Summary
Helical filamentous assembly is ubiquitous in biology, but was only recently realized to be broadly employed in the innate immune system of vertebrates. Accumulating evidence suggests that the filamentous assemblies and helical oligomerization play important roles in detection of foreign nucleic acids and activation of the signaling pathways to produce antiviral and inflammatory mediators. In this review, we focus on the helical assemblies observed in the signaling pathways of RIG-I-like receptors (RLRs) and AIM2-like receptors (ALRs). We describe ligand-dependent oligomerization of receptor, receptor-dependent oligomerization of signaling adaptor molecules, and their functional implications and regulations.
Introduction
Pattern Recognition Receptors (PRRs) in the innate immune system serves as the front line of defense against pathogen infection. PRRs detect a broad range of pathogens by recognizing conserved molecular patterns (also known as Pathogen Associated Molecular Patterns, PAMPs), and activate the inflammatory innate immune response to restrict infection. These PAMPs are bacterial cell membrane or cell wall components, or common features of viral or bacterial nucleic acids absent in the host nucleic acids. These include, but are not limited to, the endosomal localization of RNA/DNA, cytoplasmic location of DNA, duplex structure of RNA, and often times a combination of these features. While their activities are often transient and restricted to the infected state, an increasing number of studies have shown that dysregulated function of PRRs can also lead to a variety of auto-inflammatory or chronic inflammatory diseases1–3, and have led to the on-going efforts to therapeutically inhibit PRRs for the treatment of these immune disorders. Over the last decade or so, there has been a rapid progress in our understanding of the structures of PRRs in the ligand-free and ligand-bound states, and their interactions with downstream signaling adaptors, activators and regulators4–9. In doing so, one common observation made by many laboratories, including ourselves, is that PRRs and many molecules in the signaling pathway often aggregate in vitro, which has posed technical challenges in biochemical and structural characterization. The accumulating evidence, however, suggest that underlying the apparent aggregation phenomena are ordered structural assemblies, in particular helical assemblies, that are key to their functions.
In this review, we will focus on the helical assemblies observed in the signaling pathways of RIG-I-like receptors (RLRs) and AIM2-like receptors (ALRs). We will describe ligand-dependent oligomerization of receptor, receptor-dependent oligomerization of signaling adaptor molecules, and their functional implications and regulations. We will also describe our speculation and perspectives about the common occurrence of the helical oligomers or filamentous assemblies in innate immune signaling. Due to the space limitation, we will not discuss Toll-like receptors and other helical assemblies in cell death as outstanding reviews are available elsewhere7, 10.
RIG-I-like receptors (RLRs): RIG-I and MDA5
RIG-I and MDA5 are non-membrane bound, soluble receptors that recognize viral double-stranded RNAs (dsRNAs) and activate antiviral immune response by stimulating transcriptional up-regulation of type I interferons11. RIG-I and MDA5 share a high sequence similarity and the same domain architecture, consisting of the N-terminal tandem caspase activation and recruitment domain (2CARD), central DExD/H box motif helicase domain and zinc-binding C-terminal domain (CTD). As with many proteins with a similar helicase domain, RIG-I and MDA5 appear to lack the duplex unwinding activity12(unpublished data for MDA5) although there is a conflicting report on RIG-I 13. 2CARD mediates signal activation by interacting with the downstream signaling adaptor molecule, MAVS, while the helicase domain and CTD are responsible for RNA recognition (Fig. 1A). Despite the shared domain organization and the downstream signaling pathway, RIG-I and MDA5 play non-redundant functions by recognizing largely distinct types of viral RNAs14, 15. MDA5 recognizes long (>~1 kb) dsRNA that are produced in the form of replication intermediates of picornaviruses and other positive strand viruses16–18. In contrast, RIG-I recognizes relatively short duplex RNA or hairpin structure with the 5’ tri- or di-phosphate, as a sign of the lack of 5’-processing that normally occurs in cellular RNAs19–24. These RNAs are often formed from defective interfering particles or genomic single-stranded RNA of both positive and negative strand viruses19–24.
Fig. 1. dsRNA-dependent oligomerization of RIG-I and MDA5.
(A) Domain organization of RIG-I, MDA5 and MAVS. CTD refers to the C-terminal domain. TBS and TM refer to the TRAF-binding site and transmembrane domain, respectively.
(B) The electron micrographs of MDA5 filament and RIG-I filament formed on 112 bp dsRNA. The images were adapted from 26 and 32.
(C) A schematic of the filament assembly process of MDA5. The MDA5 filament nucleates on the dsRNA interior and unidirectionally propagates to the dsRNA end. Filament formation brings 2CARDs of nearby MDA5 molecules into close proximity and promotes 2CARD oligomerization, which is a pre-requisite for activation of MAVS.
(D) Schematics of the filament- and ubiquitin-dependent oligomerization of RIG-I 2CARD. (I) On > 40 bp dsRNA, RIG-I forms a signaling-competent filament that assembles from the dsRNA end and propagates to the dsRNA interior. Within this filament, 2CARD oligomerization can occur independent of K63-Ubn through the proximity-induced mechanism as with MDA5. (II) On short dsRNA (<20 bp), only a single RIG-I molecule can bind per RNA molecule, and thus 2CARD oligomerization exclusively depends on K63-Ubn. (III) In general, the two mechanisms synergize to obtain stable tetramerization of 2CARD and robust signal activation.
dsRNA-triggered receptor oligomerization of MDA5
Recognition of viral dsRNAs by MDA5 involves helical filament assembly of the RNA-binding domain (helicase-CTD), which in turn triggers the second step of oligomerization by 2CARD25. Upon dsRNA binding, the helicase-CTD cooperatively assembles into a filament that extends along the length of dsRNA26, 27 (Fig. 1B). This filament formation is required for high affinity interaction with dsRNA as binding of a monomeric MDA5 is inefficient28. The MDA5 filament nucleates on a dsRNA interior with no obvious sequence preference, and propagates to the RNA termini28 (Fig. 1C). A combination of crystallography, electron microscopy and protein-protein crosslinking studies provided a detailed model of the MDA5 filament: the helicase domain and CTD form a ring-like structure around dsRNA, and this monomeric ring stacks head-to-tail along the length of dsRNA every ~14 bp to form a filament25, 29. Mutations in the filament interface decreased MDA5’s ability to induce IFN, suggesting its importance in its signaling function25. Interestingly, the filament displays dynamic instability in that filament formation triggers ATP hydrolysis, which in turn triggers disassembly from the filament termini28. As a result of the ATP-driven end-disassembly, the stability of the MDA5 filament increases with the dsRNA length, in a manner that recapitulates the dsRNA length-dependent signaling activity of MDA5 in cells16.
How does the filament assembly of MDA5 contribute to its signaling activity? Several lines of evidence support that the filament formation of the RNA-binding domain brings 2CARD of nearby MDA5 molecules (MDA52CARD) into close proximity and promotes their oligomerization25 (Fig. 1C). The MDA52CARD oligomer can then activate MAVS by inducing its own filament formation (to be discussed more in the next section). However, our understanding of MDA5 signaling remains largely incomplete. Studies showed that additional factors, such as K63-linked polyubiquitin (K63-Ubn)30 and dephosphorylation of MDA52CARD by PP1α/γ31, play a role in MDA5 activation, and it is unclear how these factors interplay with proximity-induced oligomerization of MDA52CARD. In addition, the current structural models of the MDA52CARD oligomer and its activation of MAVS are derived from that of RIG-I (to be discussed below), and they remain to be validated.
dsRNA-triggered receptor oligomerization of RIG-I
As with MDA5, RIG-I also requires oligomerization (i.e. tetramerization) of 2CARD, in order to activate MAVS. RIG-I appears to achieve it through at least two non-mutually exclusive mechanisms: via filament formation of the helicase-CTD on relatively long dsRNA and via binding to K63-linked polyubiquitin chains (K63-Ubn) (Fig. 1D).
Filament formation of RIG-I involves a mechanism distinct from MDA532, 33. Unlike MDA5, RIG-I binds the dsRNA end with 5’ppp (or 5’pp) in a manner independent of ATP. During ATP hydrolysis, RIG-I translocates from the dsRNA end to the interior12, and accumulates near the dsRNA end forming helical oligomers that closely resemble short segments of the MDA5 filament32 (Figs. 1B & 1D). It is yet unclear how the translocation leads to oligomerization and what prevents the filament from propagating further. Regardless of the detailed mechanism, filament formation of the RNA-binding domain of RIG-I (as with MDA5) promotes proximity-induced tetramerization of RIG-I2CARD that is competent to activate MAVS32. As each RIG-I occupies ~10 bp, ~40 bp is the minimum length for tetramerization of RIG-I2CARD on dsRNA in the absence of K63-Ubn 32.
Unlike MDA5, filament formation on dsRNA is not the absolute requirement for RIG-I signaling. When stimulated with shorter (e.g. <~40 bp) dsRNA or with isolated RIG-I2CARD, RIG-I utilizes K63-Ubn to oligomerize RIG-I2CARD relies on the cofactor, K63-Ubn 30, 32, 34. RIG-I2CARD is known to be covalently conjugated with K63-Ubn 35 and at the same time can bind unanchored K63-Ubn 34. The combination of the structural and biochemical studies showed that K63-Ubn bridges between adjacent RIG-I2CARD domains through non-covalent interaction, and promote its tetramerization by wrapping around the periphery of the RIG-I2CARD tetramer36 (Fig. 1D). While the covalent conjugation of K63-Ubn is not absolutely required, it markedly enhances the tetramerization efficiency and MAVS stimulatory activity of RIG-I2CARD, as expected from the conversion of the intermolecular interaction to an intramolecular interaction36.
It is important to note that the two mechanisms described above (i.e. filament-induced and K63-Ubn-mediated mechanisms) are not mutually exclusive, and that they instead cooperate for stable tetramerization of RIG-I2CARD32, 36 (Fig. 1D). When stimulated by longer dsRNA (> 40 bp), the requirement for Ub conjugation or Ub binding decreases, consistent with the notion that filament formation on dsRNA compensates for the loss of K63-Ubn. In further support of the importance of the synergism, ~10 bp dsRNA that can accommodate only a single RIG-I molecule is generally a poor ligand for RIG-I20, 37, although conflicting reports exist 38. We suspect that this discrepancy largely stems from the different expression level of RIG-I used in different experimental settings, where a high concentration of RIG-I could potentially compensate for the lack of RNA-mediated RIG-I oligomerization. The contribution of RNA-mediated oligomerization is clearly seen from the observation that RIG-I is better stimulated by longer dsRNA (up to ~500 bp) when compared at the equivalent molar concentration, i.e. with same number of 5’ppp end33, 39. The plateau around ~500 bp, which is significantly earlier than the plateau observed with MDA5, likely reflects the inefficient propagation of the RIG-I filament along dsRNA32. Why then is RIG-I thought to be poorly stimulated by long dsRNA? One should keep in mind that this notion is true only when the comparison is made using the equivalent mass concentration of RNA16, in which case longer dsRNA samples would have fewer 5’ppp and thus would be limited in recruiting RIG-I to dsRNA32. All in all, given the requirement for the oligomerization of RIG-I2CARD for its signaling activity, it is not surprising to see the positive impact (but non-obligatory role) of RIG-I filament formation on its signaling.
Receptor-triggered oligomerization of MAVS
MAVS contains a single N-terminal CARD, the central linker of ~400 amino acid and the C-terminal transmembrane (TM) domain (Fig. 1A). TM anchors MAVS to the outer membrane of mitochondria, peroxisomes and mitochondria-associated membranes40–45. Studies showed that RIG-I and MDA5 activate MAVS by inducing its filament formation on mitochondrial surface46 (Fig. 2). The MAVS CARD (MAVSCARD) is both necessary and sufficient to interact with 2CARD of RIG-I and MDA5 and to form filament in response to these interactions46. The MAVSCARD filament can self-propagate in vitro, which led to the notion of prion-like filament. However, the MAVSCARD filament differs from amyloid filaments in that it lacks cross-beta structures46, 47, and does not involve conformational change of the protomer47, 48.
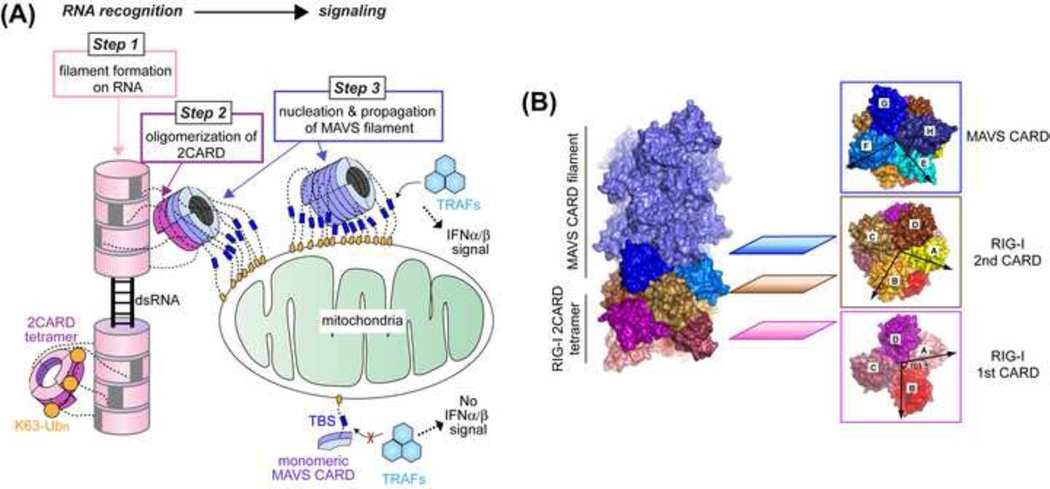
Fig. 2. Oligomerization of receptors and adaptor during RIG-I & MDA5 signal activation.
(A) Three steps of oligomerization involved during RNA recognition and signaling by RIG-I and MDA5. First, the RNA-binding domains (helicase-CTD) of RIG-I and MDA5 oligomerize on dsRNA (as shown in Fig. 1). Second, the signaling domain (2CARD) of RIGI/ MDA5 oligomerizes through proximity-induced and/or K63-Ubn-mediated mechanisms. Finally, the oligomerized 2CARD nucleates the MAVS filament through the interaction between RIG-I2CARD and MAVSCARD. The filamentous MAVS then activates the downstream signaling pathway by recruiting TRAF molecules to TRAF-binding sites (TBSs) that are clustered on the surface of mitochondria. The image was modified from Ref. 47.
(B) A composite structure obtained by overlaying the structures of the helical tetramer of the RIG-I2CARD:MAVSCARD complex and the MAVSCARD filament. These structures were reported in Ref. 47 and the image was modified from this paper.
How do the RIG-I/MDA52CARD oligomers (tetramer for RIG-I) induce MAVSCARD filament formation? CARD belongs to the Death Domain superfamily, members of which often form homotypic oligomeric structures. While their oligomerization often involves common molecular interfaces (that are conserved in location but not in sequence), their oligomeric structures vary and provide key clues to their functions7. This was also the case for RIG-I and MAVS. That is, the three oligomeric structures of RIG-I2CARD and MAVSCARD showed how RIG-I2CARD tetramer induces MAVSCARD filament formation36, 47. First, the structure of the RIG-I2CARD tetramer (in complex with K63-Ub2) showed that the tetramer forms a helical assembly36. There is no conformational change in the protomer compared to the auto-repressed, monomeric state49. However, the protomer repeats itself in a manner that the four 1st CARDs of RIG-I forms the first helical turn, which is extended by the four 2nd CARD domains36 (Fig. 2B). This helical architecture of RIG-I2CARD differs from those of other members of Death Domain superfamily7. Interestingly the structure of the MAVSCARD filament displayed a matching helical symmetry47, suggesting that the RIG-I2CARD tetramer may serve as the template to nucleate the MAVSCARD filament (Fig. 2B). In further support of this model, the structure of the RIG-I2CARD tetramer in complex with the first helical turn of MAVSCARD showed that MAVSCARD is recruited to the top of the RIG-I2CARD tetramer (on the 2nd CARD side) in a manner that extends the helical trajectory pre-defined by the RIG-I2CARD tetramer47 (Fig. 2B). While the crystallization of the RIGI2CARD: MAVSCARD complex inevitably required protein engineering to prevent the propagation of the MAVSCARD filament, functional analyses supports the validity of the structure47. The protein-engineering scheme used for this structure also provides a generalizable strategy to crystallize nucleator-filament complex.
The filamentous MAVS activates the downstream signaling pathways by recruiting the E3 ubiquitin ligases TRAF2, 5 & 6, which in turn activate cytosolic kinases, TBK1 and IKK, and subsequently the transcription factors, IRF3/7 and NF-kB46, 50. The TRAF-binding sites reside within the linker that tethers MAVSCARD to the outer membrane of mitochondria (Figs. 1A & 2A). The mechanism by which TRAFs bind to filamentous, but not monomeric MAVS, is yet unclear. Previous studies showed that the trimeric TRAF prefers pre-oligomerized substrate peptides (through the avidity effect) as binding of individual peptide is inefficient51. This observation can explain why filamentous MAVS, not monomeric MAVS, can recruit TRAFs. Recently, MAVS was shown to directly facilitate phosphorylation of IRF3 by TBK1 by recruiting both molecules to MAVS and by placing them in close proximity52. Accordingly, filamentous architecture of MAVS could play an additional role by further amplifying the clustering effect of TBK1 and IRF3. Furthermore, a recent study proposed a model where MAVS filament formation releases an auto-inhibition exerted by the cis inhibitory elements53. Although the precise nature of the auto-inhibition and the mechanism by which MAVS filament assembly releases the inhibition remain to be investigated, filament formation of MAVS appears to act at multiple steps for the signal activation.
Production of type I interferons by RIG-I/MDA5 is transient and this transiency is important to avoid chronic inflammation. As such, the MAVS filament must be resolved shortly after the activation of the downstream signaling pathways. In keeping with this, the level of MAVS was shown to decrease shortly after its activation through several distinct mechanisms. This includes Atg5-mediated autophagy of mitochondria (mitophagy)54, ubiquitin-mediated proteosomal degradation of MAVS (through the actions of E3 ligases, Smurf255, March556 and pVHL57). Other more complex regulatory mechanisms involving miniMAVS58, Trim2559 and IRTKS60 were also proposed. The relative importance of each of these regulatory mechanisms and their relationships remain to be further investigated. It would be also important to understand how the actions of these regulatory elements are restricted to post-activation (i.e. filamentous MAVS) as opposed to pre-activation (i.e. monomeric MAVS).
AIM2-like Receptors (ALRs): AIM2 and IFI16
AIM2 and IFI16 recognize dsDNA from invading viruses and bacteria (e.g. HIV, HSV-1, and F. tularensis) and activate a series of inflammatory responses61–72. AIM2 detects dsDNA in the cytoplasm and assembles into a filamentous oligomer, which then triggers the polymerization of the signaling adaptor, ASC 69, 73–75. The ASC filament in turn recruits and activates caspase-1 (converting the latent pro-caspase-1 to the active caspase-1), which processes the latent form of pro-inflammatory cytokine interleukin-1β (IL-1β) and induces pyroptosis (Fig. 3A). This ternary complex (receptor•ASC•pro-caspase-1) is also dubbed the inflammasome (Fig. 3A) 69, 73–75. Unlike AIM2, IFI16 operates in both the nucleus and cytoplasm 62–66, 76–79, and was initially reported to stimulate IFN production independent of ASC and pro-caspase-167, 69, 74. More recent studies, however, reported that IFI16 can also interact with ASC and pro-caspase-1, forming an IFI16 inflammasome and processing pro-IL-1β similarly to the AIM2 inflammasome61, 64, 66, 80. The IFI16 and AIM2 inflammasomes appear to be mutually exclusive and selectively assembled in response to different pathogenic infections. For instance, the AIM2 inflammasome assembles upon both cytosolic bacterial and viral infections (e.g. vaccinia virus and F. tularensis) 69, 73–75, whereas the IFI16 inflammasome is only found in response to nuclear viruses (e.g. HSV and KSHV) and certain retroviruses such as HIV 61, 64, 66.
Fig. 3. Activation of the ALR inflammasome pathway.
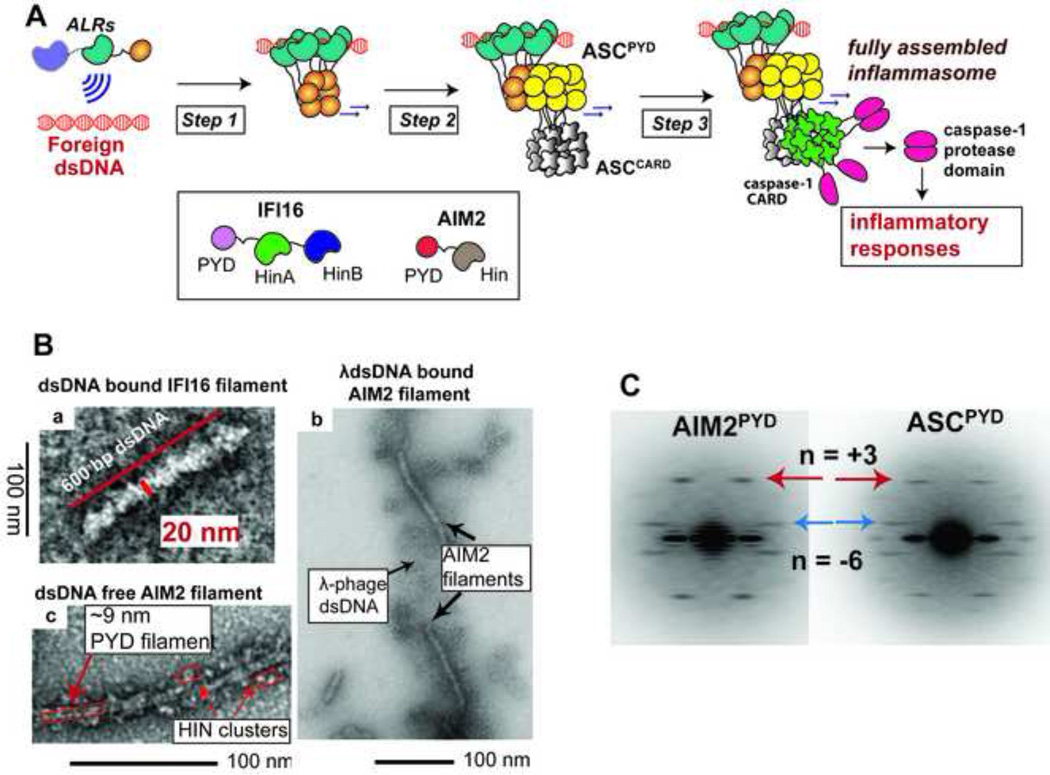
(A). The stepwise assembly of ALR inflammasomes. Step 1: ALRs assemble into receptor filaments on foreign dsDNA. Step 2: the PYD oligomers of ALRs induce the polymerization of ASCPYD by providing a structural template, which in turn induces the polymerization of ASCCARD. Finally, step 3, ASCCARD promotes the polymerization of procaspase-1, consequently activating the protease. The box shows the domain organization of ALRs. The figure is modified from Ref. 85.
(B) Electron micrographs of ALR filaments. The images were adapted from Refs. 84 and 85.
(C) The average power spectra generated from negatively stained electron micrographs of the AIM2PYD and ASCPYD filaments. The power spectrum was generated by Fourier transformation of the filament images, and it contains the full information of the helical symmetry of the filament. That is, each layer line represents periodicity of a family of helical lattice lines, and the n value refer to the Bessel order of the layer line. The figure is adapted from Ref. 85.
Both AIM2 and IFI16 contain an N-terminal pyrin domain (PYD), which is important for activation of ASC (Fig. 3A, box). AIM2 contains one dsDNA binding HIN domains (HIN: hematopoietic interferon inducible nuclear antigen), whereas IFI16 has two tandem HIN domains (Fig. 3A, box); the HIN domains are composed of two oligo-binding folds and nonspecifically bind dsDNA. As with CARD of RLRs, PYD belongs to the Death Domain superfamily and functions as the oligomerization and signaling domain. Both AIM2 and IFI16 bind dsDNA in a sequence-independent manner, and require at least ~70-bp dsDNA for robust signaling 67, 81. It is noteworthy that the host cytoplasm is normally free of dsDNA, which in turn marks any invading pathogen dsDNA to be targeted as “nonself” by both AIM2 and IFI16; however, nuclear IFI16 must distinguish between host and viral dsDNA through a mechanism to be described below.
dsDNA-triggered receptor oligomerization of IFI16
Upon invasion of viral dsDNA in the nucleus (e.g. HSV and KSHV), individual IFI16 molecules rapidly locate one another on large viral genomes and assemble into filaments61, 64, 66, while leaving the host dsDNA intact. It has been hypothesized that the chromatinization plays a key role in preventing the host DNA from being targeted, while invading viral DNA is relatively “naked” and thus susceptible to recognition by IFI1662, 64, 66, 77, 82 Indeed, Knipe and colleague have observed that transfected plasmid is more readily recognized by IFI16 than intact virons that contain additional dsDNA binding proteins62, 82. Studies showed that a “naked” stretch of dsDNA needs to be ~60-70 bp to allow stable formation of the IFI16 filament83, 84. However, longer DNA increases the assembly rate83, 84, which is likely important for facilitating foreign DNA recognition while suppressing the inadvertent assembly of the IFI16 filament on the host DNA.
How does IFI16 assemble the filament so that its assembly rate increases with the DNA length? Unlike RLRs where the dsRNA-binding domains (helicase-CTD) largely drive receptor oligomerization26, 27, the HIN domains of IFI16 one dimensionally tracks dsDNA without forming a stably bound complex 83, 84. However, the tracking ability of the HIN domains allows multiple IFI16 to dynamically associate on dsDNA and cluster into filaments via PYD-PYD interactions83, 84. Consequently, not only does IFI16 bind longer dsDNA more stably84, but it also assembles filaments faster on the longer duplexes83, 84. Importantly, we found that nucleosomes directly block these one-dimensional movements, thereby preventing IFI16 from clustering83. This tracking-assisted oligomerization mechanism would then allow IFI16 to distinguish friend from foe and assemble into signaling platforms efficiently and selectively on foreign DNA.
dsDNA-mediated oligomerization of AIM2
As with IFI16, AIM2 assembles into filaments on dsDNA 69, 73–75 (Figs. 3A step 1 and 3B-b), and its filament formation and signal activation are dependent on the oligomerization of the PYD (AIM2PYD)85. Despite the apparent similarity in the domain composition and overall filament architecture, AIM2 and IFI16 display different oligomerization properties, which may be linked to their distinct functions. While AIM2 forms the filament more efficiently with DNA, it can also self-oligomerize at high protein concentrations in the absence of DNA (Fig. 3B-c)85. This differs from IFI16, for which DNA-independent filament formation has been rarely observed even at high protein concentrations84. More detailed studies showed that unlike IFI16, the HIN of AIM2 can oligomerize in the absence of DNA, and that full-length AIM2 can self-oligomerize better than either of isolated PYD or HIN domain without DNA85. These observations suggest that AIM2 does not utilize auto-repression as previously proposed81, 86, and its cellular regulation is mediated by other mechanisms. The concentration-dependent and DNA-independent oligomerization property of AIM2 also raises the possibility that the auto-inflammatory disorders associated with high level of AIM287 could be caused by its spontaneous filament assembly rather than by detection of certain cytosolic DNA. Interestingly, mouse encodes p20288, which consists of two HIN domains and inhibits the oligomerization of AIM2 by HIN•HIN interactions. Currently, there is no known ortholog of p202 in human89. Considering that all HIN domains share high structural homology (RMSD < 2 Å) 89, the difference in oligomerization activity of the HINs of IFI16 and AIM2 suggests that the dsDNA-binding and oligomerization activities of nuclear and cytosolic ALRs have been tuned differently by amino acid variations in key areas (the sequence similarity between the HIN domains of AIM2 and IFI16 is less than 50%). Future investigation of whether the DNA-tracking property is unique to IFI16 HINs or shared with AIM2 would further provide additional insights into the functional versatility of HINs.
For both AIM2 and IFI16, the precise architecture of the filaments on dsDNA remains to be further investigated. For example, it is unclear whether the HIN domains are arranged in a helical manner on dsDNA, and whether the PYD filaments extend parallel to or co-axially with DNA. Regardless, it is noteworthy that this PYD-driven oligomerization is distinct from RLRs, of which receptor oligomerization is mainly driven by the RNA binding domain (helicase-CTD), and can also occur without 2CARD.
Receptor-induced oligomerization of ASC
How does the oligomerized ALR PYDs activate ASC, and how does the activated ASC in turn promote cleavage of pro-caspase-1 to form a mature caspase-1? Studies suggested that a series of step-wise filament assembly underlie the signal propagation from ALR to ASC (Fig. 3A step 2), and to caspase-1 (Fig. 3A step 3) 90, 91. ASC consists of an N-terminal PYD and a C-terminal CARD, whereas pro-caspase-1 consists of the N-terminal CARD and C-terminal caspase domain (Fig. 3A). The AIM2PYD filament activates ASC by nucleating the filament formation of PYD of ASC (ASCPYD) (Fig. 3A step 2). In this filamentous architecture of ASC, CARD of ASC (ASCCARD) in turn recruits pro-casepase-1 through the ASCCARD:pro-caspase-1CARD interaction and nucleates the filament assembly of pro-casepase-1 (Fig. 3A step 3). Filament assembly of pro-caspase-1 then brings together the caspase domains in close proximity, thereby promoting its dimerization and activation, likely resulting in the release of active caspase-1 from the inflammasome.
Structure of the filament assembled by the AIM2PYD and ASCPYD showed that both filaments display the same helical symmetry (~9 nm wide, six start helix with C3 symmetry) 85. This is in line with the model that AIM2PYD oligomer serves as the helical template that nucleates the ASCPYD filament (step 1), and is analogous to the mechanism by which the RIG-I2CARD tetramer nucleates the MAVSCARD filament. By contrast, the mechanism by which ASCCARD initiates the pro-casepase-1 filament formation (in step 2) is less clear. While it is tempting to suggest that AIM2CARD forms a helical template to nucleate pro-caspase-1CARD, oligomeric or filamentous structure of ASCCARD or pro-caspase-1CARD is currently unavailable to demonstrate the helical template mechanism. It is also unclear whether the signal propagation mechanism shown with AIM2 is applicable to IFI16 and other receptors that regulate the polymerization of ASC, which remains as a topic of future investigation.
The regulatory mechanisms of the ALR/ASC-signaling pathway also require further studies. In particular, how spontaneous filament assembly of ASC is suppressed in the absence of infection, and how the filament is resolved after its activation to prevent chronic inflammation. It was initially thought that ASC is autosuppressed by having an intramolecular interaction between PYD and CARD, thereby avoiding spontaneous filament formation and activation of caspase-1. However, the NMR studies of full-length ASC suggested that the PYD and CARD of ASC do not interact with each other under the experimental condition 92. It is possible that the maintenance of ASC at low concentration and having multiple steps of oligomerization to assemble the fully activated inflammasome may together provide a sufficient blockade for aberrant activation of the inflammatory response. Nevertheless, it is also likely that there could be a yet uncharacterized trans regulatory elements or post-translational modification that suppress ASC’s activity in the absence of infection. Indeed, recent studies suggest that IL-1β and IFN inducible PYD-only proteins (POPs) can suppress the oligomerization of ALRs and ASC93, 94, providing a negative feedback mechanism to prevent persistent inflammatory responses (e.g. POP3 inhibits the oligomerization of both AIM2 and IFI16, and POP1 inhibits ASC). Nonetheless, the detailed mode of actions for POPs and the mechanism by which the assembled inflammasome is resolved to prevent chronic inflammation needs future investigation.
Concluding remarks
Helical assemblies are ubiquitous in biology, ranging from structural elements (viral capsids and cytoskeletal actins) to DNA repair and recombination machineries (RecA and Rad51). It was only recently realized that helical architectures are employed in a number of innate immune signaling pathways, either during foreign nucleic acid sensing or during signal propagation. While cellular imaging of these molecules often reveals punctated or aggregated structures, rather than filaments that resemble cytoskeletons, the shape of the aggregate at the macroscale does not necessary inform about the structure of the molecular assemblies at the microscale. Our observations so far suggest that filamentous molecules can often tangle and aggregate, and that the filament propagation can be also limited in cells, both of which can lead to the appearance of non-linearity in cellular imaging. If so, how can one be confident about the in vivo relevance of filaments observed in vitro? High resolution structures of the filaments have enabled specific mutational studies, which in turn have revealed strong correlations among filament formation in vitro, puncta (or aggregate) formation in cells and the cellular signaling activity of many of these molecules47, 63, 90, 91, 95. More importantly, in vitro reconstitution of some of these systems allowed direct examination of the importance of the filament assemblies in their signaling activities25, 34, 46. We expect that recent advances in cellular imaging techniques (e.g. cryo-tomography96 and super-resolution fluorescence microscopy97) would further help defining detailed assembly architecture of these large “aggregate” in cells.
Why are these helical assemblies so common in innate immune signaling pathways? Could there be a rationale for their frequent occurrence? Helical assembly formed by the nucleic acid sensors is understandably common as it partly reflects the helical symmetry of the bound nucleic acid (i.e. dsRNA and dsDNA), and allows these sensors to measure the length of the naked nucleic acids, a common criterion for self vs non-self discrimination. Helical assembly by the signaling domains, such as CARDs and PYDs of RLRs, ALRs and their signaling adaptors, are also common despite the fact that they do not directly contact the nucleic acids. These assemblies contribute to rapid signal amplification by allowing a small number of upstream molecules to induce oligomerization of a large number of downstream molecules. Although there are foreign nucleic acid sensors, such as OAS1 98 and cGAS 99, that are currently not known form helical oligomers, our observations with RLRs and ALRs represent a growing number of receptors where helical assembly is utilized for foreign nucleic acid detection and signal propagation.
Highlights.
RIG-I and MDA5 assemble into filamentous oligomers along viral dsRNA.
They then form the helical oligomers of 2CARD to nucleate the MAVS filament.
IFI16 and AIM2 assemble into filamentous oligomers on bacterial and viral dsDNA.
The AIM2 filament provides a structural template for assembling the ASC filament.
Acknowledgments
SH is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease and is funded by NIH grants (AI106912, AI111784 and AI119880). JS is a Jerome L. Greene Scholar. JS is also supported by an American Cancer Society Research Scholar Grant (RSG-15-224-01-DMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 3.de Jesus AA, Goldbach-Mansky R. Newly recognized Mendelian disorders with rheumatic manifestations. Curr Opin Rheumatol. 2015;27:511–519. doi: 10.1097/BOR.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hauenstein A, Zhang L, Wu H. The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr Opin Struct Biol. 2015;31:75–83. doi: 10.1016/j.sbi.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawling DC, Pyle AM. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol. 2014;25:25–33. doi: 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuenchor W, Jin T, Ravilious G, Xiao TS. Structures of pattern recognition receptors reveal molecular mechanisms of autoinhibition, ligand recognition and oligomerization. Curr Opin Immunol. 2014;26:14–20. doi: 10.1016/j.coi.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. * This review article provides a summary of helical assemblies observed with Death Domains in the Myddosome, PIDDosome and the DISC complex.
- 8.del Toro Duany Y, Wu B, Hur S. MDA5-filament, dynamics and disease. Curr Opin Virol. 2015;12 doi: 10.1016/j.coviro.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu B, Hur S. How RIG-I like receptors activate MAVS. Curr Opin Virol. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci. Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 12. Myong S, et al. Cytosolic Viral Sensor RIG-I Is a 5′-Triphosphate–Dependent Translocase on Double-Stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. * This study showed that RIG-I can translocate along dsRNA during ATP hydrolysis.
- 13.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 14.Yoneyama M, et al. Shared and Unique Functions of the DExD/H-Box Helicases RIG-I, MDA5, and LGP2 in Antiviral Innate Immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 15.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q, et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Reports. 2012;29:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Triantafilou K, et al. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 19.Pichlmair A, et al. RIG-I–Mediated Antiviral Responses to Single-Stranded RNA Bearing 5'-Phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 20.Schlee M, et al. Recognition of 5'triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;108:3092. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, et al. Identification of a Natural Viral RNA Motif That Optimizes Sensing of Viral RNA by RIG-I. MBio. 2015;6:e01265–e01315. doi: 10.1128/mBio.01265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Gil L, et al. A Sendai Virus-Derived RNA Agonist of RIG-I as a Virus Vaccine Adjuvant. J. Virol. 2013;87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu B, et al. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 26. Peisley A, et al. Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral dsRNA Recognition. Proc Natl Acad Sci U S A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. ** This paper showed that human MDA5 forms filament on dsRNA and disassembles during ATP hydrolysis. The study also showed that the ATP-mediated disassembly is the key to the dsRNA-length dependent stability of the filament
- 27. Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;7:1714–1726. doi: 10.1038/emboj.2012.19. ** This article independently showed the filament formation of mouse MDA5 on dsRNA, and its ATP-dependent disassembly.
- 28. Peisley A, et al. Kinetic Mechanism for Viral dsRNA Length Discrimination by MDA5 Filament. Proc Natl Acad Sci U S A. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. * This article describes how the combination of the filament assembly and disassembly kinetics leads to the dsRNA length-sensitivity of MDA5 signaling.
- 29.Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci U S A. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X, et al. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wies E, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I Forms Signaling-Competent Filaments in an ATP-Dependent, Ubiquitin-Independent Manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. ** This article showed that RIG-I forms filamentous oligomers on dsRNA during ATP hydrolysis, and that the filamentous oligomer of RIG-I can activate MAVS in the absence of K63-Ubn.
- 33. Patel JR, et al. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. ** This article independently showed that RIG-I oligomerizes on dsRNA during ATP hydrolysis, and that oligomerization is important for efficient signaling.
- 34.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141 doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 36. Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. * This paper showed the structures of the RIG-I2CARD tetramer in complex with K63-Ub2 chains.
- 37.Anchisi S, Guerra J, Garcin D. RIG-I ATPase Activity and Discrimination of Self-RNA versus Non-Self-RNA. MBio. 2015;6:e02349-14. doi: 10.1128/mBio.02349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohlway A, Luo D, Rawling DC, Ding SC, Pyle AM. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Binder M, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid inducible gene-I. J. Biol. Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seth RB, Sun L, Ea CK, Chen Z. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kB and IRF3. Cell. 2005;122:699–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 42.Xu LG, et al. VISA Is an Adapter Protein Required for Virus-Triggered IFN-b Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 43.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 44.Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixit E, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:1–14. doi: 10.1016/j.cell.2011.06.041. ** This paper defined the filamentous state as the active state of MAVS.
- 47. Wu B, et al. Molecular imprinting as a signal activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. ** This paper showed the structures of the MAVSCARD filament and MAVSCARD in complex with the RIG-I2CARD tetramer.
- 48.Potter JA, Randall RE, Taylor GL. Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct Biol. 2008;8:1–10. doi: 10.1186/1472-6807-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 50.Liu S, et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu H. Assembly of post-receptor signaling complexes for the tumor necrosis factor receptor superfamily. Adv Protein Chem. 2004;68:225–279. doi: 10.1016/S0065-3233(04)68007-7. [DOI] [PubMed] [Google Scholar]
- 52.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, et al. An autoinhibitory mechanism modulates MAVS activity in antiviral innate immune response. Nat Commun. 2015;6:7811. doi: 10.1038/ncomms8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tal MC, et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Y, et al. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J. Immunol. 2014;192:4758–4764. doi: 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 56.Yoo YS, et al. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun. 2015;6:7910. doi: 10.1038/ncomms8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du J, et al. pVHL Negatively Regulates Antiviral Signaling by Targeting MAVS for Proteasomal Degradation. J. Immunol. 2015;195:1782–1790. doi: 10.4049/jimmunol.1500588. [DOI] [PubMed] [Google Scholar]
- 58.Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell. 2014;156:800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castanier C, et al. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012;10:44. doi: 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia P, et al. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun. 2015;6:8132. doi: 10.1038/ncomms9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monroe KM, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A. 2013;110:E4492–E4501. doi: 10.1073/pnas.1316194110. *This paper shows that IFI16 preferentially binds naked dsDNA in vivo.
- 63.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jakobsen MR, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. * This article reports that IFI16 assembles into an inflammasome in the host nucleus.
- 67. Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. **This article identifies IFI16 as a foreign dsDNA sensor.
- 68.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. *This article identifies AIM2 as a foreign dsDNA-sensing inflammasome.
- 70.Sauer JD, et al. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim S, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. *This article independently identifies AIM2 as a foreign dsDNA-sensing inflammasome.
- 74. Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. *This article independently identifies AIM2 as a foreign dsDNA-sensing inflammasome.
- 75.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 76.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. *This paper demonstrates that IFI16 operates both in the nucleus and cytoplasm.
- 78.Unterholzner L, Bowie AG. Innate DNA sensing moves to the nucleus. Cell Host Microbe. 2011;9:351–353. doi: 10.1016/j.chom.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 79.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ansari MA, et al. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J Virol. 2013;87:8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. ** This article reports the dsDNA-bound crystal structures of the HINs of IFI16 and AIM2.
- 82.Orzalli MH, Knipe DM. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Stratmann S, Morrone S, van Oijen AM, Sohn J. The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex. Elife. 2015;4 doi: 10.7554/eLife.11721. **This report shows that IFI16 tracks dsDNA and nucleosomes hamper the assembly.
- 84. Morrone SR, et al. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A. 2014;111:E62–E71. doi: 10.1073/pnas.1313577111. ** This article reports that IFI16 assembles into filaments on dsDNA.
- 85. Morrone SR, et al. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun. 2015;6:7827. doi: 10.1038/ncomms8827. ** This article reports the assembly mechanism of AIM2 filaments and the congruent helical symmetry between the PYD filaments of AIM2 and ASC.
- 86.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–13235. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shaw PJ, McDermott MF, Kanneganti TD. Inflammasomes and autoimmunity. Trends Mol Med. 2011;17:57–64. doi: 10.1016/j.molmed.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yin Q, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brunette RL, et al. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. ** This article reports the structure of ASC filament, and also identifies that filamentous assmblies underlie the inflammasome pathways.
- 91. Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. ** This article reports that filamentous assemblies underlie the inflammsome pathways.
- 92.de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. de Almeida L, et al. The PYRIN Domain-only Protein POP1 Inhibits Inflammasome Assembly and Ameliorates Inflammatory Disease. Immunity. 2015;43:264–276. doi: 10.1016/j.immuni.2015.07.018. *This article reports that POP1 inhibits ASC polymerization.
- 94. Khare S, et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol. 2014;15:343–353. doi: 10.1038/ni.2829. * This article reports that POP3 inhibits both AIM2 and IFI16 inflammasomes.
- 95.Johnson KE, et al. IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 2014;10:e1004503. doi: 10.1371/journal.ppat.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Asano S, Engel BD, Baumeister W. In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 97.Sydor AM, Czymmek KJ, Puchner EM, Mennella V. Super-Resolution Microscopy: From Single Molecules to Supramolecular Assemblies. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 98.Hovanessian AG, Brown RE, Kerr IM. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- 99.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]