Abstract
Perceived intensities of sweetness and bitterness are correlated with one another and each is influenced by genetics. The extent to which these correlations share common genetic variation, however, remains unclear. In a mainly adolescent sample (n = 1901, mean age 16.2 years), including 243 monozygotic (MZ) and 452 dizygotic (DZ) twin pairs, we estimated the covariance among the perceived intensities of 4 bitter compounds (6-n-propylthiouracil [PROP], sucrose octa-acetate, quinine, caffeine) and 4 sweeteners (the weighted mean ratings of glucose, fructose, neohesperidine dihydrochalcone, aspartame) with multivariate genetic modeling. The sweetness factor was moderately correlated with sucrose octa-acetate, quinine, and caffeine (r p = 0.35–0.40). This was mainly due to a shared genetic factor (r g = 0.46–0.51) that accounted for 17–37% of the variance in the 3 bitter compounds’ ratings and 8% of the variance in general sweetness ratings. In contrast, an association between sweetness and PROP only became evident after adjusting for the TAS2R38 diplotype (r p increased from 0.18 to 0.32) with the PROP genetic factor accounting for 6% of variance in sweetness. These genetic associations were not inflated by scale use bias, as the cross-trait correlations for both MZ and DZ twins were weak. There was also little evidence for mediation by cognition or behavioral factors. This suggests an overlap of genetic variance between perceptions of sweetness and bitterness from a variety of stimuli, which includes PROP when considering the TAS2R38 diplotype. The most likely sources of shared variation are within genes encoding post-receptor transduction mechanisms common to the various taste G protein-coupled receptors.
Key words: IQ, personality, sweetener, taste, TAS2R38, twins
Introduction
Taste perception varies greatly among individuals. For over a decade, intensity ratings of the bitter compound 6-n-propylthiouracil (PROP) have been used to distinguish an individual’s “bitter taster status” (Hayes and Keast 2011), with those rating it as extremely bitter sometimes described as “supertasters” (Bartoshuk et al. 1994). Many studies suggested these individuals are also more sensitive to other taste stimuli (Bartoshuk 1979; Tepper and Nurse 1997; Lucchina et al. 1998; Ly and Drewnowski 2001; Bajec and Pickering 2008; Fischer et al. 2014), whereas many others have failed to find such associations (Schifferstein and Frijters 1991; Drewnowski et al. 1997; Drewnowski et al. 1998; Keast and Roper 2007; Lim et al. 2008). Therefore, whether individual differences in ratings of a single compound can generalize to other taste stimuli has been questioned and, furthermore, whether there are pan-quality overarching individual differences remains unclear (Lim et al. 2008; Hayes and Keast 2011; Fischer et al. 2014).
Most of the perceptual variability in PROP is due to genetic variation within the bitter taste receptor TAS2R38 (Drayna et al. 2003; Bufe et al. 2005; Reed et al. 2010). Genetic variation in TAS2R38 does not appear to be associated with perceived intensity of other taste stimuli (e.g. quinine, and sucrose) (Hayes et al. 2008), but some evidence shows that the TAS2R38 diplotype may modify the association between PROP and other tastes (Fischer et al. 2014). In addition, the PROP response has been shown to be less predictive of overall perceived taste intensity than are collective ratings of sucrose, sodium chloride, citric acid, and quinine (Lim et al. 2008), suggesting that PROP ratings are not a sole predictor for overall taste perception. Rather, a more complex association across multiple taste classes, such as general differences in the “gain” of the taste system, appears to be at play.
A parallel body of work reveals that genetics plays a significant role in the perception of different taste qualities, accounting for over 30% of the variance in sweetness, sourness, and bitterness (Hansen et al. 2006; Wise et al. 2007; Knaapila et al. 2012; Hwang et al. 2015). Our previous studies identified a shared genetic pathway for taste perception across different bitter compounds, excluding PROP (Hansen et al. 2006), and more recently a common genetic factor for the perception of both sugars and non-caloric sweeteners (Hwang et al. 2015). Although the perception of sweetness has been weakly-to-moderately correlated with bitterness (Lim et al. 2008; Knaapila et al. 2012; Fischer et al. 2014), whether this association is due to shared genes has not been determined in humans. In addition, there is evidence that non-receptor based factors may contribute to the correlation between sweet and bitter taste perceptions. For example, prosocial (e.g. agreeableness) and antisocial (e.g. psychopathy) personalities are associated with elevated sweet and bitter taste preferences, respectively (Meier et al. 2012; Sagioglou and Greitemeyer 2016). Further, other concerns, such as psychometric properties of scale use (e.g. a tendency to rate at one side of the scale [Jewell and McCourt 2000]) and even intelligence [e.g. higher IQ is associated with less extreme rating styles (Light et al. 1965)] need to be raised when studying weak sensory associations.
The present study investigated the sources of association between multiple taste qualities using a large adolescent and young adult twin sample. Genetic covariances between perceived intensity of 4 sweet (glucose, fructose, neohesperidine dihydrochalcone [NHDC], and aspartame) and 4 bitter solutions (PROP, SOA, quinine HCl [quinine], and caffeine) were estimated using multivariate genetic modeling. In addition, this study examined the impact of the TAS2R38 diplotype on the genetic covariances, and potential confounding effects of scale use bias, general cognitive ability, and behavioral factors.
Materials and methods
Participants
Participants were adolescent and young adult Caucasian twins and their singleton siblings from the Brisbane Adolescent Twin Study (Wright and Martin 2004), also referred to as the Brisbane Longitudinal Twin Study (BLTS). They were originally recruited for a study on melanoma (Zhu et al. 2007), a common form of cancer among light-skinned people. The taste data reported here were collected between August 2002 and July 2014. The sample comprised 243 monozygotic (MZ) and 452 dizygotic (DZ) twin pairs, including 126 pairs with 1–2 singleton siblings, and 320 unpaired individuals (mean age of 16.2±2.8 years; 1023 females, 878 males) (Supplementary Table 1). This is the same sample as used previously (Hwang et al. 2015). Zygosity was determined from genotyping (92% of same sex twin pairs) or from self-report. The Queensland Institute of Medical Research Human Research Ethics Committee approved the study. Written consent was obtained from both the participants and their parents (the latter not required for those 18 years and over).
Taste test
The taste test battery has been described in detail elsewhere (Hansen et al. 2006; Hwang et al. 2015). Briefly, participants rated the intensity of 5 bitter (6.0×10–4 M PROP, 2.0×10–4 M SOA, 1.81×10–4 M quinine, 0.05M caffeine, and 4.99×10–6 M denatonium benzoate) and 4 sweet (0.60M glucose, 0.30M fructose, 8.0×10–5 M NHDC, and 1.4×10–3 M aspartame) solutions using a general labeled magnitude scale (gLMS) (Green et al. 1993). Mean intensity ratings from duplicate presentations for each of PROP, SOA, Quinine and Caffeine were used in all analyses. For the 4 sweet compounds, a weighted mean general sweet (gSweet) factor was used, as perceived intensity of the sweeteners is highly correlated at the genetic level (r g = 0.78–0.89) and most of the variance (71% for glucose, 77% for fructose, 64% for NHDC, and 59% for aspartame) is accounted for by a common genetic factor (Hwang et al. 2015). Denatonium benzoate was not included due to the violation of the normality assumption, a criterion for twin modeling (Neale et al. 2002) (see Statistical Analyses). Statistical transformation failed to overcome this problem because of its distinct bimodal distribution (Supplementary Figure 1). In addition, the mean intensity rating for denatonium benzoate was double that of other stimuli, with the most common rating being the strongest imaginable on the scale, suggesting that the concentration may have been too high to detect variation (also known as a “ceiling effect”). The intensity rating characteristics for denatonium benzoate are summarized in Supplementary Table 2.
TAS2R38 diplotype
The genotypes for the 3 TAS2R38 SNPs (rs713598, rs1726866, and rs10246939), resulting in 3 amino acid substitutions (A49P, A262V, and V296I), were available for 92% of the sample. Genotyping was done using the Illumina 610-Quad BeadChip (n = 1254) (Reed et al. 2010) or HumanCoreExome-12 v1.0 BeadChip. The frequencies of the 3 diplotypes [PAV/PAV = 17%, PAV/AVI = 52% (including 34 participants with rare diplotypes of AAV/AVI or PAV/AAV, which were shown to have similar effects on PROP perception as the PAV/AVI diplotype [Bufe et al. 2005]), and AVI/AVI = 31%] were similar to the frequencies reported previously for PROP sensitivity (i.e. a distribution of 20% high: 50% medium: 30% low-sensitivity tasters (Bartoshuk et al. 1994; Drewnowski et al. 2001).
Statistical analyses
The heritability and phenotypic correlations among the intensity scores of gSweet and the 4 bitter substances were estimated using univariate and bivariate variance components modeling. This twin method partitions the phenotypic variance or covariance into additive genetic (A), common environment (C) and unique environment (E, includes experimental error) sources by taking advantage of the differences in the genetic relatedness between MZ twins who share 100% of their genes and DZ pairs who share, on average, 50% of their genes. This method can detect genetic effects by comparing the correlations of MZ and DZ twins and without requiring the investigation of specific genes. Variance components modeling was performed using the structural equation modeling software package Mx, which utilises maximum likelihood estimation procedures (Neale et al. 2002). Prior to modeling, a square root transformation was applied to each of the 5 intensity scores to obtain a more normal distribution (Supplementary Figure 1 and Supplementary Table 3). Covariates of age, sex, and otitis media were modeled as regressions or deviation effects on the mean for all models. Damage to the chorda tympani nerve resulting from an otitis media infection can result in an increase in the number of taste buds (Bartoshuk et al. 1996), and consequently may influence taste perception. Our previous studies showed that the history of otitis media was associated with increased perceived intensity (4–9%) of the same 4 sweet and bitter solutions (Hansen et al. 2006; Hwang et al. 2015). No outlying families were identified using the %p option in Mx, which uses the Mahalanobis distance to compute a z-score for each family, with values outside the −3.5 to +3.5 range indicating excessive similarities or differences relative to other families in the sample and model expectation.
To estimate the covariance structure between the 5 traits, a multivariate Cholesky decomposition model (Neale and Cardon 1992) was used as a starting point. A series of models, including dropping A and C components, were tested to determine which pattern of covariance best fitted the data. For nested models, we assessed the comparative fit by calculating the difference in double the negative log-likelihood, which is distributed asymptotically as a χ2. For non-nested models, the Akaike’s Information Criterion (AIC), which penalizes models for increasing complexity (Burnham and Anderson 2002), was used. The effect of the TAS2R38 diplotype on the covariance structure was tested in a partial dominant model with 1 covariate for the heterozygous effect (i.e. PAV/AVI = 1 and PAV/PAV = AVI/AVI = 0) and a second covariate for the PAV homozygous effect (i.e. PAV/PAV = 1 and AVI/AVI = PAV/AVI = 0). Consequently, the genetic variance in PROP dropped from 0.72 to 0.20 (compared with an additive or dominant model where PROP genetic variance reduced to 0.25 and 0.24, respectively). This model was used to examine the covariance structure when the TAS2R38 genetic effect on intensity ratings was removed. A second model in which the low-sensitivity tasters for PROP (30% of the genotyped sample) were excluded from the analyses was also tested.
In addition, this study investigated whether intensity ratings were associated with general cognitive ability (Verbal IQ), assessed with the Multidimensional Aptitude Battery (Jackson 1998), and personality (neuroticism, extraversion, openness to experience, agreeableness, and conscientiousness) assessed with the NEO Five-Factor Inventory (FFI) (Costa and McCrae 1992). Both cognition and personality were assessed at age 16, which for the majority of twins were 2 years after assessment for sweet and bitter taste. Verbal IQ and personality were available for 1282 and 1277 participants, respectively (Loehlin et al. 2015). Further, to assure that the associations were estimated from taste perception and were not inflated by scale use bias, an emphasis score: 0 = neutral, 1 = somewhat, 2 = strongly was calculated by folding the 5-point Likert scale of the 60 responses from the NEO-FFI: strongly agree = 5, agree = 4, neutral = 3, disagree = 2, strongly disagree = 1. We tested these scale bias scores for relation with the taste ratings. Where an association was indicated, the measure was included as a covariate in the multivariate model to examine whether the genetic architecture was changed. Lastly, cross-correlations were calculated between the intensity ratings of PROP for the first born twin with those of their co-twin’s ratings for the other 4 traits, for both MZ and DZ pairs, to examine any genetic effect on scale use rather than taste perception.
Results
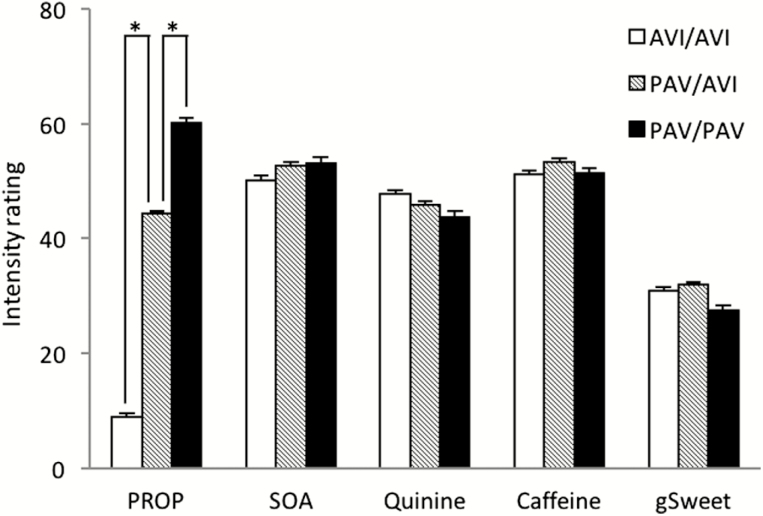
Mean ratings, standard deviations, twin correlations, and heritability estimates for the perceived intensity of PROP, SOA, quinine, caffeine, and gSweet are shown in Table 1. The mean rating was lower for PROP, and the variance was slightly larger, compared with the other bitter compounds. This was due to the distinct differences in PROP response between the 3 TAS2R38 diplotype groups and 31% of the participants being low-intensity tasters who could barely detect the bitterness in PROP (Figure 1). In contrast, there was no effect of the TAS2R38 diplotype on SOA, quinine, caffeine, or gSweet. Heritability for PROP (h 2 = 0.73) was significantly higher than that for SOA, quinine, and caffeine (h 2 = 0.35–0.4), in line with prior work (Hansen et al. 2006). The mean and heritability (h 2 = 0.36) estimates for the gSweet factor are the same as those reported previously (Hwang et al. 2015).
Table 1.
Taste intensity characteristics.
| PROP | SOA | Quinine | Caffeine | gSweet | |
|---|---|---|---|---|---|
| Mean ± SDa | 36.7±29.6 | 51.6±22.6 | 45.6±22.9 | 52.2±23.3 | 31.2±15.2 |
| Twin correlationsb | |||||
| r MZ (95% CI) | 0.72 (0.65, 0.78) | 0.34 (0.23, 0.45) | 0.40 (0.29, 0.50) | 0.29 (0.17, 0.41) | 0.36 (0.24, 0.46) |
| r DZ (95% CI) | 0.34 (0.26, 0.42) | 0.25 (0.10, 0.34) | 0.18 (0.09, 0.27) | 0.21 (0.12, 0.30) | 0.20 (0.17, 0.34) |
| Heritability (95% CI) | 0.73 (0.67, 0.77) | 0.40 (0.31, 0.49) | 0.40 (0.31, 0.49) | 0.34 (0.24, 0.43) | 0.36 (0.27, 0.45) |
| Phenotypic correlations (95% CI) | |||||
| Full sample | |||||
| SOA | 0.29 (0.25, 0.34) | — | — | — | — |
| Quinine | 0.25 (0.20, 0.29) | 0.59 (0.56, 0.62) | — | — | — |
| Caffeine | 0.31 (0.26, 0.35) | 0.64 (0.62, 0.67) | 0.63 (0.60, 0.65) | — | — |
| gSweet | 0.22 (0.18, 0.27) | 0.35 (0.30, 0.39) | 0.40 (0.37, 0.44) | 0.40 (0.36, 0.43) | — |
| TAS2R38 adjustedc | |||||
| SOA | 0.37 (0.33, 0.41) | — | — | — | — |
| Quinine | 0.44 (0.40, 0.47) | 0.60 (0.57, 0.63) | — | — | — |
| Caffeine | 0.42 (0.38, 0.46) | 0.64 (0.61, 0.67) | 0.63 (0.61, 0.66) | — | — |
| gSweet | 0.32 (0.27, 0.36) | 0.35 (0.30, 0.39) | 0.41 (0.36, 0.45) | 0.39 (0.35, 0.43) | — |
| AVI/AVI excludedd | |||||
| SOA | 0.44 (0.39, 0.48) | — | — | — | — |
| Quinine | 0.48 (0.43, 0.52) | 0.60 (0.56, 0.63) | — | — | — |
| Caffeine | 0.46 (0.41, 0.51) | 0.63 (0.60, 0.66) | 0.62 (0.59, 0.66) | — | — |
| gSweet | 0.31 (0.26, 0.36) | 0.33 (0.28, 0.38) | 0.37 (0.32, 0.42) | 0.37 (0.32, 0.42) | — |
Means and standard deviations, MZ and DZ twin correlations, heritability estimates and phenotypic correlations for perceived intensity ratings (millimeters on a labeled magnitude scale) of PROP, SOA, quinine, caffeine, and a general sweetness factor (gSweet).
a n = 1881 - 1892. The sample size (N) varies as not all participants completed the entire test.
b238–240 MZ and 446–449 DZ twin pairs; all estimates are from univariate AE models.
c TAS2R38 diplotype, available for n = 1756, was tested in a partial dominant model.
d N reduced to 1229 when TAS2R38 AVI/AVI diplotype excluded.
Figure 1.
Perceived intensity ratings (mean + standard error) for 4 bitter solutions and the general sweetness factor (a weighted mean ratings of glucose, fructose, NHDC, and aspartame). Participants grouped by TAS2R38 diplotypes (n = 527 for AVI/AVI, n = 916 for PAV/AVI, n = 313 for PAV/PAV). * indicates significant differences (Student’s t-test, P < 0.001).
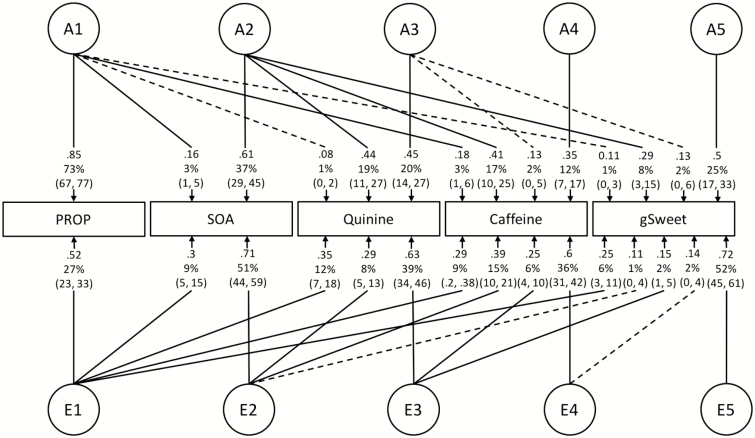
Perceived intensity for gSweet was moderately correlated with those for SOA, quinine, and caffeine (r p = 0.35–0.40) and more weakly associated with PROP (r p = 0.22) (Table 1). Multivariate model-fitting showed that the common environmental components (C) could be dropped without loss of fit (Supplementary Table 4). Multivariate AE modeling identified a genetic factor (A2 factor in Figure 2) accounting for 8% of the variance in gSweet and 17–37% of the variance in SOA, quinine, and caffeine. Only 1% of the variance in gSweet was genetically shared with PROP (A1 factor in Figure 2). There was also little shared genetic variance between PROP and the other 3 bitter compounds (i.e. 1–3%, A1 in Figure 2). Further, the association between gSweet and PROP was largely due to an environmental “PROP” factor (E1), accounting for 27% of the variance in PROP and 6% in gSweet. This environmental factor also accounted for a small amount of the variance (9–12%) in SOA, quinine, and caffeine.
Figure 2.
The Cholesky AE model showing estimates of standardized path coefficients (can be squared to get the variance) and percentage of variance with 95% CIs and covariation between perceived intensity of PROP, SOA, quinine, caffeine, and gSweet. The boxes and the circles represent observed variables (phenotypes) and latent variables, respectively. A and E are the additive genetic and non-shared environmental factors. Dash lines are insignificant estimates. See Supplementary Table 5 for absolute variance.
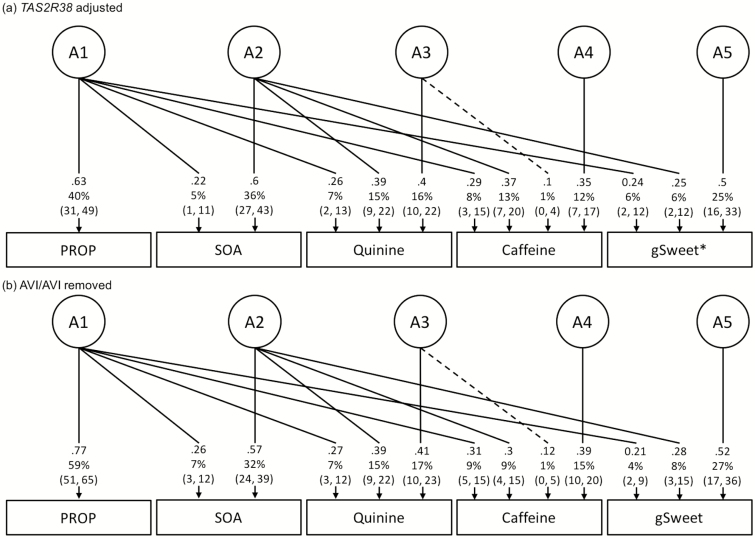
When the model was adjusted for the TAS2R38 diplotype (i.e. the TAS2R38 genetic effect was removed), the correlation between gSweet and PROP increased (r p = 0.32), as did the correlation between PROP and the other bitter compounds (r p increased from 0.25–0.31 to 0.37–0.44) (Table 1). The stronger association was due to an increase in shared genetic variance with 6% of variance in gSweet now overlapping with the genetic variance for PROP (A1 in Figure 3a; h 2 of PROP decreased to 0.40 after adjustment). Similarly, after adjusting for TAS2R38, there was an increase in shared genetic variance for SOA, quinine, and caffeine with PROP. Notably, no increase in the environmental variance (E1) shared with PROP was found for gSweet, SOA, Caffeine, or Quinine. Adjusting for TAS2R38 reduces the absolute genetic variance in PROP, with environmental variance for PROP and the genetic and environmental variances of other tastes remaining the same (Supplementary Table 5). In addition, removing PROP low-sensitivity tasters, rather than adjusting for TAS2R38, produced a similar covariance structure, even though the sample size was reduced (Figure 3b; Supplementary Table 5c). This increase in the genetic correlations after adjustment for TAS2R38 or removal of PROP low-sensitivity tasters (Table 2) contrasts with the environmental correlations that remained the same. In terms of genetic variance as a proportion of the heritability (Supplementary Table 6), 23% of the genetic variance in gSweet (8% of the variance divided by the heritability of 0.36) overlapped with 46% of genetic variance in quinine, 49% in caffeine, and 94% in SOA, whereas only 3% of the genetic variance in gSweet overlapped with PROP, which increased to 15% after adjusting for the TAS2R38 diplotype.
Figure 3.
The conditioned Cholesky AE models showing estimates of standardized path coefficients (can be squared to get the variance) and percentage of variance with 95% CIs and covariation between perceived intensity of PROP, SOA, quinine, caffeine, and gSweet. The boxes and the circles represent observed variables (phenotypes) and latent variables, respectively. Only additive genetic factors (A) are shown here because estimates of environmental factors (E) are not different from those estimated from the full sample (n = 1901). Dashed lines are insignificant estimates. (a) Adjusting for TAS2R38 diplotype (n = 1756). * Modelling results with gSweet replaced by glucose and fructose are shown in Supplementary Table 7. (b) Participants with TAS2R38 AVI/AVI diplotype removed (n = 1229). (See Supplementary Table 5 for absolute variance.)
Table 2.
Genetic (lower triangle) and environmental (upper triangle) correlations (95% confidence intervals) between perceived intensities of 4 bitter compounds and the general sweet intensity estimated from bivariate AE models.
| Genetic correlations | Environmental correlations | ||||
|---|---|---|---|---|---|
| PROP | SOA | Quinine | Caffeine | gSweet | |
| Full sample | |||||
| PROP | — | 0.38 (0.28, 0.48) | 0.45 (0.35, 0.54) | 0.36 (0.26, 0.45) | 0.32 (0.21, 0.41) |
| SOA | 0.26 (0.15, 0.37) | — | 0.52 (0.44, 0.59) | 0.58 (0.51, 0.64) | 0.24 (0.14, 0.33) |
| Quinine | 0.12 (0, 0.23) | 0.70 (0.58, 0.80) | — | 0.60 (0.53, 0.66) | 0.35 (0.25, 0.44) |
| Caffeine | 0.31 (0.19, 0.42) | 0.76 (0.65, 0.86) | 0.68 (0.55, 0.79) | — | 0.36 (0.27, 0.45) |
| gSweet | 0.18 (0.05, 0.29) | 0.51 (0.35, 0.67) | 0.50 (0.33, 0.65) | 0.46 0.27, 0.62) | — |
| TAS2R38 adjusted | |||||
| PROP | — | 0.37 (0.27, 0.47) | 0.44 (0.34, 0.52) | 0.38 (0.28, 0.46) | 0.27 (0.16, 0.37) |
| SOA | 0.36 (0.19, 0.52) | — | 0.51 (0.42, 0.58) | 0.58 (0.50, 0.64) | 0.24 (0.14, 0.33) |
| Quinine | 0.43 (0.26, 0.58) | 0.74 (0.62, 0.85) | — | 0.60 (0.52, 0.66) | 0.37 (0.27, 0.46) |
| Caffeine | 0.50 (0.32, 0.65) | 0.76 (0.64, 0.85) | 0.70 (0.57, 0.81) | — | 0.36 (0.27, 0.45) |
| gSweet | 0.40 (0.22, 0.56) | 0.52 (0.36, 0.67) | 0.47 (0.29, 0.62) | 0.45 (0.26, 0.61) | — |
| TAS2R38 AVI/AVI excluded | |||||
| PROP | — | 0.46 (0.34, 0.56) | 0.53 (0.42, 0.62) | 0.43 (0.32, 0.54) | 0.29 (0.16, 0.41) |
| SOA | 0.44 (0.27, 0.57) | — | 0.50 (0.40, 0.59) | 0.60 (0.52, 0.68) | 0.19 (0.07, 0.31) |
| Quinine | 0.44 (0.27, 0.57) | 0.75 (0.60, 0.88) | — | 0.60 (0.51, 0.68) | 0.36 (0.24, 0.47) |
| Caffeine | 0.52 (0.36, 0.66) | 0.68 (0.51, 0.80) | 0.66 (0.48, 0.79) | — | 0.34 (0.22, 0.45) |
| gSweet | 0.34 (0.17, 0.50) | 0.55 (0.36, 0.74) | 0.39 (0.17, 0.58) | 0.42 (0.19, 0.62) | — |
n = 1229. n = 1756. TAS2R38 diplotype was tested in a partial dominant model.
Since a commonly based definition of sweet taste is the oral perception of natural sugars, we used the intensity rating for glucose and fructose instead of gSweet to remove any possible bias of a weighted mean intensity rating of both the sugars and high-potency sweeteners; the results were similar (Supplementary Table 7).
Further, there was little evidence of the effect of scale use bias, IQ, or personality on intensity ratings. While some weak associations were observed (Supplementary Table 8), there was no change in the covariance structure when the emphasis score, IQ, and personality were included as covariates in the multivariate model (Supplementary Table 9). In addition, scale use had no effect on the genetic estimates; the cross-trait (e.g. PROP for twin 1 with gSweet for twin 2) correlations for both MZ and DZ twins were low and of similar magnitude (r MZ = 0–0.10, r DZ = 0.03–0.11; Supplementary Table 10).
Discussion
This study examined whether there is heritable genetic overlap between the perception of sweetness and bitterness of 8 different compounds. Using multivariate genetic modeling, we showed that up to a quarter of the genetic variance in sweet perception is shared with at least half, or more, of the genetic variance in SOA, quinine, and caffeine. Further, after adjustment of the TAS2R38 diplotype, 15% of genetic variance in sweetness is also in common with PROP perception. These results suggest that human perceptions of sweetness and bitterness are linked through shared genes and the extent of overlap depends on the specific taste stimuli.
Confirming prior work, intensity ratings for sweetness were weakly-to-moderately associated with bitterness ratings (Lim et al. 2008;Knaapila et al. 2012; Fischer et al. 2014). Although Lim et al. (2008) only found an association between sucrose and quinine and not between sucrose and PROP, their sample size was small (n = 83) compared to Fischer’s and ours (n = 1670 and 1901, respectively). This suggests that a bigger sample size is required to detect weak associations with PROP. Our finding that the associations between gSweet and SOA, quinine, and caffeine were mainly due to a shared genetic factor supports the current understanding that, in humans, the perception of both sweetness and bitterness is mediated via G protein-coupled receptors (GPCR) and other shared transduction proteins in the oral cavity (Reed et al. 2006; Temussi 2009). Genetic variation in these taste genes has been shown to link to individual differences in the perception of both sweetness (Fushan et al. 2009; Fushan et al. 2010) and bitterness (Reed et al. 2010; Ledda et al. 2014). Evidence from animal models also shows that knocking out common genes in their downstream signaling pathways, including genes that encode the G-protein alpha-gustducin, Ggamma13, the lipid enzyme phospholipase C beta2, and transient receptor potential ion channel TRPM5, leads to a reduced response to both sweet and bitter tastes (Taruno et al. 2013; Wong et al. 1996; Zhang et al. 2003).
Our prior work showed that the perception of PROP was weakly associated with the perception of other bitter compounds at the genetic level (Hansen et al. 2006). Here, using a greatly expanded sample, this study confirmed this lack of strong association, but also identified, after adjusting for the TAS2R38 diplotype, a shared genetic factor accounting for some of the genetic variance in the perception of PROP as along with other bitter and sweet tastes. This finding supports the 2-locus model (Olson et al. 1989) of the perception of TAS2R38 associated bitter compounds [i.e. PROP and its structurally related chemical propylthiocarboamide (Bufe et al. 2005)], with 1 locus controlling compound-specific tasting and the other locus controlling general taste responsiveness. This shared genetic factor (A1 factor in Figure 3) may correspond to a shared pathway at the peripheral level because the perception of PROP is believed to go through the same GPCR-based signaling elements as other bitter and sweet tastes (Margolskee 2002; Chaudhari and Roper 2010). Alternatively, shared genetic variation could also involve a shared pathway at the central neural level, supporting the hypothesis (Green and George 2004) that a central nervous system mechanism influences general responsiveness to tastes. In that study, the ability to perceive thermally induced taste predicted higher taste responses to sweet and bitter stimuli, including PROP, as well as salty, sour, and umami taste stimuli. They eliminated the potential confounders of papillae density and gustatory afferents used in signal transduction and concluded that the overall control of gain in the orosensory system is likely centrally determined.
In addition to the perception of sweetness and bitterness, the shared genetic factors identified here could also link to the perception of umami taste, especially at the peripheral level, because a glutamate taste receptor is also a member of the Class 1 Taste GPCR family (Li et al. 2002; Temussi 2009). Animal studies have shown that these taste qualities (sweetness, bitterness, and umami) are encoded by common downstream transduction components that are not believed to be used by ionic taste stimuli (sourness and saltiness) (Zhang et al. 2003; Taruno et al. 2013). Future taste genetic studies involving glutamate, salts, and acids could test this hypothesis in humans and would help tease apart which common genes are involved.
To assure that the genetic overlap between sweetness and bitterness was not due to confounding factors, we tested a series of alternative hypotheses. First, we found only subtle associations between personality traits and taste ratings, in contrast to other studies (Meier et al. 2012; Sagioglou and Greitemeyer 2016). This difference between our weak personality influence and those previously observed may be due to differences in measures of taste preference rather than taste intensity in this study. Second, individuals with higher IQ rated taste solutions as less intense. There is some evidence that people with higher IQ are less likely to give extreme ratings (Light et al. 1965) and we also observed a negative correlation between IQ and emphasis scores (r p = −0.24) in this study. Therefore, these people could be more conservative in rating the intensity of taste solutions. Alternatively, the association may be due to pleiotropic effects with shared genes influencing both taste perception and the development of intelligence. Nevertheless, neither personality traits nor IQ modifies the genetic architecture between sweetness and bitterness. Third, the emphasis scores were not associated with any taste ratings, so an individual’s response style is not likely a concern. Lastly, scale use style is not influenced by genetics and did not inflate or deflate the estimates of heritability or genetic covariances. The last test supports that a gLMS can be a valid instrument to study taste genetics. These 4 tests of alternative hypotheses allow us to conclude more strongly that the genetic correlation between sweetness and bitterness results from true correlates of taste perception and that these estimates of genetic covariances are valid rather than an artifact of scale use.
There are some limitations of this study. First, partitioning trait variance using Cholesky decomposition is restricted by the trait order in the model, with only the last trait in the model including a trait specific factor. For all other traits, the trait-specific variance is pushed to a group factor(s), which can, therefore, elevate covariances between traits. We tested these models in different orders, however, and obtained similar results. Second, although adjusting for the TAS2R38 diplotype increased the shared genetic variance between PROP and other taste stimuli, the increase was small and the confidence intervals overlapped [e.g. gSweet increased from 3% (confidence intervals: 0 and 9%) to 15% (confidence intervals: 5 and 30%)]. Future studies with larger samples could provide more distinct results. Lastly, whether there is a common genetic pathway for the overall taste perception cannot be answered from this study. Although we showed a genetic factor for the perception of both sweet and bitter tastes, even including PROP, it might simply be a specific taste transduction factor that does not also account for the perception of umami, sourness, and saltiness.
In conclusion, this study examined the associations among perceived intensities of 4 bitter compounds—PROP, SOA, quinine, and caffeine—and an average sweet factor from 4 sweeteners, and modeling results identified 2 latent genetic factors suggesting 2 shared genetic pathways. We speculate that there are genes responsible for the recognition of a GPCR-taste signal or a general taste signal. Genetic covariation of downstream signaling elements could involve peripheral and/or central mechanisms, including genes encoding molecules that transduce information from the taste receptors to the nerves and neural elements in the gustatory brain circuits. Future genome-wide association analysis in humans could identify common genes across different taste modalities, which have implications for understanding the molecular mechanism of GPCR-transduced taste perception as well as the taste-based metabolic signals from the gastrointestinal tract, pancreas, liver, thyroid, and elsewhere.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was supported by the National Institute of Health [DC02995 to PASB and DC004698 to DRR] and the Australian NHMRC [241944 and 1031119].
Supplementary Material
Acknowledgements
We thank Kirsten J Mascioli, Christopher Tharp, Fujiko Duke, Deborah Lee and Corrine Mansfield from the Monell Chemical Senses Center for manufacturing the taste tests; and from the QIMR Berghofer, Marlene Grace, Ann Eldridge, Natalie Garden, Kerrie McAloney for project co-ordination, data collection, and data entry, and David Smyth and Anthony Conciatore for computer support. In particular, thanks go to twins and their families for their participation.
References
- Bajec MR, Pickering GJ. 2008. Thermal taste, PROP responsiveness, and perception of oral sensations. Physiol Behav. 95:581–590. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM. 1979. Bitter taste of saccharin related to the genetic ability to taste the bitter substance 6-n-propylthiouracil. Science. 205:934–935. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Duffy VB, Miller IJ. 1994. PTC/PROP tasting: anatomy, psychophysics, and sex effects. Physiol Behav. 56:1165–1171. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Duffy VB, Reed D, Williams A. 1996. Supertasting, earaches and head injury: genetic and pathology atler our taste. Neurosci Biobehav Rev. 20:79–87. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. 2005. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 15:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media. [Google Scholar]
- Chaudhari N, Roper SD. 2010. The cell biology of taste. J Cell Biol. 190:285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PT, McCrae RR. 1992. Revised NEO personality inventory (NEO PI-R) and NEO Five-Factor Inventory (NEO-FFI): Professional manual. Psychological Assessment Resources. [Google Scholar]
- Drayna D, Coon H, Kim UK, Elsner T, Cromer K, Otterud B, Baird L, Peiffer AP, Leppert M; Utah Genetic Reference Project 2003. Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum Genet. 112:567–572. [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Barratt-Fornell A. 1998. Genetic sensitivity to 6-n-propylthiouracil and sensory responses to sugar and fat mixtures. Physiol Behav. 63:771–777. [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Barratt-Fornell A. 2001. Genetic taste markers and food preferences. Drug Metab Dispos. 29(4 Pt 2):535–538. [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Shore AB, Barratt-Fornell A. 1997. Nontasters, tasters, and supertasters of 6-n-propylthiouracil (PROP) and hedonic response to sweet. Physiol Behav. 62:649–655. [DOI] [PubMed] [Google Scholar]
- Fischer ME, Cruickshanks KJ, Pankow JS, Pankratz N, Schubert CR, Huang GH, Klein BE, Klein R, Pinto A. 2014. The associations between 6-n-propylthiouracil (PROP) intensity and taste intensities differ by TAS2R38 haplotype. J Nutrigenet Nutrigenomics. 7:143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Drayna D. 2010. Association between common variation in genes encoding sweet taste signaling components and human sucrose perception. Chem Senses. 35:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. 2009. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr Biol. 19:1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BG, George P. 2004. ‘Thermal taste’ predicts higher responsiveness to chemical taste and flavor. Chem Senses. 29:617–628. [DOI] [PubMed] [Google Scholar]
- Green BG, Shaffer GS, Gilmore M. 1993. Derivation and evaluation of a semantic scale of oral sensation magnitude with apparent ratio properties. Chem Senses. 18: 683–702. [Google Scholar]
- Hansen JL, Reed DR, Wright MJ, Martin NG, Breslin PA. 2006. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCl, and caffeine. Chem Senses. 31:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Bartoshuk LM, Kidd JR, Duffy VB. 2008. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem Senses. 33:255–265. [DOI] [PubMed] [Google Scholar]
- Hayes JE, Keast RS. 2011. Two decades of supertasting: where do we stand? Physiol Behav. 104:1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LD, Zhu G, Breslin PA, Reed DR, Martin NG, Wright MJ. 2015. A common genetic influence on human intensity ratings of sugars and high-potency sweeteners. Twin Res Hum Genet. 18:361–367. [DOI] [PubMed] [Google Scholar]
- Jackson DN. 1998. Multidimensional aptitude battery II. Sigma Assessment Systems. [Google Scholar]
- Jewell G, McCourt ME. 2000. Pseudoneglect: a review and meta-analysis of performance factors in line bisection tasks. Neuropsychologia. 38:93–110. [DOI] [PubMed] [Google Scholar]
- Keast RS, Roper J. 2007. A complex relationship among chemical concentration, detection threshold, and suprathreshold intensity of bitter compounds. Chem Senses. 32:245–253. [DOI] [PubMed] [Google Scholar]
- Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, James RS, Wysocki CJ, Rhyu M, Tordoff MG, et al. 2012. Genetic analysis of chemosensory traits in human twins. Chem Senses. 37:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda M, Kutalik Z, Souza Destito MC, Souza MM, Cirillo CA, Zamboni A, Martin N, Morya E, Sameshima K, Beckmann JS, le Coutre J, Bergmann S, Genick UK. 2014. GWAS of human bitter taste perception identifies new loci and reveals additional complexity of bitter taste genetics. Hum Mol Genet. 23:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. 2002. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 99:4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light CS, Zax M, Gardiner DH. 1965. Relationship of age, sex, and intelligence level to extreme response style. J Pers Soc Psychol. 2:907–909. [DOI] [PubMed] [Google Scholar]
- Lim J, Urban L, Green BG. 2008. Measures of individual differences in taste and creaminess perception. Chem Senses. 33:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin JC, Bartels M, Boomsma DI, Bratko D, Martin NG, Nichols RC, Wright MJ. 2015. Is there a genetic correlation between general factors of intelligence and personality? Twin Res Hum Genet. 18:234–242. [DOI] [PubMed] [Google Scholar]
- Lucchina LA, Curtis OF, 5th, Putnam P, Drewnowski A, Prutkin JM, Bartoshuk LM. 1998. Psychophysical measurement of 6-n-propylthiouracil (PROP) taste perception. Ann N Y Acad Sci. 855:816–819. [DOI] [PubMed] [Google Scholar]
- Ly A, Drewnowski A. 2001. PROP (6-n-Propylthiouracil) tasting and sensory responses to caffeine,sucrose, neohesperidin dihydrochalcone and chocolate. Chem Senses. 26:41–47. [DOI] [PubMed] [Google Scholar]
- Margolskee RF. 2002. Molecular mechanisms of bitter and sweet taste transduction. J Biol Chem. 277:1–4. [DOI] [PubMed] [Google Scholar]
- Meier BP, Moeller SK, Riemer-Peltz M, Robinson MD. 2012. Sweet taste preferences and experiences predict prosocial inferences, personalities, and behaviors. J Pers Soc Psychol. 102:163–174. [DOI] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. 2002. Mx: statistical modeling (6th edn). Richmond: (VA: ): Department of Psychiatry, Virginia Commonwelth University. [Google Scholar]
- Neale MC, Cardon L. 1992. Methodology for genetic studies of twins and families. Dordrecht, Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Olson JM, Boehnke M, Neiswanger K, Roche AF, Siervogel RM. 1989. Alternative genetic models for the inheritance of the phenylthiocarbamide taste deficiency. Genet Epidemiol. 6:423–434. [DOI] [PubMed] [Google Scholar]
- Reed DR, Tanaka T, McDaniel AH. 2006. Diverse tastes: Genetics of sweet and bitter perception. Physiol Behav. 88:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Zhu G, Breslin PA, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ. 2010. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 19:4278–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagioglou C, Greitemeyer T. 2016. Individual differences in bitter taste preferences are associated with antisocial personality traits. Appetite. 96:299–308. [DOI] [PubMed] [Google Scholar]
- Schifferstein HN, Frijters JE. 1991. The perception of the taste of KCl, NaCl and quinineHCl is not related to PROP-sensitivity. Chem Senses. 16: 303–317. [Google Scholar]
- Taruno A, Vingtdeux V, Ohmoto M, Ma Z, Dvoryanchikov G, Li A, Adrien L, Zhao H, Leung S, Abernethy M, et al. 2013. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 495: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temussi PA. 2009. Sweet, bitter and umami receptors: a complex relationship. Trends Biochem Sci. 34:296–302. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Nurse RJ. 1997. Fat perception is related to PROP taster status. Physiol Behav. 61:949–954. [DOI] [PubMed] [Google Scholar]
- Wise PM, Hansen JL, Reed DR, Breslin PA. 2007. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem Senses. 32:749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF. 1996. Transduction of bitter and sweet taste by gustducin. Nature. 381:796–800. [DOI] [PubMed] [Google Scholar]
- Wright MJ, Martin NG. 2004. Brisbane Adolescent Twin Study: Outline of study methods and research projects. Austral J Psychol 56:65–78. [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. 2003. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 112:293–301. [DOI] [PubMed] [Google Scholar]
- Zhu G, Montgomery GW, James MR, Trent JM, Hayward NK, Martin NG, Duffy DL. 2007. A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur J Hum Genet. 15:94–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.