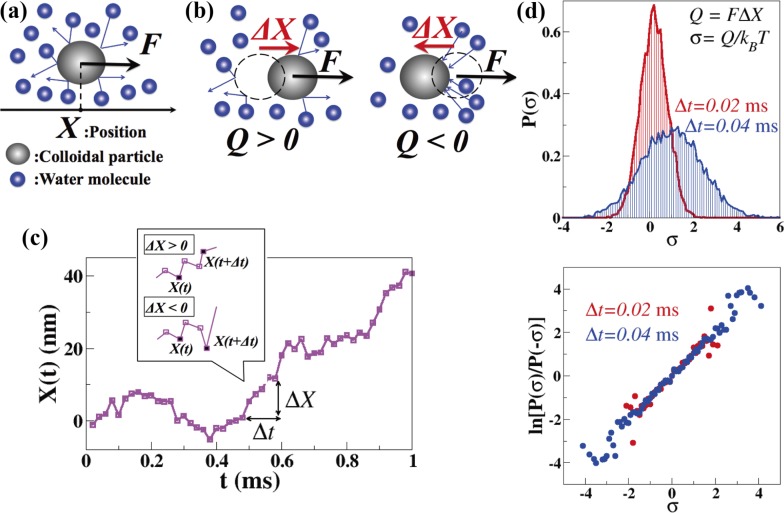
Figure 1.
FT verified using simulation. (a) Schematic of a charged colloidal particle driven by an electric field. (b) Heat, Q, generated during Δt. (c) A sample time course, X(t), obtained by the simulation. (d) (top) Probability distribution, P(σ), of the entropy production, σ, in the cases Δt=0.02 ms (red) and Δt=0.04 ms (blue). (bottom) ln[P(σ)/P(−σ)] is plotted as a function of σ in the cases Δt=0.02 ms (red) and Δt=0.04 ms (blue). It is seen that ln[P(σ)/P(−σ)]=σ.
Theory and Simulation
To check Eq. (1), we performed a simulation of the colloidal particle system depicted in Figure 1a. Because the effect of the inertia is small in our colloidal particle system, the time evolution of X(t) is described by the overdamped Langevin equation:
| (2) |
where Γ is the friction coefficient of the particle, and ξ is Gaussian noise of the intensity , which represents the effect of thermal noise. Γ=6πηa (Stokes law) where η is the viscosity of water, and a is the radius of the particle5. In Figure 1c, a sample time course of X(t) is plotted for the case a=1.5 μm, T=25°C, η =0.89×10−9 pNs/nm2, and F=1 pN. From X(t), we calculated the probability distributions, P(σ), of the entropy production, σ = FΔX/kBT (where ΔX=X(t+Δt)−X(t)), for the cases Δt=0.02 ms and 0.04 ms (Fig. 1d, left). In Figure 1d (right), ln[P(σ)/P(−σ)] is plotted as a function of σ. We can see that ln[P(σ)/P(−σ)]=σ.
Next, we theoretically derive Eq. (1) using the model (2). Setting X(t)=X1 and X(t+Δt)=X2, the detailed balance condition in equilibrium5 is written as
| (3) |
where Tr(X1 → X2) (Tr(X2 → X1)) is the transition probability from X1 to X2 (from X2 to X1). peq(X) ∝ e−E(X)/kBT (the Boltzmann distribution), where E represents the energy of the system. Then, Eq. (3) can be rewritten as
| (4) |
where ΔE=E(X2) − E(X1). In non-equilibrium in which F ≠ 0 (Fig. 1a), E(X) is modified into E(X)−FX considering the energy injected into the system by the external driving force, F. Eq. (4) is then written as
| (5) |
Eq. (5), which is the extended form in non-equilibrium of the detailed balance condition (Eq. (3)), is called the local detailed balance condition (LDB). See Hayashi and Sasa6 and Hayashi7 for more details on the LDB.
Because ΔE=0 in our model (Eq. (2)), Eq. (5) is further rewritten as
| (6) |
When we consider the path [X] = X1 → X2 → X3 → ... XN, Eq. (6) is extended to
| (7) |
where [X̃]=XN → XN−1 → ... → X1, ΔX=XN − X1, TR([X])=Tr(X1 → X2)Tr(X2 → X3) ... Tr(XN−1 → XN) and TR([X̃ ])=Tr(XN → XN−1)Tr(XN−1 → XN−2) ... Tr(X2 → X1). TR([X]) and TR([X̃]) are transition probabilities for the paths [X] and [X ˜ ], respectively. Then the probability distribution of the entropy production, σ = FΔX/kBT, is defined by
| (8) |
With Eq. (7), Eq. (8) is rewritten as
| (9) |
where ΔX̃ =−ΔX. Eq. (9) represents Eq. (1). Physical interpretations of Eq. (9) seem difficult because Eq. (9) is obtained formally from the mathematical identity transformation using the LDB (Eq. (5)). In this paper, we do not consider the physical interpretations of Eq. (9), but consider how to use Eq. (9) in single-molecule experiments. See Hayashi and Sasa6 for details, and Hayashi7 for a brief summary on Eq. (9).
Using FT to Measure the Driving Forces of Bio-motors
In the case of a linear motor protein that moves in one direction, the driving force, F, and thermal noise act on a cargo (Fig. 2, left). For this model, we can derive the following expression of the fluctuation theorem:
| (10) |
where X(t) is the position of the cargo, ΔX=X(t+Δt)−X(t) and, P(ΔX) is the probability distribution of ΔX. We use ΔX instead of σ in Eq. (9) to derive Eq. (10). In the case of a rotary motor protein that rotates in one direction, the rotary torque, N, and thermal noise act on a bead (Fig. 2, right). Similarly to Eq. (10), we obtain,
| (11) |
where θ(t) is the rotary angle of the bead, Δθ=θ(t+Δt)−θ(t), and P(Δθ) is the probability distribution of Δθ. In Eqs. (10) and (11), we assume that F and N are constant. Such an assumption is valid when the variations in a driving force are much smaller than the mean value of the driving force.
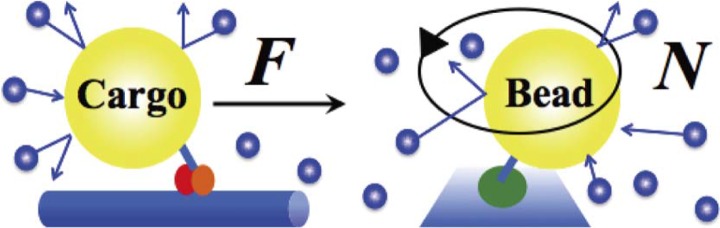
Figure 2.
Driving forces of motor proteins. (left) Schematic of a linear motor protein. A driving force, F, exerted by a motor and thermal noise act on a cargo. (right) Schematic of a rotary motor protein. A rotary torque, N, exerted by a motor and thermal noise act on a bead.
We can obtain X(t) (θ(t)) from single-molecule experiments. Measuring P(ΔX) (P(Δθ)), when we plot ln[P(ΔX)/P(−ΔX)] (ln[P(Δθ)/P(−Δθ)]) as a function of ΔX/kBT (Δθ/kBT), the slope of the graph corresponds to F(N). See the next section for an example.
Application of Eq. (11) to F1-ATPase
F1 is a rotary motor protein and a part of FoF1-ATPase/synthase8–14. The minimum complex that can act as a motor is the α3β3γ subcomplex, in which the γ subunit rotates in the α3β3 ring (Fig. 5a, top, left) upon ATP hydrolysis. The three catalytic β subunits hydrolyze ATP sequentially and cooperatively. Three ATP molecules are hydrolyzed per turn, or in other words, the free energy obtained from the hydrolysis of a single ATP molecule is used for a 120° rotation. The conformations of the β subunits change as the elementary chemical steps, such as the ATP binding, the ATP hydrolysis (cleavage of the covalent bond), and the product releases (ADP and inorganic phosphate) proceed (Fig. 3a). The coordinated push-pull motion of the C-terminal domains of the β subunits produces torque for the γ subunit to rotate9,10.
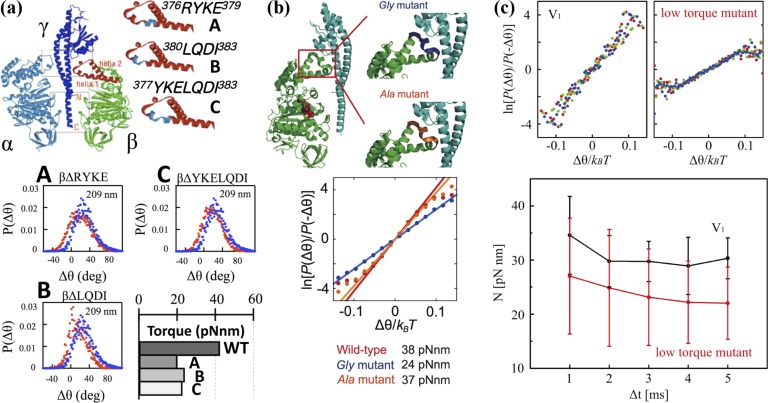
Figure 5.
FT applied to mutants F1 and V1. (a) (top) The structures of α, β and γ subunits of the F1. Truncated sequences are shown in blue19. (bottom) P(Δθ) for the wild-type F1 (blue) and the mutants (red) probed by 209-nm sized duplex beads in the case Δt=4-ms at [ATP]=2 mM. Torque was calculated from P(Δθ) using Eq. (11). (b) (top) The structures of ß and γ subunits of the F1. Residues replaced by glycine (Gly mutant) and alanine (Ala mutant) are shown in blue and orange, respectively.(bottom) ln[P(Δθ)/P(−Δθ)] as a function of Δθ/kBT for the stepping rotations probed by magnetic beads in the case Δt=5 ms at [ATPγS]=1 mM. The recording rate was 1000 fps. (c) (top) Comparison between V1 and a rotor-modulated mutant of ln[P(Δθ)/P(−Δθ)] as a function of Δθ/kBT for the stepping rotations probed by magnetic beads in the cases Δt=3 ms (red), Δt=4 ms (green), and Δt=5 ms (blue) at [ATP]=10 μM. The recording rate was 1000 fps. (bottom) Δt dependence of N measured using Eq. (11) for V1 (black) and the mutant V1 (red). Results are the average from 5 molecules (black) and 4 molecules (red), respectively.
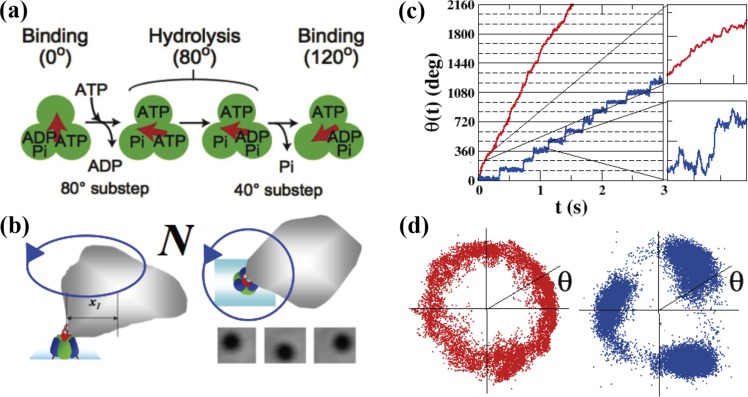
Figure 3.
Rotary motor protein, F1. (a) Reaction scheme of F1. The green circles and the red arrow represent the ß subunits and γ subunit of F1, respectively. F1 performs a 120° step rotation upon ATP hydrolysis consisting of 80° and 40° substeps. (b) Schematic diagrams of our experimental system (not to scale). The rotation of F1 is probed by an irregularly shaped magnetic bead (see Methods in Hayashi et al.11 for this irregular shape). The size of F1 is about 10 nm and the size of the bead is about 300–500 nm. (c) ATP-driven rotations of F1 probed by the magnetic beads at 1 mM ATP (red) and 100 nM ATP (blue). The recording rate was 2000 fps. (d) The center of mass of the bead was calculated from the recorded images for the case of 1 mM ATP (red) and 100 nM ATP (blue).
In our single-molecule assay (see Methods in Hayashi et al.11), the rotation of the γ subunit was observed as the rotation of a bead attached to it (Fig. 3b) because the size of the γ subunit itself is too small (∼2 nm) for its rotation to be observed directly under an optical microscope. The rotational angle, θ(t), was calculated from the recorded images of the bead (Figs. 3c and 3d). In Figure 3c, θ(t) is plotted for the cases of 1 mM ATP and 100 nM ATP. While the rotation was continuous for 1 mM ATP, it became stepwise for 100 nM ATP, pausing every 120°. The ATP-binding dwell (the time that a catalytic site waits for ATP binding) is very short at high [ATP] (<0.1 ms at 1 mM), and the time constants of the dwells for ATP hydrolysis and that for the product releases are also short (about 1 ms independent of [ATP] for the wild-type F1). The rotation appears continuous when the response time of a bead is longer than these time periods.
In previous studies12–15, the rotary torque, N, was estimated using the equation
| (12) |
where ω and Γ are the mean angular velocity and friction coefficient of a probe (e.g. a bead), respectively. From the calculation of fluid mechanics, the functional forms of Γ are known as
| (13) |
where a and xi (i=1, 2) are the radius and rotation radius of each bead, and η is the viscosity of water (η=8.9×10−10 pNs/nm2 at 25°C). Note that Eq. (13) is derived under the assumption that the rotation of a bead or a duplex of beads occurs in bulk. This causes inaccuracy in estimating the friction coefficients of beads in the case of the single-molecule experiment on F115. Because F1 attaches a glass slide and rotates the beads near the glass surface (Fig. 2b), an interaction between the beads and the glass makes the estimation using Eq. (13) diffierent from the real value. In fact, the real value of Γ is larger than the value estimated by Eq. (13). In single-molecule experiments in general, it is difficult to obtain accurate values of the friction coefficients of probes, because the probe sizes are distributed and their exact shapes are unknown, and because the motion of probes is often observed close to the surface of a glass slide, where motor proteins attach, rather than in bulk. In fact, the difficulty in estimating Γ is a common problem for force measurements of biological motors such as a bacterial flagella16 which rotates a bead near the surface of a cell, and an RNA polymerase17 which moves near the surface of a glass slide. To overcome this problem, we use FT (Eqs. (10) and (11)) that can estimate driving forces without using the value of Γ.
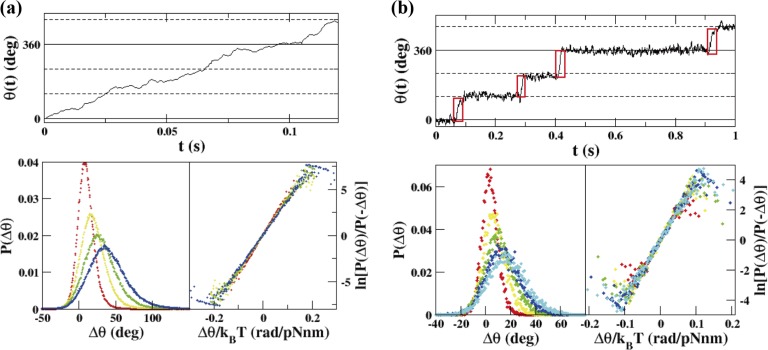
For the continuous rotation of F1 probed by a bead (Fig. 4a, top), P(Δθ) and ln[P(Δθ)/P(−Δθ)] are plotted for the cases Δt=2.5–10 ms (Fig. 4a, bottom). The slopes of the graphs in Figure 4a (bottom, right) are almost the same for all cases Δt=2.5–10 ms, and the value of the slope in the case Δt=10 ms is 38 pNnm, which corresponds to N according to Eq. (11). This value coincides with previous observations12,13. See Hayashi et al.11 for details about the application of Eq. (11), e.g., the recording rate dependence, the bead-size dependence, Δt dependence and the comparison between Eqs. (11) and (12).
Figure 4.
FT applied to the wild-type F1. (a) (top) ATP-driven rotation of F1 at 1 mM ATP. (bottom, left) Probability distribution, P(Δθ), for the cases Δt=2.5 ms (red), 5.0 ms (yellow), 7.5 ms (green) and 10 ms (blue) for the particular F1 probed by the magnetic bead. (bottom, right) ln[P(Δθ)/P(−Δθ)] as a function of Δθ/kBT. The slope was 38 pNnm in the case Δt=10 ms. The recording rate was 2000 fps. (b) (top) ATP-driven rotation of F1 at 100 nM ATP. We analyzed the θ(t) encircled in red (we used about 100–300 steps). The steps were determined by eye. (bottom, left) Probability distributions, P(Δθ), for the cases Δt=0.5 ms (red), 1.0 ms (yellow), 1.5 ms (green), 2.0 ms (blue), and 2.5 ms (aqua) for the particular F1 probed by the magnetic bead. (bottom, right) ln[P(Δθ)/P(−Δθ)] as a function of Δθ/kBT. The slope was 40 pNnm in the case Δt=2.5 ms. The recording rate was 2000 fps.
For the wild-type F1, the rotation became stepwise at 100 nM ATP (Figs. 3c and 4b, top). In this case, Δθ was calculated using the 120° steps encircled in red (Fig. 4b, top). In Figure 4b (bottom), P(Δθ) and ln[P(Δθ)/P(−Δθ)] are plotted for the cases Δt=0.5–2.5 ms. From the slope of the graph, we obtained 40 pNnm. Note that the values of Δt are smaller than those used at 1 mM ATP because the P(Δθ) for Δt larger than 2.5 ms was hard to measure precisely for the stepping rotations due to a small number of samples. See Hayashi et al.11 for the stepping rotations of the mutant F1 (βE190D), whose torque was also about 40 pNnm.
Application of (11) to V1-ATPase
We applied FT (Eq. (11)) to another rotary motor V1, which is a part of VoV1-ATPase18 (see also Supplementary Material of Hayashi et al.11 for the rotation assay of V1). In V1, the D subunit rotates in the A3B3 ring and 120° steps are observed at low [ATP] (in our case [ATP] = 10 μM). Using Eq. (11), we obtained 33±2.2 pNnm as the rotary torque of V1, which was smaller than the rotary torque of the wildtype F1 (38±2.7 pNnm)11.
Further Applications to Lower-Torque Mutants
To obtain lower-torque mutants, contact regions in F1 between the catalytic β subunits and the rotor γ subunit were mutated. The torque generation of these mutants F1 was investigated (Fig. 5a, top)19. By the analysis of FT (Eq. (11)), the torque value is directly obtained from the fluctuation of a rotating bead, without needing to know the value of the friction coefficient, which requires the exact size of the bead, the rotational radius, and the viscosity of the medium near the surface of a glass slide19. Taking advantage of Eq. (11), the torques of the mutants F1 were accurately compared with that of the wild-type F1 (Fig. 5a, bottom). We also investigated other lower-torque mutants of F1 (Fig. 5b) and a lower-torque mutant of V1 (Fig. 5c) by using Eq. (11).
Discussion
Besides the rotary motor proteins studied here, FT (Eqs. (10) and (11)) may be applied to other biological systems, including bacterial flagella and linear motor proteins such as kinesins and myosins.We hope that FT, which is a nondestructive force measurement method using fluctuation, will be applied to a wide range of biological systems in future.
When we consider applying FT to more complex biological systems, we need to note the limitations of using Eqs. (10) and (11). The temperature of the environment, T, appears in these equations because we assumed that fluctuation in the system were attributed to thermal noise. The application of FT to cell motion, which is affected by spontaneous fluctuation of a cell in addition to thermal noise, explains how the spontaneous fluctuation causes problems to use Eqs. (10) and (11). See Hayashi and Takagi20 for details.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research from the MEXT (No. 23107703) to K.H., and by Honjo International Scholarship Foundation to M.T. We thank Dr. R. Watanabe for the analysis of the mutant F1, and A. Nakanishi for the experiment on the mutant V1.
References
- 1.Evans DJ, Cohen EGD, Morriss GP. Probability of second law violations in shearing steady states. Phys. Rev. Lett. 1993;71:2401–2404. doi: 10.1103/PhysRevLett.71.2401. [DOI] [PubMed] [Google Scholar]
- 2.Wang GM, Sevick EM, Mittag E, Searles DJ, Evans DJ. Experimental demonstration of violations of the second law of thermodynamics for small systems and short time scales. Phys. Rev. Lett. 2002;89:050601. doi: 10.1103/PhysRevLett.89.050601. [DOI] [PubMed] [Google Scholar]
- 3.Ciliberto S, Joubaud S, Petrosyan A. Fluctuation in out of equilibrium systems: from theory to experiment. J. Stat. Mech. 2010:P12003. [Google Scholar]
- 4.Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco I, Jr, Bustamante C. Verification of the Crook fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates Inc; 2001. [Google Scholar]
- 6.Hayashi K, Sasa S. Linear response theory in stochastic many-body systems revisited. Physica A. 2006;370:407–429. [Google Scholar]
- 7.Hayashi K. Fluctuation theorem applied to bio-motors (review in Japanese) Seibutsu Butsuri. 2011;51:188–189. [Google Scholar]
- 8.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Oster G. Energy transduction in the F1 motor of ATP systhase. Nature. 1998;396:279–282. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 10.Masaike T, Koyama-Horibe F, Oiwa K, Yoshida M, Nishizaka T. Cooperative threestep motions in catalytic subunits of F1-ATPase correlate with 80° and 40° substep rotation. Nat. Struct. Mol. Biol. 2008;15:1326–1333. doi: 10.1038/nsmb.1510. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi K, Ueno H, Iino R, Noji H. Fluctuation theorem applied to F1-ATPase. Phys. Lett. 2010;104:218103. doi: 10.1103/PhysRevLett.104.218103. [DOI] [PubMed] [Google Scholar]
- 12.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 13.Noji H, Blad D, Yasuda R, Itoh H, Yoshida M, Kinosita K., Jr Purine but not pyrimidine nucleotides support rotation of F1-ATPase. J. Biol. Chem. 2001;276:25480–25486. doi: 10.1074/jbc.M102200200. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 15.Pänke O, Cherepanov DA, Gumbiowski K, Engelbrecht S, Junge W. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 2001;81:1220–1233. doi: 10.1016/S0006-3495(01)75780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sowa Y, Berry RM. Bacterial flagellar motor. Q. Rev. Biophys. 2008;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 17.Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K., Jr Direct observation of DNA rotation during transcription by Escherichia coil RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 18.Imamura H, Takeda M, Funamoto S, Shimabukuro K, Yoshida M, Yokoyama K. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. USA. 2005;102:17929–17933. doi: 10.1073/pnas.0507764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Usukura E, Suzuki T, Furuike S, Soga N, Saita E, Hisabori T, Kinosita K, Jr, Yoshida M. Torque generation and utilization in the motor enzyme F0F1-ATP synthase: half torque F1 with short-sized pushrod helix and reduced ATP synthesis by half-torque F0F1. J. Biol. Chem. 2012;287:1885–1891. doi: 10.1074/jbc.M111.305938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi K, Takagi H. Fluctuation theorem applied to Dictyostelium discoideum systems. Phys. Soc. Jpn. 2007;76:105001. [Google Scholar]