This study shows a novel and safe method for inducing β-cell differentiation from expanded human pancreatic duct-derived cells by using MAFA synthetic modified mRNA-based reprogramming. The resulting cells showed glucose-dependent insulin secretion both in vitro and after transplantation into diabetic animals, where they led to significant and prompt reduction of blood glucose levels.
Keywords: Diabetes, Insulin-producing cells, SCID-beige mice, Synthetic modified mRNA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A (MAFA)
Abstract
β-Cell replacement therapy represents the most promising approach to restore β-cell mass and glucose homeostasis in patients with type 1 diabetes. Safety and ethical issues associated with pluripotent stem cells stimulated the search for adult progenitor cells with endocrine differentiation capacities. We have already described a model for expansion and differentiation of human pancreatic duct-derived cells (HDDCs) into insulin-producing cells. Here we show an innovative and robust in vitro system for large-scale production of β-like cells from HDDCs using a nonintegrative RNA-based reprogramming technique. Synthetic modified RNAs for pancreatic transcription factors (pancreatic duodenal homeobox 1, neurogenin3, and V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A [MAFA]) were manufactured and daily transfected in HDDCs without strongly affecting immune response and cell viability. MAFA overexpression was efficient and sufficient to induce β-cell differentiation of HDDCs, which acquired a broad repertoire of mature β-cell markers while downregulating characteristic epithelial-mesenchymal transition markers. Within 7 days, MAFA-reprogrammed HDDC populations contained 37% insulin-positive cells and a proportion of endocrine cells expressing somatostatin and pancreatic polypeptide. Ultrastructure analysis of differentiated HDDCs showed both immature and mature insulin granules with light-backscattering properties. Furthermore, in vitro HDDC-derived β cells (called β-HDDCs) secreted human insulin and C-peptide in response to glucose, KCl, 3-isobutyl-1-methylxanthine, and tolbutamide stimulation. Transplantation of β-HDDCs into diabetic SCID-beige mice confirmed their functional glucose-responsive insulin secretion and their capacity to mitigate hyperglycemia. Our data describe a new, reliable, and fast procedure in adult human pancreatic cells to generate clinically relevant amounts of new β cells with potential to reverse diabetes.
Significance
β-Cell replacement therapy represents the most promising approach to restore glucose homeostasis in patients with type 1 diabetes. This study shows an innovative and robust in vitro system for large-scale production of β-like cells from human pancreatic duct-derived cells (HDDCs) using a nonintegrative RNA-based reprogramming technique. V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A overexpression was efficient and sufficient to induce β-cell differentiation and insulin secretion from HDDCs in response to glucose stimulation, allowing the cells to mitigate hyperglycemia in diabetic SCID-beige mice. The data describe a new, reliable, and fast procedure in adult human pancreatic cells to generate clinically relevant amounts of new β cells with the potential to reverse diabetes.
Introduction
Type 1 diabetes (T1D) is a chronic autoimmune disease characterized by T cell-mediated destruction of insulin-producing β cells and subsequent imbalance of blood glucose homeostasis [1]. Proof-of-concept for β-cell replacement therapy was established with whole-organ or islet transplantation [2, 3], but organ shortage and midterm graft dysfunction are recurrent barriers to clinical use. Reprogramming of progenitor cells represents an attractive alternative, with embryonic and induced pluripotent stem cells (PSCs) being closest to clinical grade because of unlimited supply and in vivo glucose-regulated insulin secretion capacities [4–6]. Recent data suggest the possibility to foster in vitro maturation of PSCs [7], which per se might improve diabetes outcome after transplantation. Unfortunately, the use of PSCs remains associated with ethically sensitive sampling and/or costly propagation procedures. Moreover, the risk for overgrowth or metastasis of undifferentiated cells may not be totally avoided by selection strategies [8], which encouraged the development of macro-encapsulation techniques in the setting of clinical trials (NCT02239354). There is thus a need to identify new sources of cells that have β-cell differentiation potential and expansion capacities and that maintain a stable differentiated phenotype in vivo.
In the past decade, somatic cells in the pancreas (e.g., duct cells [DCs]), acinar cells, and α cells [9–11]) were shown to have capacities to generate insulin-producing cells. Among those lineages, DCs are attractive because they give rise to all pancreatic epithelial cells during embryonic development [12] and because they participate in β-cell turnover after injury [13]. Recent data on α-to-β transdifferentiation revealed a crucial role of the pancreatic duct lining in the recruitment of newly formed β cells [14] and the involvement of a preliminary duct-to-α switch occurring after a transitory state of epithelial-mesenchymal transition (EMT). Because human DCs—like all epithelial lineages—undergo early loss of proliferation and of epithelial phenotype in vitro, we previously developed a new strategy to culture and expand human DCs via the forcing of a partial EMT [15]. The resulting cells, called human pancreatic duct-derived cells (HDDCs), had potential for β-cell differentiation, with glucose-unresponsive insulin secretion detected in populations containing 2%–3% insulin-positive (insulin+) cells.
Murine loss-of-function models revealed the essential role of transcription factors (TFs) in controlling pancreas specification, endocrine differentiation, and β-cell maturation. The three prominent TFs implicated in these milestones are pancreatic duodenal homeobox 1 (PDX1), neurogenin3 (NGN3), and V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A (MAFA) [9, 16–20]. MAFA, a basic-leucine zipper TF expressed in mature β cells only, regulates insulin secretion by specifically binding the RIPE3b/C1 insulin promoter region [21].
To improve the timing and efficiency of β-cell differentiation of HDDCs, we developed a TF-based reprogramming strategy using synthetic modified mRNAs (smRNAs). The smRNA technology was used to induce pluripotency or transdifferentiation in various settings [22–26] and showed its superiority to virus-based protocols [27]. Concerns about smRNA degradation, stability problems, and activation of immune mechanisms against single-stranded RNAs were addressed, and current synthesis protocols allow robust protein expression and maintenance of cell viability after consecutive transfections [28, 29]. In our study, we show a novel and safe method to induce β-cell differentiation from expanded HDDCs using MAFA smRNA-based reprogramming. The resulting cells showed glucose-dependent insulin secretion both in vitro and after transplantation into diabetic animals, where they lead to significant and prompt reduction of blood glucose levels. To our knowledge, this is the first demonstration of efficient smRNA-based β-cell reprogramming using an adult human primary cell model.
Materials and Methods
Cell Isolation and Culture
Human pancreatic DCs were isolated from 32 cadaveric donors age 1 month to 68 years. The exocrine tissue was obtained through the collaboration with the Diabetes Research Institute, IRCCS San Raffaele Scientific Institute, Milan, Italy, within a human islet distribution program for basic research supported by the Juvenile Diabetes Research Foundation [30]. DCs were isolated within 48 hours using MACS Separation columns to purify CA19-9+ DCs as previously described [15]. CA19-9+ DCs were initially plated at 3 × 105 cells per cm2 in EGM-2-MV medium (Lonza, Allendale, NJ, http://www.lonza.com) without hydrocortisone. The medium was changed every 72 hours and the cells were cultured in 37°C humidified atmosphere containing 5% CO2. When the confluence reached 80%, DCs and HDDCs were passaged using 0.05% trypsin (CellGro; CellGenix, Freiburg, Germany, http://www.cellgenix.com) and seeded at 5,000 cells per cm2 into culture-treated plates. HDDCs were cryopreserved at each passage in aliquots containing 1 × 106 cells with fetal bovine serum (FBS; Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com) containing 10% dimethyl sulfoxide (Sigma-Aldrich).
In Vitro Production of Synthetic Modified mRNA
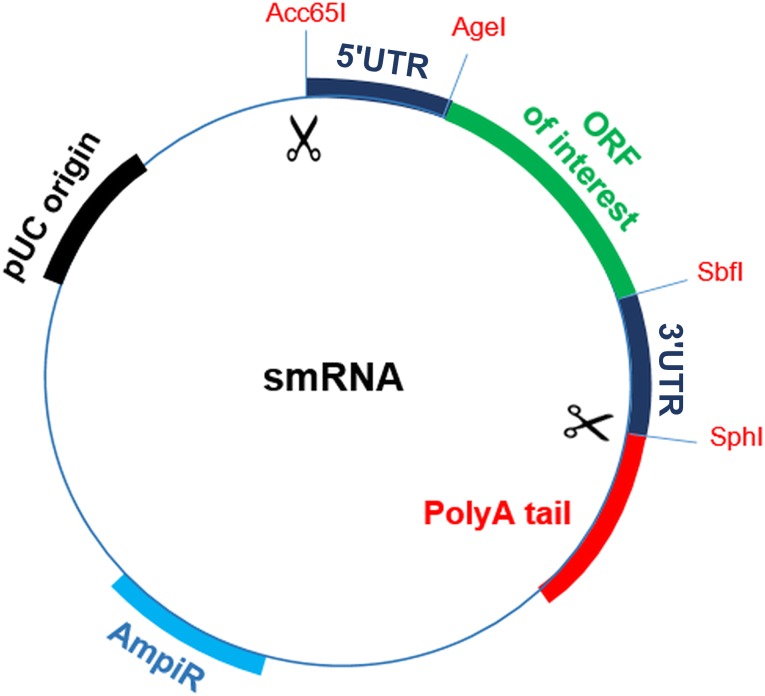
A “ready-to-use” plasmid (pRTU) containing 5′ and 3′ untranslated regions (UTRs) and a cloning site in a pIDTSmart Amp (IDT) backbone (Figure 1) was designed to generate the templates for in vitro transcription (IVT). The 5′ UTR incorporated a T7 promoter and a strong Kozak site to improve translation efficiency, whereas the 3′ UTR contained a murine α-globin oligo(dT) sequence. The open reading frames (ORFs) of interest (Addgene, Cambridge, MA, https://www.addgene.org) were cloned into the pRTU and digested using SbfI and AgeI restriction enzymes (Thermo Fisher Scientific Life Sciences). Subsequently, the linearized templates were amplified by polymerase chain reaction (PCR) using tailed primers to generate polyA sequences. IVTs were performed using a Megascript T7 kit (Ambion, Thermo Fisher Scientific Life Sciences) and 1.6 µg of PCR products that were capped with 15 mM of cap analog (New England Biolabs, Ipswich, MA, https://www.neb.com) to increase the stability of synthetic mRNAs. Complete substitution of 5-methyl cytidine bases for cytidine triphosphate and of pseudouridine for uridine-5′-triphosphate was performed to reduce immunogenicity of the molecules. Synthesized RNAs were purified using genomic DNA eliminator columns (RNA extraction kit, Qiagen, Hilden, Germany, https://www.qiagen.com) and then treated with Antarctic Phosphatase (New England Biolabs) for 30 minutes at 37°C to remove immunogenic 5′-triphosphates. RNAs were quantified by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Life Sciences) and aliquoted in volumes of 100 ng/µl in Tris-EDTA endotoxin-free water (Qiagen).
Figure 1.
Plasmid template for generation of synthetic modified RNAs. Structural diagram shows “ready-to-use” plasmid containing 5′ and 3′ UTR sequences used to generate smRNAs of interest. The ORFs for PDX1, NGN3, and MAFA were cloned and subsequently excised outwardly to the UTR regions using Acc65I and SphI enzymes. A polyA tail was added to the template with a polymerase chain reaction, and the amplicons were then used for in vitro transcription. Abbreviations: ORF, open reading frame; pUC, origin of replication; smRNA, synthetic modified mRNA; UTR, untranslated region.
smRNA Transfection
Differentiation experiments were performed by five to seven daily smRNA transfections using jetPEI (Polyplus Transfection, Illkirch, France, http://www.polyplus-transfection.com). After each transfection, HDDCs were incubated with the reprogramming medium for 24 hours. Transfection solution consisted of 1.2 µg smRNA diluted in 100 µl Opti-MEM basal medium (Thermo Fisher Scientific Life Sciences) containing 1.2 µl of jetPEI reagent. The mix was incubated for 15 minutes at room temperature (RT) before being dispended in 1 ml differentiation medium composed by DMEM low glucose (Thermo Fisher Scientific Life Sciences) without antibiotics supplemented by 10% FBS, 5 µM A83-01 (Tocris, Britsol, UK, https://www.tocris.com/), 1 mM valproic acid (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), 100 ng/ml bFGF (PeproTech, Rocky Hill, NJ, https://www.peprotech.com), and 200 ng/ml B18R (eBioscience, San Diego, CA, http://www.ebioscience.com). At the end of incubation, HDDCs were collected using 0.05% trypsin/EDTA (CellGro) for processing.
RNA Extraction, Reverse Transcription, and Quantitative PCR Analysis
Total RNA was extracted from cultured cells using Tripure isolation reagent (Roche, Basel, Switzerland, http://www.roche.com). The concentration was determined by using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Life Sciences). Reverse transcription was obtained by using a High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific Life Sciences). The quantitative PCR was carried out using primers of interest (supplemental online Table 1), Power SYBR Green PCR master mix (Thermo Fisher Scientific Life Sciences) and StepOnePlus Real-Time PCR System (Thermo Fisher Scientific Life Sciences). Primers for detection of exogenous smRNAs were designed to recognize the modified UTR regions of synthetic molecules. Results were analyzed by the comparative ∆CT method [31] and normalized to TATA-binding protein.
Immunofluorescence and Immunohistochemistry
Differentiated HDDCs were fixed in 4% paraformaldehyde at RT for 20 minutes and then permeabilized by using 1% Triton X-100 (Sigma-Aldrich) in phosphate-buffered saline (PBS) for 15 minutes. Nonspecific sites were blocked using 1% lamb serum (Immunosource, Schilde, Belgium, http://www.immunosource.com) for 1 hour at RT. Subsequently, cells were washed and incubated at 4°C overnight with primary antibodies at desired concentrations (supplemental online Table 2). After washing, cells were incubated at RT for 2 hours with secondary antibodies and with 1:5000 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Kidney tissues were fixed overnight at RT with 4% formaldehyde, embedded in paraffin, and sectioned (5 µm) for histological analysis. Before staining, paraffin was removed from the sections, rehydrated, and subjected to antigen retrieval by treatment with Dako target retrieval buffer (catalog no. S1699, Dako, Glostrup, Denmark, http://www.dako.com) 20 minutes in microwave at 350 W. The antibodies concentrations are described in supplemental online Table 2. The images were acquired in fluorescent mode using a Zeiss Axiovision A1 microscope (Carl Zeiss Inc., Thornwood, NY, http://www.zeiss.com) and a Cell Observer Spinning Disk confocal microscope. Cell frequencies were evaluated by counting positive cells in four to six different random fields at ×40 magnification (supplemental online Table 3).
Annexin V Assay
Cells were trypsinized and analyzed by flow cytometry to evaluate cell viability after seven daily transfections. About 105 cells were suspended in 100 µl Annexin V binding buffer (Pharmingen Annexin V-FITC Apoptosis Detection kit, BD Biosciences, Franklin Lakes, NJ, http://www.bdbiosciences.com/) containing 5 µl Annexin V antibody and 5 µl DAPI solutions. The mix was incubated for 15 minutes at RT before addition of 400 µl Annexin V binding buffer. The analyses were performed by using a BD FACS Canto II cytometer (BD Biosciences) and BD FACSDiva Software.
Intracellular MAFA smRNA detection
Cells transfected with smRNA for 5 consecutive days were incubated overnight for 16 hours with MAFA Hu-Cy5 SmartFlare RNA Detection Probe (SF-3118, EMD Millipore, Billerica, MA, http://www.emdmillipore.com) in 10% FBS-containing DMEM low glucose without antibiotics. After incubation, differentiated HDDCs were analyzed by using Zeiss Axiovision A1 microscope.
Luciferase Assay
HEK293 and MIN6 cell lines were plated into white 96-well plates before being transfected using jetPEI reagent at day 1 with 100 ng of a vector expressing luciferase under the control of the insulin promoter or with an empty control vector to measure background signal (SwitchGear Genomics, Menlo Park, CA, http://www.switchgeargenomics.com). At day 2, HEK293 were transfected with 100 ng of MAFA smRNA. At day 3, both HEK293 and MIN6 cells were incubated in dark condition for 30 minutes at RT with 100 µl Light Switch Assay Solution (SwitchGear Genomics) containing the luciferase substrate. Luciferase activity was measured for 10 seconds by using a Victor ×4 plate reader (PerkinElmer, Waltham, MA, http://www.perkinelmer.com).
Electron Microscopy
Ultrastructure analysis was performed on differentiated HDDCs fixed with 1% glutaraldehyde in 0.1 M sodium cacodylate buffer for 2 hours at 4°C, pH 7.4. The cells were then washed in 0.1 M sodium cacodylate and postfixed for 2 hours at 4°C in 1% osmium tetroxide. After a further washing, the cells were dehydrated in series of graded ethanol and embedded in Spurr. Cut sections of the samples were analyzed, colored with lead citrate, and uranyl acetate for 10 minutes each at RT, and observed in the transmission electron microscope at 80 kV (CM12, FEI, Eindhoven, The Netherlands, http://www.fei.com).
Laser Scanning Confocal Microscopy
HDDCs were plated into µ-slides glass bottom (Ibidi, Planegg/Martinsried, Germany, http://www.ibidi.com) and transfected with MAFA smRNA for 5 consecutive days. After 24 hours from the last transfection, the cells were analyzed with the LSM510 laser scanning confocal. The backscatter signal was obtained by using a 633-nm laser. Scanning speed and laser intensity were adjusted to minimize the labeling in the nontransfected cells.
Calcium Signaling Detection
The smRNA transfections were performed on HDDCs plated onto eight-well glass chamber slides. After treatment, cells were washed in Krebs Ringer Bicarbonate HEPES buffer (137 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO40.7H2O, 2.5 mM CaCl20.2H2O, 5 mM NaHCO3, 16 mM HEPES, 0.1% bovine serum albumin, pH 7.4) and incubated with Fluo4-AM (Thermo Fisher Scientific Life Sciences) for 60 minutes at 37°C. Cells were then washed in Krebs buffer for an additional 15 minutes to remove nonspecific dye. The images were acquired by using a Zeiss Axiovision A1 microscope after consecutive incubations of HDDCs with low (2.8 mM) glucose, high (20.2 mM) glucose, and low-glucose Krebs buffer containing 30 mM KCl for 30 minutes.
Glucose-Stimulated Insulin Secretion Assay
The cells were plated onto a 6-well plate and transfected with smRNA for 7 consecutive days. Subsequently, the cells were washed with PBS and incubated in 500 µl Krebs Ringer bicarbonate HEPES solution for 24 hours in basal and stimulated conditions (2.8 mM and 20.2 mM glucose, respectively). The supernatant was collected and stored at −80°C until analysis. Insulin secretion was measured by using the Bio-Plex Pro Assays kit (Bio-Rad, Hercules, CA, http://www.bio-rad.com). Fluorescence was measured by using a Bio-Plex 200 system (BioRad).
Insulin Content Measurement
CelLytic M reagent (Sigma-Aldrich) was used to extract insulin content. Protein content was consecutively measured using Bio-Plex Pro Assays kit (Bio-Rad). Fluorescence was measured using a Bio-Plex 200 system (BioRad).
Transplantation Studies
Experimental diabetes was induced in immunosuppressed SCID-beige male mice aged 8–10 weeks by intraperitoneal injection of streptozotocin (STZ) (180 mg/kg body weight). Animals were considered diabetic only if morning-fed blood glucose exceeded 350 mg/dl. A week after STZ injection, 1 × 106 of differentiated or undifferentiated HDDCs were transplanted under kidney capsules of the animals under isoflurane anesthesia. Animals were maintained on a 12 hours light/dark cycle during all the experimental procedure. Glycemia and body weight were monitored every 24 hours during follow-up period. At specific time points, intraperitoneal glucose tolerance assay was performed by IP injection of 2 g/kg body weight d-(+)-glucose and glycemic levels of the animals were measured at time 0 and 30 minutes after injection, whereas serum was collected to measure human insulin at 0, 15, and 30 minutes from glucose administration. Serum human insulin were measured by using Bio-Plex Pro Assays kit (Bio-Rad), with fluorescence read on a Bio-Plex 200 system (BioRad). At selected times, animals were sacrificed and kidneys were removed for graft analysis. All experiments were performed under approval of the ethical committee of our institution.
Statistical Analysis
Results are expressed as mean ± SEM, and statistically significant differences were assessed using an unpaired Student’s t test. All data were analyzed by Prism 5 software (GraphPad, San Diego, CA, http://www.graphpad.com).
Results
Messenger RNAs for PDX1, NGN3, and MAFA were manufactured using a ready-to-use plasmid (pRTU) to avoid the need for splint reactions (Fig. 1). After digestion, UTR-flanked ORFs were polyA-tailed to generate templates for IVT. Purified smRNAs were then treated with phosphatase to remove immunogenic residues. Although our initial HDDC populations were devoid of insulin or amylase expression from passage (P) 1 onward (data not shown), transfection experiments were carried out on HDDCs from P 2 to 5 to avoid the possibility of contamination with β or acinar cells. As described elsewhere [15], HDDCs are a uniform lineage of cells arising from purified CA19-9+ human pancreatic ductal cells and presenting partial EMT features with coexpression of mesenchymal and ductal epithelium markers. HDDCs have advantageous characteristics in terms of proliferation [15] and resistance to cryopreservation, as we calculated a recovery rate of 73.2% ± 9.9% (n = 5) after one cycle of cryopreservation at −196°C.
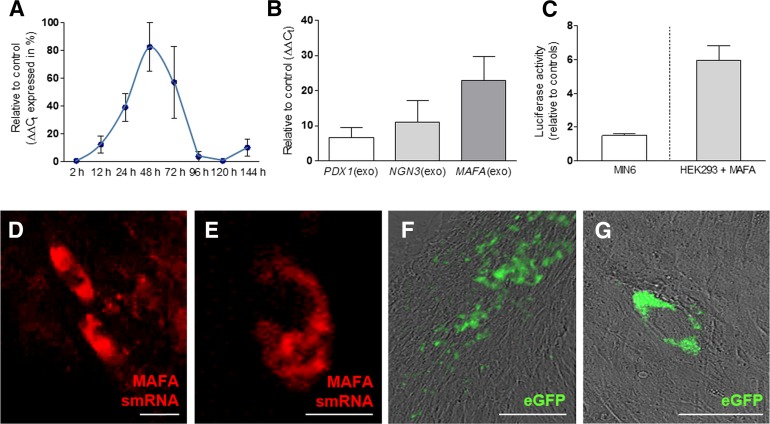
For transfection experiments, the jetPEI cationic vehicle was chosen from among other reagents (RNAiMAX and Lipofectamine 2000 [Thermo Fisher Scientific Life Sciences], X-treme GENE [Roche], polyethylenimine; data not shown) for its superior efficacy and minor toxicity. Reprogramming protocols were assayed after daily transfections (n = 7) to ensure sustained ectopic RNA and protein expression [32]. Study of the uptake kinetics (Fig. 2A, supplemental online Fig. 1) showed that peak cytoplasmic concentrations of smRNA occurred 48 hours after single transfections before being progressively degraded and undetectable after 5 days. Dose-response experiments were performed to evaluate the amounts of daily transfections required for differentiation to occur while maintaining viable cells. Our data showed that smRNAs for PDX1, NGN3, and MAFA were detected without variation of expression levels after 5, 7, or 10 daily transfections (respectively, 6.7 ± 3.2-fold, 11 ± 6.5-fold, 22.9 ± 7.0-fold untransfected controls; n = 5-11), suggesting a noncumulative effect of consecutive transfections after 5 days, when HDDCs achieved confluence. However, stability of MAFA smRNA was higher in HDDCs after 7 days as compared with PDX1 and NGN3 smRNAs (Fig. 2B).
Figure 2.
Validation of the smRNA system. (A): Time course showing kinetics and stability of MAFA smRNA after single transfection on human pancreatic duct-derived cells (HDDCs) at passages 2–5 (n = 3). (B): Quantitative real-time polymerase chain reaction shows the detection of exogenous PDX1, NGN3, and MAFA smRNAs on HDDCs (passages 2–5) after 7 consecutive daily transfections (n = 5–11). (C): Luciferase assays showed the binding of exogenous MAFA protein to human insulin promoter after transfection on HEK293. MIN6 cells were used as negative control (n = 3–5). (D, E): Fluorescent hybridization probes allowed detection of cytoplasmic MAFA smRNA (red) in 7.6% ± 1.2% of HDDCs after 5 transfections. (F, G): After 5 daily transfections with EGFP smRNA, 47.6% ± 27.4% of HDDCs sustained green fluorescence emission. Scale bars = 400 µm (F), 50 µm (D, G), and 25 µm (E). Abbreviations: eGFP, enhanced green fluorescent protein; MAFA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A; smRNA, synthetic modified mRNA.
Further experiments validated the efficiency of the smRNA system and the biological activity of cytoplasmic smRNAs. First, we studied the integrity of MAFA smRNA by evaluating its capacity to hybridize with a complementary base pair sequence of human MAFA gene. For this purpose, we used the Smart-Flare technology (EMD Millipore), which uncovered hybridization of the probes with MAFA by emission of cyanine 5 fluorescence in HDDCs (7.6% ± 1.2%) after a series of 5 daily smRNA transfections (Fig. 2D and 2E). Second, we evaluated protein translation of exogenous smRNA via the production of a template encoding for enhanced green fluorescent protein (EGFP), and we observed fluorescence in 45.3% ± 18.4% of HDDCs (n = 3) after 5 daily transfections (Fig. 2F and 2G). In addition, MAFA protein expression was detected with nuclear localization in 39.3% ± 6.2% HDDCs after 5 daily transfections (Figs. 3D, 3E, 4F–4I). Third, to verify whether the overexpressed MAFA protein had an insulin promoter binding capacity, we performed a luciferase assay using the HEK293 cell line. After a single MAFA smRNA transfection, we observed a 5.9 ± 0.8-fold increased luciferase activity as compared with untransfected cells, whereas no luciferase activity could be detected with MIN6 cells (Fig. 2C), suggesting specificity to exogenous human MAFA protein. Fourth, in all experiments involving PDX1, NGN3, and MAFA smRNA transfections, HDDCs showed upregulation of endogenous biological targets related to each TF. Endogenous NGN3 and MAFA were respectively upregulated after PDX1 and NGN3 smRNA overexpression (data not shown), whereas endogenous pyruvate carboxylase (PC) and prolactin receptor (PRLR) expression increased after MAFA smRNA transfection (Fig. 4B).
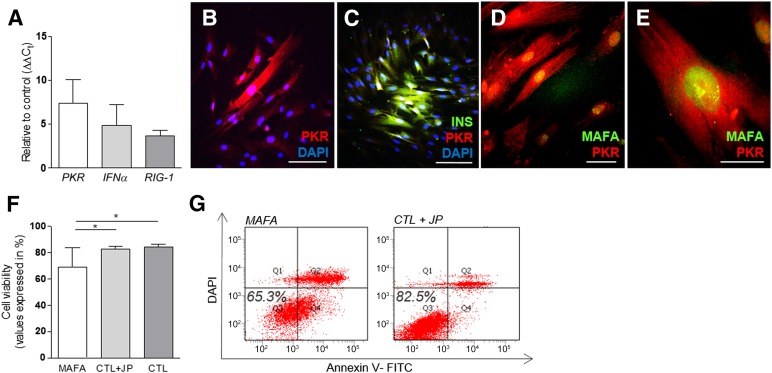
Figure 3.
Effects of synthetic modified mRNAs (smRNAs) on cellular innate immune response. (A): Increased levels of PKR, IFNα, and RIG-1 gene expression were detected on human pancreatic duct-derived cells (HDDCs) after 7 transfections (n = 3–6). (B–E): PKR protein expression was detected after smRNA transfection. (F, G): Flow cytometry analysis showed similar cell viability in smRNA-transfected HDDCs and control HDDCs incubated with jetPEI (n = 3). ∗, p < .05 compared with control HDDCs. Scale bars = 100 µm (B–D) and 25 µm (E). Abbreviations: CTL + JP, control HDDCs incubated with transfection reagent only (jetPEI); DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanateins; INS, insulin; MAFA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A; PKR, protein kinase R.
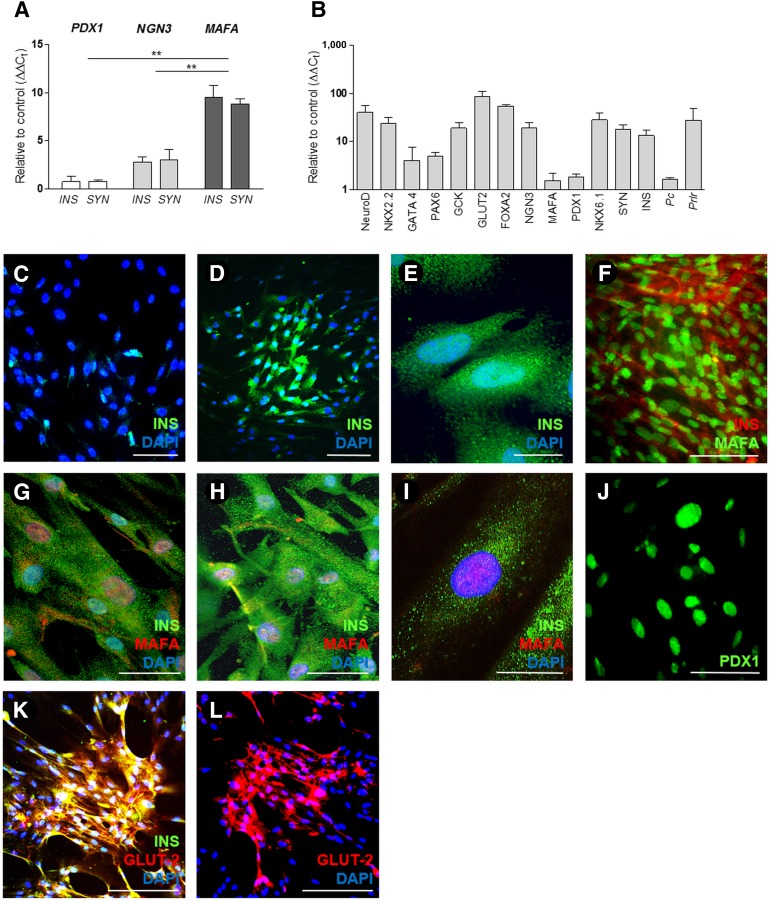
Figure 4.
In vitro β-cell differentiation of human pancreatic duct-derived cells (HDDCs) . (A): HDDCs individually transfected with PDX1, NGN3, and MAFA show upregulation of INS and SYN gene expression with highest detection levels observed in MAFA transfection conditions (n = 3–6). ∗∗, p < .01 compared with PDX1 and NGN3 conditions. (B): Real-time polymerase chain reaction analysis also shows upregulation of several β-cell markers (NeuroD, NKX2.2, GATA4, PAX6, GCK, GLUT-2, FOXA2, NGN3, MAFA, PDX1, NKX6.1) and MAFA biological target genes (Pc, Prlr) after transfections with MAFA synthetic modified mRNA (n = 3–11). Logarithmic scale is presented as powers of 10. (C): INS expression on HDDCs after 5 transfections with NGN3 smRNA. (D, E): Immunostaining for INS (green) in HDDCs after 5 transfections with MAFA smRNA. (F–I): After MAFA differentiation, HDDCs coexpressed insulin and MAFA localized into the nucleus. (J–L): Positive staining for PDX1 (J) and coexpression of INS (green) and GLUT-2 (red) (K and L) were observed on HDDCs after 5 transfections with MAFA smRNA. Staining images are representative of 3–5 different experiments. Scale bars = 200 µm (D, F, J–L), 100 µm (C), 50 µm (G, H), and 20 µm (E, I). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; INS, insulin; MAFA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A; PDX1, pancreatic duodenal homeobox 1.
Because cytoplasmic delivery of single-strand RNAs in mammalian cells may trigger cellular immune response with induction of apoptosis pathways, we evaluated whether smRNA transfection affected HDDCs expression profile and viability. When analyzed after 7 daily MAFA smRNA transfections, HDDCs activated the expression of interferon-α (IFNα), protein kinase R (PKR), and retinoic acid-inducible gene 1 (RIG-1) at low levels compared with control cells incubated with transfection reagent only (CTL+JP condition) (Fig. 3A). By immunostaining, we observed that high proportions of HDDCs coexpressed PKR and MAFA proteins (44.9% ± 16.1%, n = 3), whereas only 20.5% ± 4.5% (n = 3) of MAFA+/PKR− HDDCs were observed (Fig. 3B–3E). No evidence was found of PKR+/MAFA− cells. Although the innate immune response was triggered in HDDCs by smRNAs, we detected acceptable levels of cell toxicity after transfections. Using flow cytometry, we detected 65.3% ± 13.8% (n = 3) of viable HDDCs after 7 daily MAFA smRNA transfections, compared with 82.5% ± 1.9% (n = 3) and 83.0% ± 0.4% (n = 3), respectively, in CTL+JP condition or control cells without any treatment (CTL) (Fig. 3F, 3G). B18R was added in our reprogramming media, although its omission did not significantly alter cell viability (supplemental online Fig. 2A).
For differentiation purposes, we first evaluated the common potential of PDX1, NGN3, and MAFA smRNAs to induce β-cell reprogramming and observed that protocols where TFs were combined or sequentially dispensed failed to modify HDDCs expression profile, without significantly affecting cell viability (data not shown). In contrast, HDDCs transfected with single TFs acquired some characteristics of β cell-like derivatives, with the highest expression levels obtained after MAFA overexpression (Fig. 4A). MAFA smRNA transfection induced the expression of a repertoire of β-cell markers in HDDCs, including NEUROD, NKX2.2, NKX6.1, GATA4, PAX6, glucokinase (GCK), GLUT2 (SLCA2), and FOXA2 (Fig. 4B). Control experiments were performed to distinguish the effects of MAFA-induced differentiation from those provoked by the inflammatory response to smRNA transfection. Gene expression analysis showed the absence of significant β-cell differentiation of HDDCs after 7 daily consecutive transfections with EGFP smRNA (1.3 ± 0.2-fold and 1.9 ± 0.3-fold undifferentiated HDDCs [n = 3], respectively, for insulin and synaptophysin expression, ΔΔCt) or after 72-hour incubation with IFNα (0.8 ± 0.3-fold and 1.1 ± 0.6-fold undifferentiated HDDCs [n = 3], respectively, for insulin and synaptophysin expression, ΔΔCt) (supplemental online Fig. 2B).
Because undifferentiated HDDCs showed mesenchymal and EMT features [15], we evaluated whether these were affected by MAFA-induced reprogramming. After 7 daily MAFA transfections, HDDCs showed significant downregulation of EMT markers, such as N-cadherin (CDH2) and Snail1, and upregulation of fibroblast-specific protein 1 (FSP-1) (supplemental online Fig. 3A). Characteristic partial expression of mesenchymal markers (vimentin, αSMA, and CD73/CD90/CD105 triad) was unchanged (supplemental online Fig. 3A). Furthermore, HDDCs resulting from MAFA-induced differentiation (called β-HDDCs) globally decreased their expression of mesenchymal proteins as compared with undifferentiated HDDCs: whereas only a trend toward decreased αSMA expression was observed (from 71% ± 13.4% to 50.5% ± 24.7%, n = 3–4; p = .4), nestin was significantly downregulated (from 98.4% ± 1.6% to 73.9% ± 17.6%, n = 3; p ˂ .05) and vimentin expression was lost in β-HDDCs (supplemental online Fig. 3C–3I).
By immunostaining, we observed that β-HDDC populations contained 37.3% ± 4.8% insulin+ cells (n = 5) (Fig. 4D, 4E), whereas control populations were devoid of insulin expression. High proportions of differentiated HDDCs showed coexpression of β-cell proteins: after MAFA-induced reprogramming, we found 31.8% ± 8.2% insulin+/MAFA+ cells (n = 5) (Fig. 4F–4I), 46.0% ± 5.3% PDX1+ cells (n = 3) (Fig. 4L), and 36.3% ± 2.16% insulin+/GLUT2+ cells (n = 3) (Fig. 4M). Moreover, 50.3% ± 6.6% (n = 3) of insulin+ β-HDDCs coexpressed Ki67 (supplemental online Fig. 3J), confirming cell proliferation in these cells during MAFA-induced endocrine reprogramming. All insulin-positive cells coexpressed PKR (31.0% ± 3.18% insulin+/PKR+ cells, n = 3) (Fig. 3C) in our differentiated population, whereas no insulin−/PKR+ and insulin+/PKR− cells were detected.
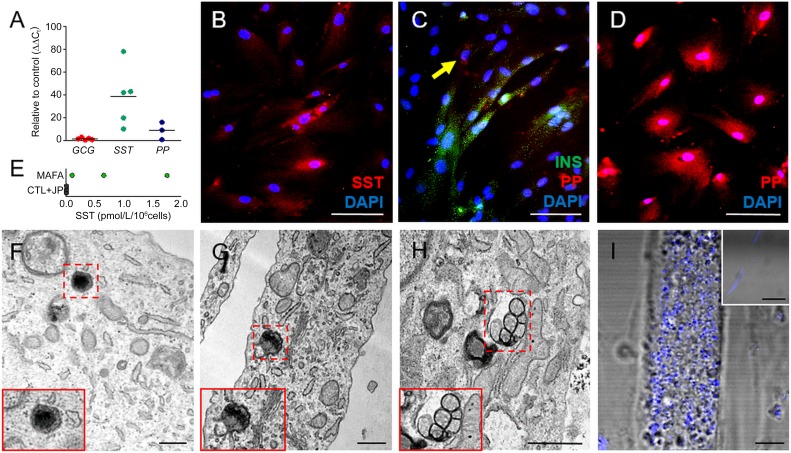
According to previous studies [33], β cell-like derivatives obtained after reprogramming or transdifferentiation may differ from bona fide β cells by expression and/or secretion of multiple pancreatic endocrine hormones. Similarly, our data suggested that β-HDDCs acquired somatostatin (SST) (38.5 ± 11.7-fold controls, n = 5) and pancreatic polypeptide (PP) (8.66 ± 4.3-fold controls, n = 3) gene expression (Fig. 5A). β-HDDC populations contained 13.3% ± 7.2% SST+ cells (Fig. 5B), and colocalization of islet hormones was observed by immunostaining with 25.2% ± 5.9% of β-HDDCs coexpressing insulin and PP (n = 3) (Fig. 5C, 5D). Only negligible quantities of human SST (0.8 ± 0.4 pmol/106 cells, n = 3 [Fig. 5E]) were secreted by β-HDDCs. Furthermore, our β-like cells did not show any significant glucagon (GCG) gene (1.5 ± 0.44-fold, n = 5) (Fig. 5A) or protein (data not shown) expression.
Figure 5.
Polyhormonal feature and insulin granules of differentiated human pancreatic duct-derived cells (HDDCs). (A): MAFA-transfected HDDCs showed gene expression of SST and PP, but not GCG (n = 3–5). (B–D): Immunostaining for SST (B) and PP (D) on β-like HDDCs confirmed the quantitative polymerase chain reaction data (n = 3). (C): β-Like HDDCs coexpressed INS and PP as shown by immunofluorescence. Only 10.3% ± 3.3% of the cells showed exclusive expression pattern (arrow). (E): Enzyme-linked immunosorbent assay showing negligible quantities of human SST secreted by β-HDDCs compared with control group (CTL + JP). (F–H): Electron microscopic images of both mature (F, G) and immature granules (H) in differentiated HDDCs. (I): Increased signal of intrinsic light backscattering induced by insulin content on HDDCs after MAFA overexpression compared with undifferentiated cells (CTL + JP condition in small box). Scale bars = 500 nm (F–H), 100 µm (B–D), 10 µm (I), and 50 µm (I, small box). Abbreviations: CTL + JP, control HDDCs incubated with transfection reagent only (jetPEI); DAPI, 4′,6-diamidino-2-phenylindole; GCG, glucagon; INS, insulin; MAFA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A; PP, pancreatic polypeptide; SST, somatostatin.
Structural changes were observed in HDDCs after MAFA-induced differentiation. Using flow cytometry, we observed that MAFA overexpression led to morphological switch of HDDCs with increased cell size and granularity, as compared with HDDCs transfected with EGFP or untreated cells (supplemental online Fig. 4). Ultrastructure analysis showed presence of cytoplasmic insulin secretory granules in differentiated HDDCs. Both mature crystallized insulin granules and immature light vesicles without dense core were observed (Fig. 5F–5H). In contrast, no abnormal endocrine granules were found in our cell population. Additionally, we investigated the presence of β-cells and insulin granules evaluating the intrinsic capacity of light scattering of the cells. This property in pancreatic islets depends on the abundance of hormone-containing secretory granules, which are composed of dense crystal structures [34]. When assessed for their scattering feature, HDDCs showed increased signal after MAFA differentiation (Fig. 5I), whereas in control populations the backscatter capacity was negligible.
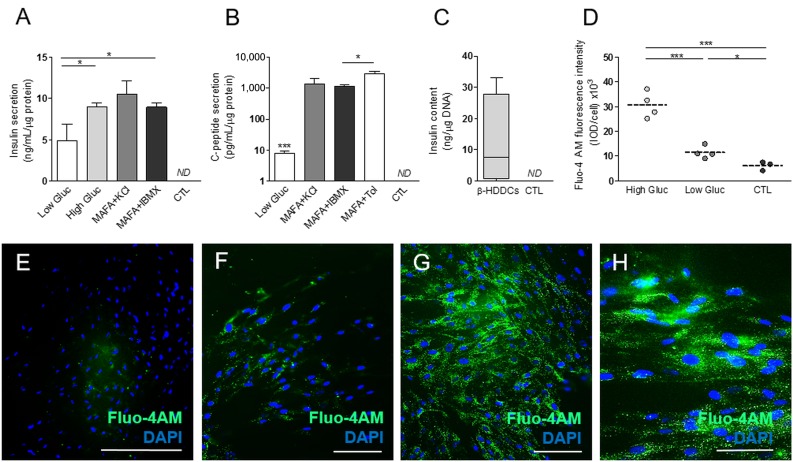
To determine whether β-HDDCs secreted insulin and C-peptide in response to stimulated conditions, we performed glucose stimulated insulin secretion assays showing that our β-like cells released significant amounts of human insulin in the media respectively in basal (2.8 mM of glucose) (4.9 ± 1.9 ng/ml/µg protein, n = 4) and stimulated conditions using high (20.2 mM) glucose (8.9 ± 0.4 ng/ml/µg protein, n = 4), KCl (10.4 ± 1.6 ng/ml/µg protein, n = 5), and 3-isobutyl-1-methylxanthine (IBMX) (8.9 ± 0.4 ng/ml/µg protein, n = 4) (Fig. 6A). β-HDDCs secreted C-peptide in basal (7.5 ± 1.5 pg/ml/µg protein, n = 4) and stimulated conditions with KCl (1417.5 ± 606.8 pg/ml/µg protein, n = 4), IBMX (1170.8 ± 164.2 pg/ml/µg protein, n = 3), and tolbutamide (3039.5 ± 493.0 pg/ml/µg protein, n = 3) (Fig. 6B). Surprisingly, diazoxide did not decrease insulin and C-peptide secretion levels (data not shown). Total insulin content of β-HDDCs was 12.0 ± 7.4 ng/µg DNA (n = 4) (Fig. 6C), which is slightly lower than the expected range for human islet preparations (from 75 to 454 ng/μg DNA [35]).
Figure 6.
MAFA overexpression drives reprogramming into functional insulin-producing cells. (A): Bioplex analysis in culture media of β-like HDDCs showed a significant increase of insulin output between low (2.8 mM) and high (20.2 mM) glucose, KCl, or IBMX conditions (n = 4–5). ∗, p < .05 comparing high glucose and IBMX to low-glucose condition. (B): Secretion of C-peptide was detected in basal and stimulated conditions with KCl, IBMX, or Tol. ∗∗∗, p < .001 comparing low glucose to KCl, IBMX, and Tol; ∗, p < .05 comparing tolbutamide to IBMX condition. (C): Total insulin content observed on β-HDDCs compared with control group. (D): Analysis of fluorescence microscopic images after Fluo-4 AM staining showing significant increased levels of cytoplasmic calcium concentration in stimulated (20.2 mM) conditions and lower signal in low-glucose (2.8 mM) and control conditions. (E–H): Fluorescence images of Fluo-4 AM staining in control (E), low-glucose (F), and high-glucose conditions (G, H). Scale bars = 400 µm (E), 200 µm (F, G), and 100 µm (H). Abbreviations: CTL, control; DAPI, 4′,6-diamidino-2-phenylindole; Fluo, fluorescent; Fluo-4AM, nonfluorescent acetoxymethyl ester; Gluc, glucose; HDDC, human pancreatic duct-derived cell; IBMX, 3-isobutyl-1-methylxanthine; IOD, integrated optical density; MAFA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A; ND, not determined; Tol, tolbutamide.
Consistent with the insulin secretion analysis, increased levels of intracellular calcium concentrations were observed by monitoring cytoplasmic influx. Although low levels of calcium ions were detected in basal (2.8 mM) condition, differentiated HDDCs showed increased intracellular calcium concentration after incubation in high glucose (20.2 mM) (Figs. 6D–6H). These data suggest that differentiated HDDCs secreted insulin in low-glucose conditions and significantly responded to increased levels of glucose stimulation.
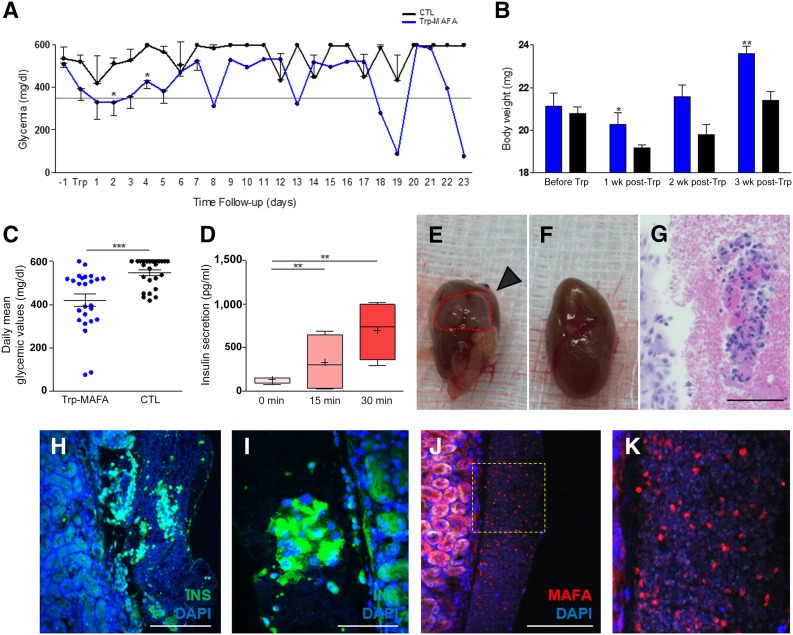
To assess the function of β-HDDCs in vivo, we transplanted 1 × 106 differentiated cells under the kidney capsule of STZ-treated SCID-beige mice. Within the first week after transplantation, β-HDDCs induced a significant reduction of blood glucose levels in nonfasted condition as compared with control mice transplanted with 1 × 106 undifferentiated HDDCs (Fig. 7A). Body weight of β-HDDC-transplanted mice was significantly improved 1 and 3 weeks after transplantation (Fig. 7B). During a follow-up period of 23 days, blood glucose values varied whereas mean blood glucose values were globally significantly reduced in the β-HDDC-transplanted group (Fig. 7C). Human insulin secretion was detected at day 7 after transplantation in the bloodstream of β-HDDC-transplanted animals (131.1 ± 19.2 pg/ml at time 0, n = 4) and showed a response to acute glucose challenge after 15 (327.4 ± 166.3 pg/ml, n = 4) and 30 (696.8 ± 167.7 pg/ml, n = 4) minutes (Fig. 7D). Graft analyses showed cell clusters located under the fibrous capsule that expressed human insulin and human MAFA (Fig. 7H–7M) without reactivity to antiglucagon antibodies. These data suggested the in vivo maintenance of a mature phenotype by β-HDDCs, which allowed fast glycemic improvements in transplanted animals.
Figure 7.
β-Human pancreatic duct-derived cell function after transplantation into diabetic mice. In vivo function of β-HDDCs was assessed by transplanting 1 × 106 differentiated cells under the kidney capsule of streptozotocin-treated SCID-beige mice (n = 8). (A): Within the first week after transplantation, β-HDDCs induced a significant reduction of blood glucose levels in nonfasted condition as compared with control mice transplanted with 1 × 106 undifferentiated HDDCs (n = 2–8). ∗, p < .05 comparing Trp-MAFA to control group. (B): Animals treated with β-HDDCs (n = 8) showed significant restoration of their initial body weight at 1 and 3 weeks after transplantation compared with control animals (n = 3–7). ∗, p < .05 and ∗∗, p < .01 comparing Trp-MAFA group to control group, respectively, at 1 week and 3 weeks after transplantation. (C): During a follow-up period of 23 days, daily mean blood glucose values were significantly reduced in the β-HDDCs transplanted group. ∗∗∗, p < .001 comparing Trp-MAFA group to control group. (D): Human insulin secretion was detected at day 7 after transplantation in the bloodstream of β-HDDC-transplanted animals, showing a response to acute glucose challenge after 15 and 30 minutes (n = 4). ∗∗, p < .01 comparing insulin secretion at 15 and 30 minutes to 0 minutes. (E–K): Graft analyses showed cell clusters located under the fibrous capsule that expressed human insulin and human MAFA. Scale bars = 400 µm (H, J), 200 µm (G), and 100 µm (I). Abbreviations: CTL, control; DAPI, 4′,6-diamidino-2-phenylindole; INS, insulin; MAFA, V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A; Trp, transplanted.
Discussion
The limited supply of cadaveric donors for islet or pancreas transplantation stimulated the search for new sources of functional insulin-producing cells with potential for translational studies. We previously showed the capacity of HDDCs to secrete human insulin [15], and although differentiated HDDCs did not respond to glucose challenges, several features make HDDCs appropriate candidates for cell therapy (e.g., ease of isolation, high proliferation rate, absence of phenotypic alteration after thawing). In the current study, we developed a new RNA-based technology that allows fast, reliable, and robust β-cell differentiation of HDDCs to functional levels with glucose-responsive insulin and C-peptide secretion properties.
RNA-based reprogramming is regarded as a strong alternative to DNA-based techniques to generate defined cell types for clinical application [24, 36, 37]. In 2010, Warren et al. demonstrated the successful smRNA-mediated reprogramming of human fibroblast to induced pluripotent stem cells (iPSCs) in vitro [27] using transcripts for KLF4, c-MYC, OCT4, and SOX2. Their protocol was reportedly 36-fold more effective in deriving iPSCs than a similar retroviral-based procedure. The reliability of the smRNA reprogramming method was also assessed in vivo. In a mouse model of myocardial infarction, heart function and survival of animals were improved after a direct and single intramyocardial injection of human vascular endothelial growth factor A (VEGF-A) smRNA [23]. After VEGF-A overexpression, histology showed reduced infarct size and concomitant expansion and differentiation of endogenous heart progenitors cells. For clinical perspectives, RNA-based reprogramming bears the advantage over DNA-based techniques to be devoid of risk for insertional mutagenesis and genomic integration that may disrupt cell cycle control and lead to outgrowth of transplanted cells [24, 36, 37].
On the basis of this evidence, we evaluated the potential of smRNA-based protocols to drive HDDCs toward functional insulin production. Synthetic mRNAs encoding for PDX1, NGN3, and MAFA TFs were synthesized using a ready-to-use plasmid because this strategy allows fast and large-scale production of transcripts. Transfections were dispensed daily on HDDCs to ensure sustained ectopic protein expression [25, 38, 39]. We observed peak exogenous smRNAs concentrations at 48 hours after transfection without cumulative effect on cytoplasmic smRNA levels during daily transfection protocols. Similarly, rapid decay of EGFP protein expression was demonstrated elsewhere [27] after single smRNA transfections, motivating the use of 24-hour transfection regimens. The delay observed in the smRNA uptake in our system could be partly related to the nature of our cell population (expanded cells generated after partial EMT) and to the transfection reagent (i.e., jetPEI) used, which could lead to different patterns of endosomal escape rates. Indeed, a temporal diversity was observed in the way endosomes release their lipocomplex into the cytosol [40]. This diversity might be fostered by the specific combinations of our reprogramming protocol.
In our system, efficient smRNA-to-protein translation was shown in HDDCs by detection of cytoplasmic fluorescence induced by EGFP smRNA and by nuclear localization of MAFA protein after MAFA smRNA transfection. According to protein expression levels, we estimate our transfection efficiency to be between 35% and 50% with EGFP and MAFA smRNAs, which is slightly lower than other studies reporting higher transfection rates (50%–100%) [23, 27, 37, 41, 42], although performed either in rodent adult or human fetal tissues. MAFA protein was able to bind to human insulin promoter in HEK293 cells that were used for their capacity to efficiently overexpress ectopic and recombinant proteins [43]. Furthermore, functionality of PDX1, NGN3, and MAFA proteins was suggested by the activation of specific target genes after transfection of corresponding smRNAs into HDDCs.
During IVT of smRNAs, ribonucleotides were substituted by modified nucleotides and 5′ triphosphates were capped according to published protocols [44–46] to escape from cellular antiviral responses. Exogenous single-stranded RNA were shown to activate IFN and nuclear factor κB pathways [47]. Therefore, the B18R molecule—a vaccinia virus decoy type I IFN receptor—was supplemented to media during smRNA transfection to capture IFN molecules, although we could not confirm that adding B18R improved cell viability. In our system, HDDCs showed evidence for activation of innate immune response with expression of IFNα, PKR, and RIG-1 markers while maintaining acceptable levels of cell viability (Fig. 3F and 3G), as compared with other reports showing 70%–80% viable cells after RNA transfection [23, 27, 41, 45].
Our investigations showed that MAFA smRNA was superior to PDX1 and NGN3 for inducing β cell-specific gene expression in HDDCs. Although many pancreatic TFs may be used to engineer β-cell surrogates [48, 49], MAFA is the lead regulator of β-cell function [50–53] and is critical to maintain glycemic control in mice [54, 55]. Moreover, MAFA itself is responsible for β-cell maturation of endocrine progenitors or neonatal β cells, as shown by overexpression or knockdown studies in rodent [56, 57] or human fetal islet [58] cells. Also, MAFA synergizes with other pancreas-specific TFs (e.g., PDX1, NGN3, PAX4) for β-cell transdifferentiation from mouse pancreatic [18, 59] or liver [39, 60] progenitors. For example, temporal and hierarchical overexpression of PDX1, PAX4, and MAFA induced β cell-like characteristics in human liver cells in vitro [61], and interestingly, the omission of MAFA from the last stage of differentiation protocols ablated β cell-like maturation. In our system, HDDCs did not show evidence for transdifferentiation when MAFA, NGN3, and PDX1 transcripts were concomitantly or sequentially transfected, although cell viability was preserved. This particularity may be partly accounted to the specific phenotype of HDDCs that display partial EMT features [15].
MAFA-transfected HDDC populations contained up to 37% insulin+, 36% GLUT-2+, and 46% PDX1+ cells. By comparison, state-of-the-art β-cell differentiation strategies with embryonic stem cells report 25%–50% insulin- or C-peptide-expressing cells [4, 6, 62, 63] after a 14- to 18-day in vitro procedure. In these studies, an important proportion (15%–70%) of insulin+ cells coexpressed GCG and/or SST. Recent refinements in the 2 weeklong stepwise protocols [7, 64] led to enrichment of monohormonal insulin+ populations (up to 50%) and to consecutive improvements of in vivo insulin secretion capacities [7]. In our system, β-HDDC populations did not contain GCG but yielded 13% insulin+/SST+ cells and 25% insulin+/PP+ cells. These polyhormonal cells only secreted low amounts of SST in media (not significantly different from control groups), suggesting only negligible influences on insulin secretion or glucose homeostasis. Although MAFA restricts SST expression during normal pancreas development [54], stem cell- or progenitor-derived β-like cells may harbor unexpected phenotypes that resemble immature endocrine cells to some extent [62]. For example, in Zhou's group [65] 2014 study on adult mouse acinar cell reprogramming, they observed that Ad-Ngn3 alone induced the acquisition of SST expression by 40% of infected cells. When combined with Ad-MAFA, both GCG+ and SST+ cells were obtained in culture [65]. Similarly, Rezania et al. [66] found insulin+/SST+-coexpressing cells in their NKX6.1+-enriched fraction of human embryonic stem cell-derived endocrine lineages. In our system, HDDCs derive from human ductal cells that are known to express PDX1 at low levels [67], and expanded HDDCs contain PDX1 mRNA at detectable levels. Furthermore, MAFA overexpression in HDDCs led to upregulation of PDX1 expression. One hypothesis might be that these basal PDX1 levels temporally combined to an increase of MAFA-induced PDX1 expression and led to SST gene and protein induction in HDDCs. However, this hypothesis—as it is for PP expression—would be difficult to confirm experimentally because little is known about δ cell development. By comparison, population of HDDCs differentiated with growth factors [15] did not show expression of GCG, SST, or PP. MAFA thus induces a deeper involvement of HDDCs into the endocrine lineage, which is also suggested by the EMT (i.e., N-cadherin, Snail1) and mesenchymal (e.g., vimentin) marker downregulation shown by MAFA-transfected cells, although differentiated HDDCs maintained the expression of several mesenchymal markers (e.g., αSMA, fibronectin, nestin) and upregulated FSP-1. The increased levels of FSP-1, which is known as a cytoplasmic calcium-binding protein [68], may partly be explained by the increased intracellular calcium concentrations observed in differentiated HDDCs.
A mature β cell-like phenotype was generated in β-HDDCs with insulin content up to 33.0 ng/µg DNA and glucose-responsive insulin (and C-peptide) secretion reaching up to 16.8 ng/ml/µg protein in stimulating conditions. Our limited increase of insulin secretion in stimulated conditions was also described elsewhere with ESCs [69] and may represent the requirement for an in vivo environment for further maturation of insulin secretion patterns. Compared with previous studies that showed β-cell generation in vitro after differentiation of ESCs [4, 6, 7, 33], we observed about fourfold lower insulin release with HDDCs. However, the difference in insulin secretion between HDDCs and ESCs is not significant anymore when we normalize for the amount of insulin-positive cells in both systems.
When β-HDDCs were transplanted under the kidney capsule of immunosuppressed diabetic mice, we observed a general decrease of blood glycemia in transplanted animals compared with control group during a short-term follow-up of 23 days. We demonstrated a rapid glucose-responsive insulin secretion by β-HDDCs with levels being similar to those observed in stem cell-derived β cells [7]. Furthermore, histological analysis of the graft confirmed the insulin+/MAFA+/GCG− phenotype of β-HDDCs.
Conclusion
Our study confirms the potential of HDDCs to engineer β-like cells with insulin secretion capacities both in vitro and in vivo. This feature, together with favorable in vitro characteristics, strengthens the role of HDDCs as candidates for diabetes cell therapy. Moreover, our data show the efficiency of an smRNA-based overexpression system for reprogramming of human primary cells with differentiation occurring in a timely fashion (5-7 days) and via cost-effective protocols. Although further work is required to bring smRNA-reprogramming to clinical grade procedures, our HDDC RNA-based differentiation model shows the opportunity for translational studies in diabetes and may lead to applications for other diseases.
Supplementary Material
Acknowledgments
We thank J. Ravau and F. André (Pôle Pédiatrie) for their expert technical assistance. This study was supported by grants from the Belgian Society for Pediatric Endocrinology and Diabetology (BESPEED), Institut de Recherche Clinique et Expérimentale (IREC), Fonds National de la Recherche Scientifique (FNRS), and International Society for Pediatric and Adolescent Diabetes (ISPAD).
Author Contributions
E.C.: collection and/or assembly of data, data analysis and interpretation, manuscript writing; Y.-S.L.: collection and/or assembly of data, data analysis and interpretation; V.P. and D.L.: provision of study material or patients; M.-J.H.: collection and/or assembly of data; C.A.L.: technical support, review of the manuscript; P.V.D.S.: technical support, manuscript writing; A.V. and S.B.-W.: technical support; L.P.: provision of study material or patients, critical revision of the manuscript for important intellectual content; E.S.: financial support, administrative support; P.A.L.: conception and design, data analysis and interpretation, manuscript writing, administrative support, financial support, study supervision.
Disclosure of Potential Conflicts of Interest
L.P. is a compensated consultant for and has received compensated research funding from Dompe Spa and has received honoraria from Novartis. The other authors indicated no potential conflicts of interest.
References
- 1.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: Etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 2.Barton FB, Rickels MR, Alejandro R, et al. Improvement in outcomes of clinical islet transplantation: 1999-2010. Diabetes Care. 2012;35:1436–1445. doi: 10.2337/dc12-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orlando G, Gianello P, Salvatori M, et al. Cell replacement strategies aimed at reconstitution of the β-cell compartment in type 1 diabetes. Diabetes. 2014;63:1433–1444. doi: 10.2337/db13-1742. [DOI] [PubMed] [Google Scholar]
- 4.Kroon E, Martinson LA, Kadoya K, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 5.Lima MJ, Docherty HM, Chen Y, et al. Pancreatic transcription factors containing protein transduction domains drive mouse embryonic stem cells towards endocrine pancreas. PLoS One. 2012;7:e36481. doi: 10.1371/journal.pone.0036481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rezania A, Bruin JE, Riedel MJ, et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61:2016–2029. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagliuca FW, Millman JR, Gürtler M, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C, Flatt CC, McClenaghan NH. Stem cell-based approaches for the treatment of diabetes. Stem Cells Int 2011;2011:424986. [DOI] [PMC free article] [PubMed]
- 9.Zhou Q, Brown J, Kanarek A, et al. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lysy PA, Weir GC, Bonner-Weir S. Making β cells from adult cells within the pancreas. Curr Diab Rep. 2013;13:695–703. doi: 10.1007/s11892-013-0400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemper M, Leuckx G, Heremans Y, et al. Reprogramming of human pancreatic exocrine cells to β-like cells. Cell Death Differ. 2015;22:1117–1130. doi: 10.1038/cdd.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan FC, Wright C. Pancreas organogenesis: From bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 13.Inada A, Nienaber C, Katsuta H, et al. Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc Natl Acad Sci USA. 2008;105:19915–19919. doi: 10.1073/pnas.0805803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Hasani K, Pfeifer A, Courtney M, et al. Adult duct-lining cells can reprogram into β-like cells able to counter repeated cycles of toxin-induced diabetes. Dev Cell. 2013;26:86–100. doi: 10.1016/j.devcel.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Corritore E, Dugnani E, Pasquale V, et al. β-Cell differentiation of human pancreatic duct-derived cells after in vitro expansion. Cell Reprogram. 2014;16:456–466. doi: 10.1089/cell.2014.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artner I, Hang Y, Mazur M, et al. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59:2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguayo-Mazzucato C, Zavacki AM, Marinelarena A, et al. Thyroid hormone promotes postnatal rat pancreatic β-cell development and glucose-responsive insulin secretion through MAFA. Diabetes. 2013;62:1569–1580. doi: 10.2337/db12-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada T, Cavelti-Weder C, Caballero F, et al. Reprogramming mouse cells with a pancreatic duct phenotype to insulin-producing β-like cells. Endocrinology. 2015;156:2029–2038. doi: 10.1210/en.2014-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heremans Y, Van De Casteele M, in’t Veld P, et al. Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J Cell Biol. 2002;159:303–312. doi: 10.1083/jcb.200203074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meivar-Levy I, Sapir T, Gefen-Halevi S, et al. Pancreatic and duodenal homeobox gene 1 induces hepatic dedifferentiation by suppressing the expression of CCAAT/enhancer-binding protein beta. Hepatology. 2007;46:898–905. doi: 10.1002/hep.21766. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Brun T, Kataoka K, et al. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50:348–358. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavernier G, Wolfrum K, Demeester J, et al. Activation of pluripotency-associated genes in mouse embryonic fibroblasts by non-viral transfection with in vitro-derived mRNAs encoding Oct4, Sox2, Klf4 and cMyc. Biomaterials. 2012;33:412–417. doi: 10.1016/j.biomaterials.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 23.Zangi L, Lui KO, von Gise A, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernal JA. RNA-based tools for nuclear reprogramming and lineage-conversion: towards clinical applications. J Cardiovasc Transl Res. 2013;6:956–968. doi: 10.1007/s12265-013-9494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Pham P, Thi-My Nguyen P, Thai-Quynh Nguyen A, et al. Improved differentiation of umbilical cord blood-derived mesenchymal stem cells into insulin-producing cells by PDX-1 mRNA transfection. Differentiation. 2014;87:200–208. doi: 10.1016/j.diff.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Poleganov MA, Eminli S, Beissert T, et al. Efficient reprogramming of human fibroblasts and blood-derived endothelial progenitor cells using non-modified RNA for reprogramming and immune evasion. Hum Gene Ther. 2015;26:751–766. doi: 10.1089/hum.2015.045. [DOI] [PubMed] [Google Scholar]
- 27.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karikó K, Muramatsu H, Ludwig J, et al. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kormann MS, Hasenpusch G, Aneja MK, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 30.Nano R, Bosco D, Kerr-Conte JA, et al. Human islet distribution programme for basic research: Activity over the last 5 years. Diabetologia. 2015;58:1138–1140. doi: 10.1007/s00125-015-3536-5. [DOI] [PubMed] [Google Scholar]
- 31.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu M, Fabrega C, Liu SW, et al. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol Cell. 2004;14:67–80. doi: 10.1016/s1097-2765(04)00180-7. [DOI] [PubMed] [Google Scholar]
- 33.D’Amour KA, Bang AG, Eliazer S, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 34.Ilegems E, van Krieken PP, Edlund PK, et al. Light scattering as an intrinsic indicator for pancreatic islet cell mass and secretion. Sci Rep. 2015;5:10740. doi: 10.1038/srep10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayton NS, Poffenberger G, Henske J, et al. Human islet preparations distributed for research exhibit a variety of insulin-secretory profiles. Am J Physiol Endocrinol Metab. 2015;308:E592–E602. doi: 10.1152/ajpendo.00437.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quabius ES, Krupp G. Synthetic mRNAs for manipulating cellular phenotypes: An overview. N Biotechnol. 2015;32:229–235. doi: 10.1016/j.nbt.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Avci-Adali M, Behring A, Steinle H, et al. In vitro synthesis of modified mRNA for induction of protein expression in human cells. J Vis Exp. 2014:e51943. doi: 10.3791/51943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandal PK, Rossi DJ. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Protoc. 2013;8:568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- 39.Simeonov KP, Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS One. 2014;9:e100134. doi: 10.1371/journal.pone.0100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonhardt C, Schwake G, Stögbauer TR, et al. Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine (Lond) 2014;10:679–688. doi: 10.1016/j.nano.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Drews K, Tavernier G, Demeester J, et al. The cytotoxic and immunogenic hurdles associated with non-viral mRNA-mediated reprogramming of human fibroblasts. Biomaterials. 2012;33:4059–4068. doi: 10.1016/j.biomaterials.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 42.Wang XL, Hu P, Guo XR, et al. Reprogramming human umbilical cord mesenchymal stromal cells to islet-like cells with the use of in vitro-synthesized pancreatic-duodenal homebox 1 messenger RNA. Cytotherapy. 2014;16:1519–1527. doi: 10.1016/j.jcyt.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Thomas P, Smart TG. HEK293 cell line: A vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods. 2005;51:187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Karikó K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: Implication for therapeutic RNA development. Curr Opin Drug Discov Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 45.Anderson BR, Muramatsu H, Nallagatla SR, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang CL, Leblond AL, Turner EC, et al. Synthetic chemically modified mrna-based delivery of cytoprotective factor promotes early cardiomyocyte survival post-acute myocardial infarction. Mol Pharm. 2015;12:991–996. doi: 10.1021/mp5006239. [DOI] [PubMed] [Google Scholar]
- 47.Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 48.Piccand J, Strasser P, Hodson DJ, et al. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Reports. 2014;9:2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerace D, Martiniello-Wilks R, O’Brien BA, et al. The use of β-cell transcription factors in engineering artificial β cells from non-pancreatic tissue. Gene Ther. 2015;22:1–8. doi: 10.1038/gt.2014.93. [DOI] [PubMed] [Google Scholar]
- 50.Kaneto H, Matsuoka TA, Nakatani Y, et al. A crucial role of MafA as a novel therapeutic target for diabetes. J Biol Chem. 2005;280:15047–15052. doi: 10.1074/jbc.M412013200. [DOI] [PubMed] [Google Scholar]
- 51.Nishimura W, Bonner-Weir S, Sharma A. Expression of MafA in pancreatic progenitors is detrimental for pancreatic development. Dev Biol. 2009;333:108–120. doi: 10.1016/j.ydbio.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hang Y, Yamamoto T, Benninger RK, et al. The MafA transcription factor becomes essential to islet β-cells soon after birth. Diabetes. 2014;63:1994–2005. doi: 10.2337/db13-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaneto H, Matsuoka TA. Role of pancreatic transcription factors in maintenance of mature β-cell function. Int J Mol Sci. 2015;16:6281–6297. doi: 10.3390/ijms16036281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishimura W, Takahashi S, Yasuda K. MafA is critical for maintenance of the mature beta cell phenotype in mice. Diabetologia. 2015;58:566–574. doi: 10.1007/s00125-014-3464-9. [DOI] [PubMed] [Google Scholar]
- 55.Matsuoka TA, Kaneto H, Kawashima S, et al. Preserving Mafa expression in diabetic islet β-cells improves glycemic control in vivo. J Biol Chem. 2015;290:7647–7657. doi: 10.1074/jbc.M114.595579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu He K, Juhl K, Karadimos M, et al. Differentiation of pancreatic endocrine progenitors reversibly blocked by premature induction of MafA. Dev Biol. 2014;385:2–12. doi: 10.1016/j.ydbio.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aguayo-Mazzucato C, Koh A, El Khattabi I, et al. Mafa expression enhances glucose-responsive insulin secretion in neonatal rat beta cells. Diabetologia. 2011;54:583–593. doi: 10.1007/s00125-010-2026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aguayo-Mazzucato C, DiIenno A, Hollister-Lock J, et al. MAFA and T3 drive maturation of both fetal human islets and insulin-producing cells differentiated from hESC. J Clin Endocrinol Metab. 2015;100:3651–3659. doi: 10.1210/jc.2015-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akinci E, Banga A, Greder LV, et al. Reprogramming of pancreatic exocrine cells towards a beta (β) cell character using Pdx1, Ngn3 and MafA. Biochem J. 2012;442:539–550. doi: 10.1042/BJ20111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagasaki H, Katsumata T, Oishi H, et al. Generation of insulin-producing cells from the mouse liver using β cell-related gene transfer including Mafa and Mafb. PLoS One. 2014;9:e113022. doi: 10.1371/journal.pone.0113022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berneman-Zeitouni D, Molakandov K, Elgart M, et al. The temporal and hierarchical control of transcription factors-induced liver to pancreas transdifferentiation. PLoS One. 2014;9:e87812. doi: 10.1371/journal.pone.0087812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hrvatin S, O’Donnell CW, Deng F, et al. Differentiated human stem cells resemble fetal, not adult, β cells. Proc Natl Acad Sci USA. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruin JE, Erener S, Vela J, et al. Characterization of polyhormonal insulin-producing cells derived in vitro from human embryonic stem cells. Stem Cell Res (Amst) 2014;12:194–208. doi: 10.1016/j.scr.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Agulnick AD, Ambruzs DM, Moorman MA, et al. Insulin-producing endocrine cells differentiated in vitro from human embryonic stem cells function in macroencapsulation devices in vivo. Stem Cells Translational Medicine. 2015;4:1214–1222. doi: 10.5966/sctm.2015-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, Nakanishi M, Zumsteg A, et al. In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. eLife. 2014;3:e01846. doi: 10.7554/eLife.01846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rezania A, Bruin JE, Xu J, et al. Enrichment of human embryonic stem cell-derived NKX6.1-expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells. 2013;31:2432–2442. doi: 10.1002/stem.1489. [DOI] [PubMed] [Google Scholar]
- 67.Guo L, Inada A, Aguayo-Mazzucato C, et al. PDX1 in ducts is not required for postnatal formation of β-cells but is necessary for their subsequent maturation. Diabetes. 2013;62:3459–3468. doi: 10.2337/db12-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Österreicher CH, Penz-Österreicher M, Grivennikov SI, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proc Natl Acad Sci USA. 2011;108:308–313. doi: 10.1073/pnas.1017547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rezania A, Bruin JE, Arora P, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.