In the present study, functional skeletal myotubes were efficiently generated from human induced pluripotent stem cells using a small molecule-based approach. Myotubes derived from patients carrying the C9orf72 repeat expansion show no change in differentiation efficiency and normal TDP-43 localization after as many as 120 days in vitro when compared to unaffected controls. The protocol described in this study for the generation of skeletal myotubes from human induced pluripotent stem cells may serve as a valuable tool in drug discovery and modeling of musculoskeletal and neuromuscular diseases.
Keywords: Induced pluripotent stem cells, Skeletal muscle, C9orf72, Amyotrophic lateral sclerosis, Frontotemporal dementia
Abstract
Induced pluripotent stem cells (iPSCs) offer an unlimited resource of cells to be used for the study of underlying molecular biology of disease, therapeutic drug screening, and transplant-based regenerative medicine. However, methods for the directed differentiation of skeletal muscle for these purposes remain scarce and incomplete. Here, we present a novel, small molecule-based protocol for the generation of multinucleated skeletal myotubes using eight independent iPSC lines. Through combinatorial inhibition of phosphoinositide 3-kinase (PI3K) and glycogen synthase kinase 3β (GSK3β) with addition of bone morphogenic protein 4 (BMP4) and fibroblast growth factor 2 (FGF2), we report up to 64% conversion of iPSCs into the myogenic program by day 36 as indicated by MYOG+ cell populations. These cells began to exhibit spontaneous contractions as early as 34 days in vitro in the presence of a serum-free medium formulation. We used this protocol to obtain iPSC-derived muscle cells from frontotemporal dementia (FTD) patients harboring C9orf72 hexanucleotide repeat expansions (rGGGGCC), sporadic FTD, and unaffected controls. iPSCs derived from rGGGGCC carriers contained RNA foci but did not vary in differentiation efficiency when compared to unaffected controls nor display mislocalized TDP-43 after as many as 120 days in vitro. This study presents a rapid, efficient, and transgene-free method for generating multinucleated skeletal myotubes from iPSCs and a resource for further modeling the role of skeletal muscle in amyotrophic lateral sclerosis and other motor neuron diseases.
Significance
Protocols to produce skeletal myotubes for disease modeling or therapy are scarce and incomplete. The present study efficiently generates functional skeletal myotubes from human induced pluripotent stem cells using a small molecule-based approach. Using this strategy, terminal myogenic induction of up to 64% in 36 days and spontaneously contractile myotubes within 34 days were achieved. Myotubes derived from patients carrying the C9orf72 repeat expansion show no change in differentiation efficiency and normal TDP-43 localization after as many as 120 days in vitro when compared to unaffected controls. This study provides an efficient, novel protocol for the generation of skeletal myotubes from human induced pluripotent stem cells that may serve as a valuable tool in drug discovery and modeling of musculoskeletal and neuromuscular diseases.
Introduction
Human embryonic stem cells and, more recently, induced pluripotent stem cells (iPSCs) have been heralded for their potential to generate all cell types of any germ layer, providing avenues for disease research, drug discovery, and transplantation [1, 2]. A variety of successful protocols for directed differentiation of iPSCs into different cell types have emerged in recent years; however, the generation of skeletal myotubes has proven to be more challenging. The generation of PAX3+ and PAX7+ paraxial mesoderm progenitors from iPSCs would serve as a valuable tool for repopulating the satellite cell niche in patients with degenerative disorders such as Duchenne muscular dystrophy, and the generation of skeletal myotubes offers options for drug screening and disease modeling in disorders such as amyotrophic lateral sclerosis (ALS), inclusion body myopathy with Paget’s disease and early-onset frontotemporal dementia (IBMPFD), and spinal muscular atrophy (SMA).
Initial studies in human and mouse showed that directed differentiation followed by isolation of paraxial mesoderm progenitors by flow cytometry are viable for engraftment in vivo [3–5]. Differentiation efficiencies were later improved by transient expression of myogenic genes such as PAX3, PAX7, or MYOD1 [6–12]. In regenerative medicine, however, it is desirable to produce cells absent of genetic modification, which more accurately replicate development and can be suitable for future clinical applications [13]. As such, recent protocols have focused on recapitulating development via chemically defined, small molecule-based methods for the generation of paraxial mesoderm progenitors and mature skeletal myotubes [14–18].
Here we build on previous studies to report an efficient protocol for the derivation of skeletal myotubes from patient-derived iPSCs. This protocol uses dimethylsulfoxide (DMSO) and phosphoinositide 3-kinase (PI3K) inhibition to enhance differentiation efficiency toward the mesodermal lineage in the presence of bone morphogenetic protein 4 (BMP4) and fibroblast growth factor 2 (FGF2) and promotion of Wnt signaling via glycogen synthase kinase 3β (GSK3β) inhibition, yielding up to 90% of myogenic cells by exclusive PAX7+/MYOG+ populations after 36 days in vitro. These cells fuse to form spontaneously contractile, multinucleated myotubes in the presence of serum-free medium as early as 34 days in vitro, thus providing a viable tool for modeling skeletal muscle diseases.
Given that the contribution of skeletal muscle and the neuromuscular junction in motor neuron diseases such as ALS and SMA remains largely unexplored, we aimed to study whether pathologic phenotypes described in neurons derived from individuals carrying a pathogenic expansion in C9orf72, the most common genetic mutation in ALS and frontotemporal dementia (FTD) [19], could be present in skeletal muscle. Mutations seen in IBMPFD patients such as VCP and hnRNPA1 overlap with mutations seen in ALS patients, suggesting shared affected pathways [20]. Interestingly, TDP-43 and ubiquitin pathology observed in the brains of repeat expansion carriers is also present in the brains and skeletal muscle of IBMPFD patients [21]. We report that C9orf72 is expressed in iPSC-derived skeletal muscle, which also contains nuclear RNA foci, one of the hallmarks of pathology in the brain [19]. We also explored potential skeletal muscle pathology in iPSCs derived from GGGGCC expansion carriers and controls but observed no aberrant changes in differentiation efficiencies, TDP-43 localization, nuclear envelope breakdown via RanGAP1 and lamin A/C (LMNA) immunocytochemistry, and ubiquitin or p62 pathology.
Materials and Methods
Human iPSCs
Fibroblasts were obtained with patient consent and institutional review board approval (#10-000574) from skin biopsies of patients diagnosed with C9orf72-related FTD or sporadic FTD and healthy controls from the University of California, San Francisco, Memory and Aging Center. Fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and 10% fetal bovine serum (FBS). Reprogramming was performed with the STEMCCA lentiviral vector containing OCT4, c-MYC, SOX2, and KLF4 (EMD Millipore, Billerica, MA, http://www.emdmillipore.com). Pluripotency was assessed via Pluritest [22] and immunocytochemical staining of NANOG, OCT4, SSEA4, and TRA-181. Karyotyping was performed by Cell Line Genetics in passages 5–25 after reprogramming. iPSCs were cultured at 37°C and 5% CO2 on plates coated with vitronectin (Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) in TeSR-E8 medium (Stem Cell Technologies) and passaged every 4–6 days using ReLeSR reagent (Stem Cell Technologies) according to the manufacturer’s instructions. Supplemental online Table 1 lists the iPSC lines used and relevant patient information.
Skeletal Muscle Differentiation
When iPSC colonies were ∼250–400 µm in diameter, 1.5% DMSO (Sigma-Aldrich, St. Louis, MO, http://www.sigamaaldrich.com) in TeSR-E8 medium was added. On day 0, cells were cultured in CDM FLyBC for 36 hours. Chemically defined medium (CDM) consists of a 1:1 mixture of Iscove’s modified Dulbecco’s medium and Ham’s F12 medium (Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com) with the addition of bovine serum albumin (5 mg/ml; Thermo Fisher Scientific Life Sciences), 100× lipids (1×; Thermo Fisher Scientific Life Sciences), transferrin (15 µg/ml; Sigma-Aldrich), 1-thioglycerol (450 µM; Sigma-Aldrich), and insulin (7 µg/ml; Thermo Fisher Scientific Life Sciences). FLyBC consists of FGF2 (20 ng/ml; Stem Cell Technologies), LY294002 (10 µM; Stem Cell Technologies), BMP4 (10 ng/ml; Stem Cell Technologies), and CHIR99021 (10 µM; Sigma-Aldrich). After 36 hours, cells were cultured in CDM FLy for an additional 5.5 days. On day 7, cells were cultured in MB-1 and 15% FBS (three-quarters daily medium change) donated from the laboratory of Dr. Jacques Tremblay (Université Laval, Ville de Québec, QC, Canada) and prepared per that laboratory’s instructions. On day 12, cells were cultured in fusion medium (replaced on alternating days) consisting of 2% horse serum (Sigma-Aldrich) in DMEM (Thermo Fisher Scientific Life Sciences). For terminal differentiation, cells were changed to N-2 medium (replaced on alternating days) consisting of DMEM F12 (Thermo Fisher Scientific Life Sciences) supplemented with 1% N-2 supplement (Thermo Fisher Scientific Life Sciences) and 1% insulin-transferrin-selenium solution (Thermo Fisher Scientific Life Sciences).
Neuronal Differentiation
iPSC colonies were grown to confluence and collected with TrypLE enzyme (Thermo Fisher Scientific Life Sciences). Neural progenitor cells (NPCs) were obtained using AggreWell 800 plates (Stem Cell Technologies) according to the manufacturer’s protocol for NPC generation in STEMdiff neural induction medium. Briefly, 3 million iPSCs were cultured in neural induction medium and Y-27632 (Stem Cell Technologies) per AggreWell for 5 days. Neural aggregates were harvested in a 37-µm strainer and plated onto dishes coated with poly-l-ornithine (PLO) and laminin (Thermo Fisher Scientific Life Sciences) in neural induction medium for 7 days. Neural rosettes were isolated via Rosette Selection Reagent (Stem Cell Technologies) and replated onto Matrigel (Thermo Fisher Scientific Life Sciences) or PLO/laminin-coated plates as NPCs. NPCs were maintained in DMEM F12 supplemented with 0.5% N-2 supplement, 0.5% B-27 supplement (Thermo Fisher Scientific Life Sciences), 1× sodium pyruvate (Thermo Fisher Scientific Life Sciences), 1× nonessential amino acids (Thermo Fisher Scientific Life Sciences), epidermal growth factor (EGF) (10 ng/ml; Stem Cell Technologies), and FGF2 (10 ng/ml). Differentiation into cortical neurons was performed when NPCs reached ∼40% confluence, replacing EGF and FGF with brain-derived neurotrophic factor (10 ng/ml; Stem Cell Technologies).
Immunocytochemistry
Cells were fixed in cold 4% paraformaldehyde (PFA) (Electron Microscopy Services) for 15 minutes, rinsed three times for 5 minutes in phosphate-buffered saline (PBS), and stored at 4°C until staining. Cells were permeabilized using 0.25% Triton-X in PBS for 15 minutes at room temperature, washed three times for 5 minutes in PBST (0.1% Tween-20 in PBS), blocked for 1 hour at room temperature using 10% goat serum and 1% BSA in PBST, and incubated in primary antibodies at 4°C overnight. Cells were then washed three times for 5 minutes in PBST and incubated in Alexa Fluor-conjugated secondary antibodies for 1 hour at room temperature in the dark. Cells were washed in PBST and incubated in 4′,6-diamidino-2-phenylindole (DAPI) (1:1,000) for 10 minutes before being washed in PBST and stored at 4°C in the dark until imaging. Antibody information can be found in supplemental online Table 3.
Cell nucleus counts were performed manually by the investigators. In each experiment, multiple images were taken in a nonbiased fashion across multiple independent wells under ×10 magnification and sectioned into frames of 675 × 1,000 pixels for counting.
RNA Fluorescent In Situ Hybridization
Fluorescent in situ hybridization (FISH) was performed as described [23]. Briefly, cells grown on coverslips were fixed in cold 4% PFA for 15 minutes and rinsed three times for 5 minutes in RNase-free PBS (pH 7.4; Thermo Fisher Scientific Life Sciences). Cells were permeabilized using 0.25% Triton-X (Thermo Fisher Scientific Life Sciences) in 1× RNase-Free PBS for 20 minutes at room temperature. Cells were then washed in RNase-Free PBS. Hybridization buffer consisting of 50% formamide (Thermo Fisher Scientific Life Sciences), 2× SSC buffer (300 mM NaCl and 30 mM sodium citrate in nuclease-free water, pH 7.0), 10% dextran sulfate (Sigma-Aldrich), and sodium phosphate (50 mM, pH 7.0) was added to cells for 30 minutes at 66°C. The hybridization buffer was removed and replaced with hybridization buffer containing a (/5TYE563/CCCCGGCCCCGGCCC) probe (batch #609686, Exiqon, Woburn, MA, http://www.exiqon.com) at 50 nM for 3 hours at 66°C in a dark, humidified chamber. Cells were rinsed with 2× SSC/0.1% Tween (Thermo Fisher Scientific Life Sciences) at room temperature followed by two rinses for 5 minutes in 0.2% SSC at 66°C. Coverslips were mounted using DAPI with Vectashield reagent (Vector Laboratories, Burlingame, CA, http://vectorlabs.com) and stored in the dark at 4°C until imaging. For coimmunocytochemistry experiments, RNA FISH was performed as above. In darkness, primary antibodies were added in 1× RNase-free PBS at concentrations listed above for 2 hours at room temperature. Cells were washed in 1× PBS, and secondary antibody was added for 1 hour at room temperature. Cells were washed in 1× PBS and mounted as mentioned previously.
Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated from frozen cell pellets using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA, http://www.qiagen.com) according to the manufacturer’s protocol. Complementary DNA (cDNA) synthesis was performed using the SuperScript III First-Strand Synthesis kit (Thermo Fisher Scientific Life Sciences). Relative gene expression levels were determined in triplicate reactions of 25 ng cDNA per reaction using the SensiFAST SyBR kit (Bioline, Luckenwalde, Germany, http://www.bioline.com) on a LightCycler 480 (Roche, Basel, Switzerland, http://www.roche.com). Quantification was performed using the –ΔΔCt method. GAPDH was used as a housekeeping gene with all iPSC-derived cells compared with their undifferentiated parent line. Total RNA from human skeletal muscle was obtained from a 20-year-old male (Clontech, Mountain View, CA, http://www.clontech.com) and used as a control. Primer sets are listed in supplemental online Table 3.
Repeat Primed Polymerase Chain Reaction
C9orf72 repeat expansion mutations were determined using the repeat-primed polymerase chain reaction (PCR) as described by DeJesus-Hernandez et al. [24]. PCR products were run on an ABI3730 DNA Analyzer and analyzed using Peak Scanner software. The characteristic sawtooth pattern is indicative of the presence of a repeat expansion.
Fluorescence-Activated Cell Sorting
Cells were collected on day 5 of differentiation using TrypLE enzyme, counted, and incubated in fluorescence-activated cell sorting (FACS) buffer (filtered salt-free PBS, 1 mM EDTA, 25 nM HEPES, pH 7.0, and 1% FBS) with CD309 (20 µL/5 million cells, phycoerythrin conjugated; BD, Franklin Lakes, NJ, http://www.bd.com) and CD140a (25 µL/5 million cells, Alexa Fluor 647 conjugated; BD) antibodies for 30 minutes on ice in the dark. Sorting was performed at the UCLA Broad Stem Cell Research Center Flow Cytometry Core on a FACSAria II cell sorter (BD).
Image and Video Acquisition
Images were taken on an Olympus iX53 microscope with a Retiga EXi Fast 1394 camera (QImaging, Surrey, BC, Canada, http://www.qimaging.com) using Q-Capture Pro 7 software. Videos were taken as 75-frame sequences with maximum frame speed enabled and converted in ImageJ at 7 frames per second, totaling 10 seconds. Confocal microscopy for RNA FISH was performed at the California NanoSystems Institute Advanced Light Microscopy Core on a Leica SP2-1P-FCS microscope. Z-stacks were averaged by frame and merged together using native Leica Microsystems Confocal software. Confocal images were taken on a Zeiss LSM 310. Z-stacks were merged together using ImageJ software.
Individual myotube contractions per second were quantified manually by a blinded observer. Only distinct individual myotubes were included for quantification, as movements of densely packed fibers could not reliably be quantified. Only contracting myotubes were counted.
Statistical Analysis
Statistics were performed in RStudio (version 0.98.978). Data were resampled 10,000 times using bootstrapping methodology [25]. Bootstrapping was performed on differences in the median between test samples. One-way, rank-based analysis of variance was performed for comparisons of three or more groups.
Results
iPSC Derivation
We obtained skin fibroblasts from four healthy control individuals, one patient diagnosed with sporadic FTD, and three FTD patients harboring a GGGGCC repeat expansion in the C9orf72 gene. Multiple iPSC lines from each patient were generated via lentiviral reprogramming vectors containing the canonical factors OCT4, SOX2, KLF4, and c-MYC and characterized via karyotyping, immunocytochemical staining, and Pluritest analysis (supplemental online Fig. 1 [22]). Fibroblasts derived from patients with a C9orf72 repeat expansion were confirmed to contain expanded GGGGCC repeats via repeat primed PCR and characteristic RNA foci (supplemental online Fig. 2).
Combinatorial Inhibition of PI3K and GSK3β With BMP4 and FGF2 Drives iPSCs to a Mesodermal Fate
During embryonic development, somitogenesis and eventual separation of paraxial, intermediate, and lateral plate mesoderm is controlled in part by morphogen gradients of BMPs, Wnts, FGF, and retinoic acid [26]. BMP4, in tandem with high concentrations of FGF and Wnt signaling, promotes development of the paraxial mesoderm along the primitive streak into the dermomyotome, which eventually forms the skeletal musculature of the body [26–28]. Studies have shown that inhibition of PI3K enhances the differentiation of human embryonic stem cells into mesodermal lineages through the promotion of activin A and nodal signaling [29]. Likewise, DMSO has been previously shown to improve differentiation efficiencies of pluripotent stem cells into terminal cell lineages through activation of the retinoblastoma protein [30]. Thus, we combined these two differentiation enhancement strategies with developmental morphogens to drive iPSCs into a mesodermal lineage (Fig. 1A).
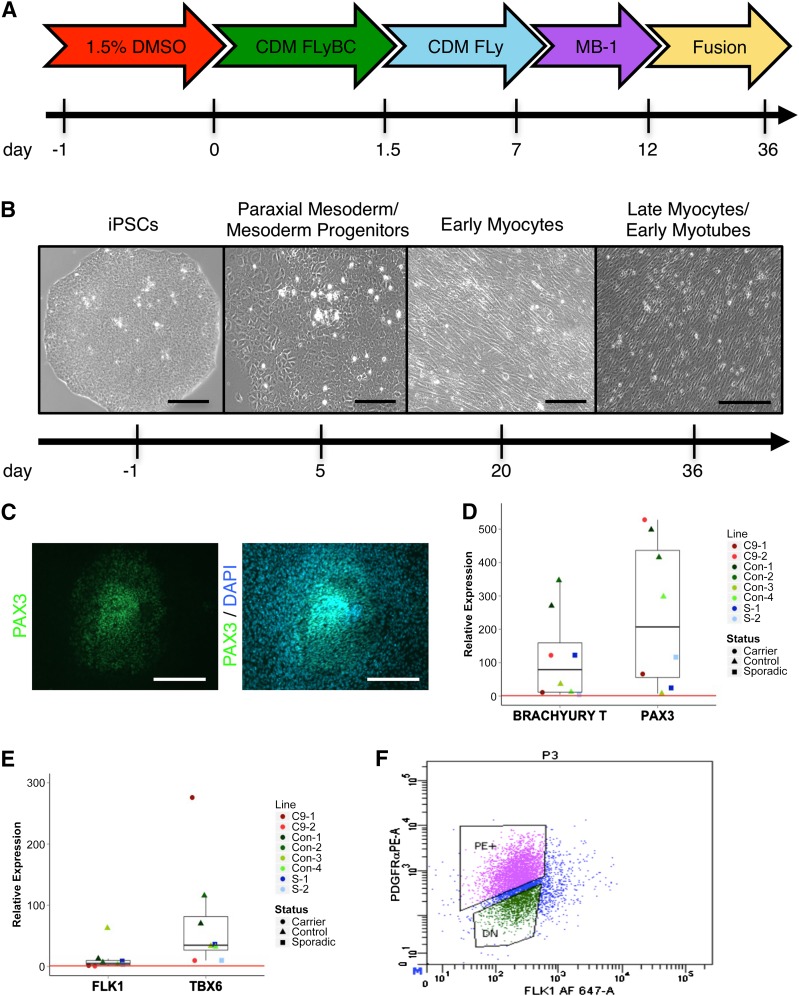
Figure 1.
Small molecule-directed differentiation. (A): Timeline schematic of medium formulations over 36-day protocol. (B): Representative phase contrast images of myogenic differentiation state over time. Scale bar = 100 µm, 250 µm (far right). (C): Representative immunostaining shows PAX3 expression beginning at the center of colonies and spreading radially outward at day 5. Scale bar = 250 µm. (D): Gene expression via qPCR at day 5 reveals high expression of early myogenic markers PAX3 and BRACHYURY T. (E): Gene expression shows preferential induction of paraxial mesoderm marker TBX6 compared with lateral plate marker FLK1 at day 5 (p = .138). Data shown are single differentiation trials of eight individual iPSC lines. Lines were derived from clones of a single rGGGGCC carrier diagnosed with FTD (C9-1, C9-2), four control individuals (Con-1, Con-2, Con-3, Con-4), and clones of a single patient diagnosed with sporadic FTD (S-1, S-2). Red line represents relative gene expression equal to 1, representative of undifferentiated iPSCs. (F): FACS sorting on day 5 reveals a cell population that is 48% PDGFRα+/FLK1−, representing prospective paraxial mesoderm progenitors. Line shown is Con-2. Abbreviations: CDM, chemically defined medium; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethylsulfoxide; DN, double-negative; FACS, fluorescence-activated cell sorting; FTD, frontotemporal dementia; iPSC, induced pluripotent stem cell; PE, phycoerythrin; qPCR, quantitative polymerase chain reaction.
Differentiation of sparsely plated iPSC colonies on vitronectin-coated plates or coverslips was initiated 1–3 days after passaging, when colonies were of small to medium size (approximately 250–400 µm). We exposed iPSCs to 1.5% DMSO in TeSR-E8 medium for 24 hours to prime cells for differentiation [30]. After DMSO treatment, cells were exposed to a chemically defined medium supplemented with FGF2, the PI3K inhibitor LY294002, and BMP4, termed CDM FLyB, as previously described [31]. Inhibition of GSK3β promotes Wnt/β-catenin signaling, which in turn promotes cellular differentiation to a mesoderm or endoderm fate [14, 15, 32]. Based on these studies, we added the GSK3β inhibitor, CHIR99021, to CDM FLyB during the initial 36 hours of differentiation (FLyBC) followed by withdrawal of BMP4 and CHIR99021 for 5.5 days.
At day 5, we observed PAX3+ mesodermal progenitors present at the center of differentiating iPSC colonies, making up <5% of the total cell population; however, we did not detect any PAX7+ cells at that time point (Fig. 1C). Analysis by quantitative PCR (qPCR) showed a significant induction of mesodermal genes BRACHYURY T and PAX3, with a concomitant loss of pluripotency markers NANOG and OCT4 (Fig. 1D, supplemental online Fig. 3A, 3B). Additionally, we observed increased expression of TBX6 compared with FLK1, indicating a favored differentiation into paraxial versus lateral plate mesoderm (Fig. 1E). We did not detect any significant difference in gene expression at day 5 based on carrier status between 8 total lines tested (supplemental online Fig. 3C).
To gauge the efficiency of paraxial mesoderm production, we used FACS to sort prospective paraxial mesoderm progenitors based on a PDGFRα+/FLK1− population [5, 16]. We first observed that 1.5% DMSO-treated cells contained 92% more PDGFRα+/FLK1− cells than those left untreated (supplemental online Fig. 4). We observed an overall cell population of up to 48% PDGFRα+/Flk1−, a proportion substantially higher than previously published protocols using embryoid bodies (Fig. 1F) [33]. Attempted recovery and propagation of the sorted population failed in every trial after replating cells in CDM FLy onto vitronectin-, Matrigel-, or gelatin-coated plates at various densities, even in the presence of the Rho kinase (ROCK) inhibitor Y-27632.
MB-1 Medium Followed by DMEM and Low Serum Promotes Skeletal Myotube Formation
Based on our day-5 analysis, we sought to find medium formulations and time frames that allowed paraxial mesoderm progenitors to continue to propagate in vitro while retaining their cell identity after 7 days. We first prepared myogenic induction medium, as in Darabi et al. [7]; however, the mesoderm progenitors failed to propagate in this formulation (data not shown). We then tested a serum-containing medium, MB-1, which allows for continued propagation of myoblasts and initiation of the myogenic program via TBX1 and TBX4 activation upstream of MYF5 and MYOD1 [10]. After 7 days in vitro and beyond, iPSC-derived mesodermal progenitors continued to propagate in MB-1 medium, and cellular morphology began to show elongation and alignment of progenitor myocytes, suggestive of maturation (Fig. 1B).
Before the addition of CHIR99021 to our protocol, we sought to test multiple timelines of selected medium formulations that best yield myogenic induction based on gene expression of PAX3, MYOG, MYH2 (myosin heavy chain 2, referred to hereafter as MHC), and MYOD1 after 36 days in vitro (supplemental online Table 2). The best-performing protocol, group E, showed high induction of PAX3, MYOG, and MYOD1, indicative of a cell population of myocytes in the mid-stages of differentiation together with a pool of PAX3+ progenitors and was selected for further study (supplemental online Fig. 5) [26]. The expression of MHC in group E cells was lower than in skeletal muscle RNA obtained from an adult, indicating that iPSC-derived myocytes are likely immature at day 36. This was supported by qPCR of acetylcholine receptor subunits, which showed high expression of the fetal γ subunit compared with the adult ε subunit, and immunostaining of MHC, which displayed both embryonic and adult isoforms (supplemental online Fig. 6).
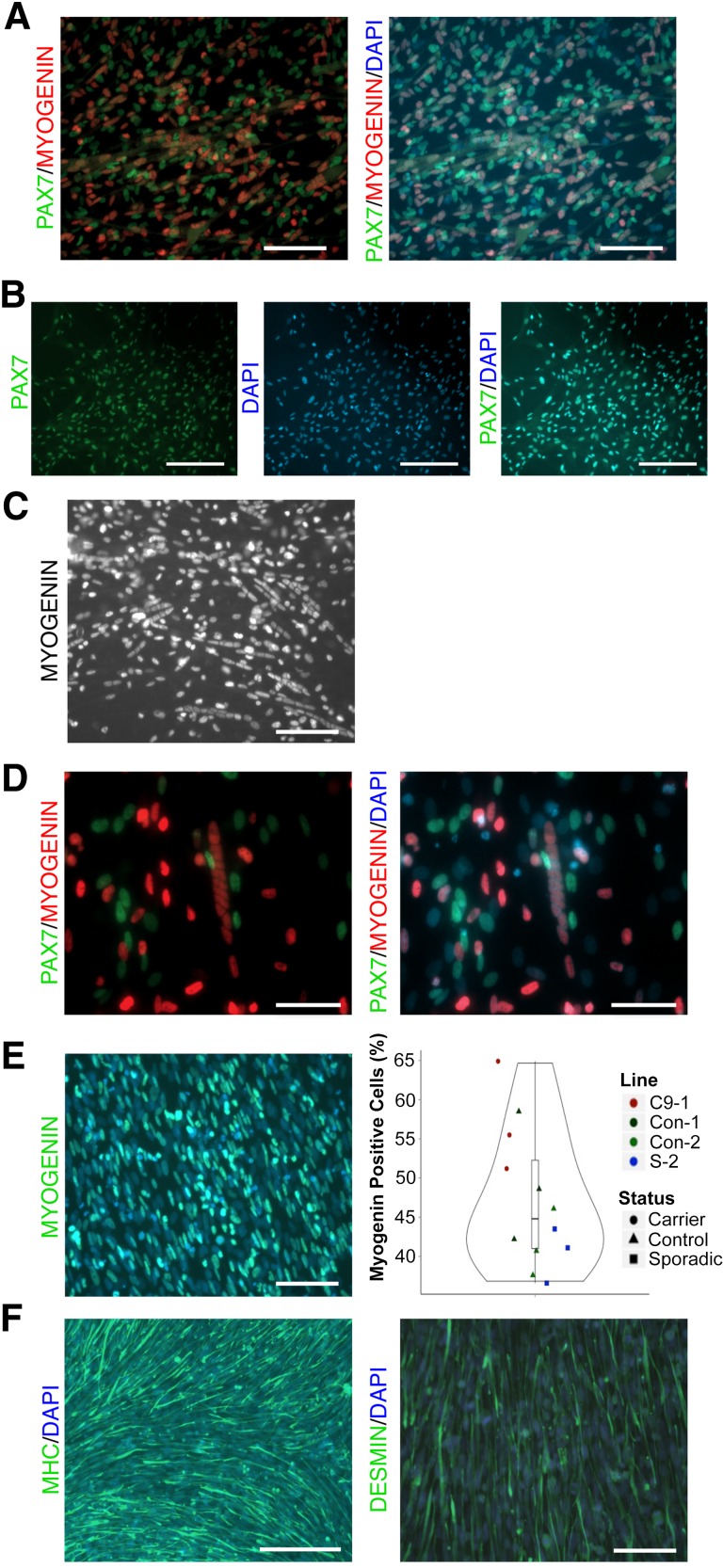
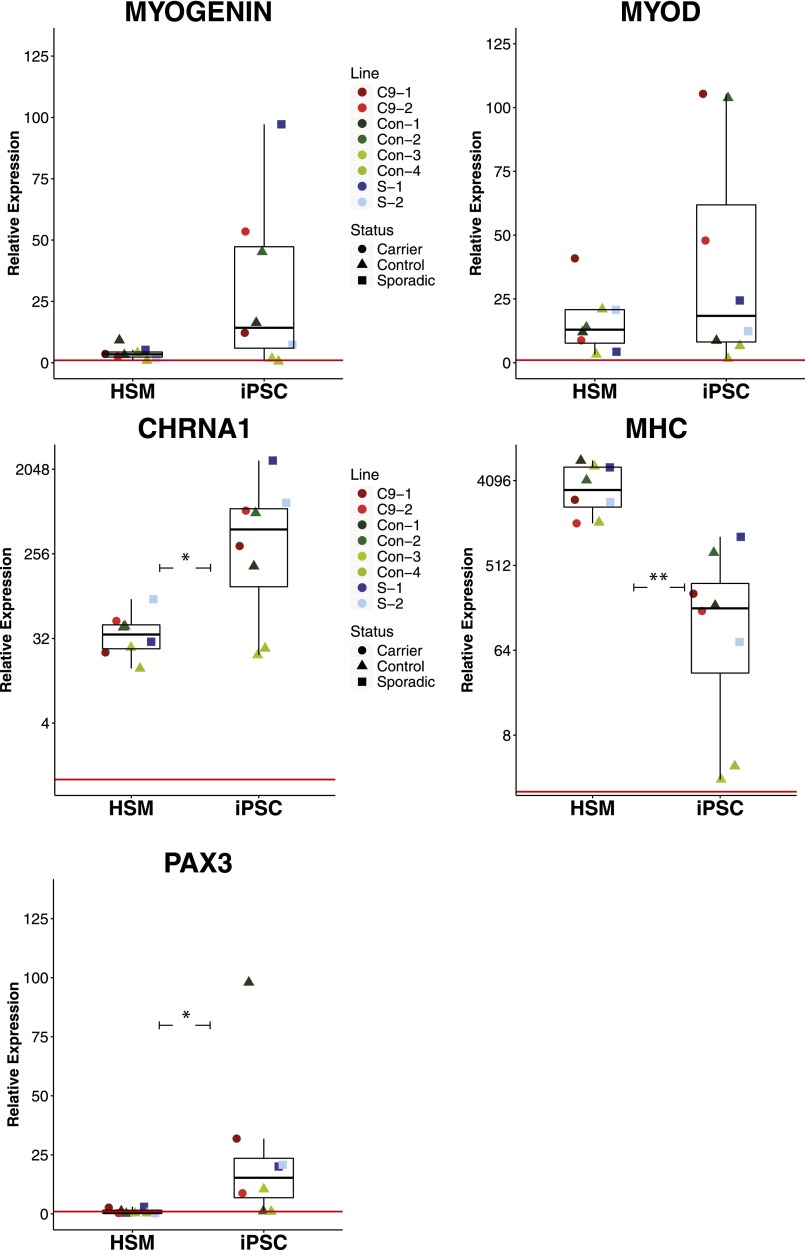
During development, myogenesis is governed by the progressive expression of transcription factor genes such as PAX3, PAX7, MYF5, MYOD1, and MYOG, representing distinct genetic landmarks of maturing myotubes [26]. During the switch from the PAX7+ myoblast stage to terminal differentiation, PAX7 expression is suppressed and MYOG is upregulated, marking distinct cell populations [34, 35]. We observed this phenomenon in vitro, as PAX7 and MYOG marked mutually exclusive cell populations, indicating a mixed population of cells at different differentiation stages (Fig. 2A). Interestingly, we observed areas of nearly 100% PAX7+ cells, whereas other regions displayed a mixture of cells at various stages of nuclear fusion (as defined by ≥3 concurrent nuclei), perhaps indicative of unique microsignaling environments (Fig. 2B, 2C). In all cases observed, multinucleated cells were MYOG+/PAX7− (Fig. 2D). Across four cell lines at day 36, we observed up to 64% of nuclei expressing MYOG (median = 44.8%), indicating highly efficient differentiation toward a skeletal muscle fate (Fig. 2E). We also observed a mix of intermediate- and late-stage skeletal muscle as shown by desmin (DES) and MHC staining, respectively (Fig. 2F). Gene expression via qPCR showed high upregulation of late-stage skeletal muscle transcription factors MYOG and MYOD1, as well as upregulation of MHC and the cholinergic receptor CHRNA1 compared with undifferentiated iPSCs (Fig. 3). Using RNA obtained from a 20-year-old adult’s skeletal muscle as a comparison for gene expression, we observed a significant increase in iPSC-derived muscle expression of CHRNA1 and PAX3 and decreased expression of MHC, supporting an immature phenotype of iPSC-derived muscle. PAX3 continued to be expressed, although at lower levels than on day 5, suggestive of the existence of a progenitor or satellite-cell pool within the population.
Figure 2.
iPSCs form multinucleated, MYOG+ myotubes by day 36. (A): Immunostaining of myogenic cells at day 36 reveals mutually exclusive PAX7+ and MYOG+ cell populations representing unique satellite cell progenitor and terminally committed populations. Scale bar = 100 µm. (B): Immunostaining for PAX7 on day 36 reveals certain regions with near-100% PAX7+ cells, possibly because of unique microsignaling environments. Scale bar = 250 µm. (C): Immunostaining for MYOG on day 36 shows certain regions with many myotubes actively undergoing nuclear fusion. Scale bar = 100 µm. (D): Multinucleated myotubes undergoing nuclear fusion on day 36 are always MYOG+. Scale bar = 50 µm. (E): Quantification of myogenic induction by MYOG+ cells across four individual iPSC lines. Each data point represents >125 cell nuclei counted in separate wells. Scale bar = 100 µm. (F): Immunostaining at day 36 shows large areas of cytoskeletal MHC+ and DES+ myogenic cells, representing terminally differentiated and intermediate cell populations, respectively. Scale bar = 250 µm (left), 100 µm (right). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; iPSC, induced pluripotent stem cell; MHC, myosin heavy chain.
Figure 3.
iPSC-derived skeletal muscle cells express key myogenic genes at day 36. Myogenic genes PAX3, MYOG, MYOD1, CHRNA1, and MHC all revealed high expression levels compared with undifferentiated iPSCs. CHRNA1 (p = .0391) and PAX3 (p = .0381) were upregulated compared with HSM, whereas MHC was downregulated (p = .0017). Data shown are single differentiation trials of eight individual iPSC lines. Lines were derived from clones of a single repeat expansion carrier diagnosed with FTD (C9-1, C9-2), four control individuals (Con-1, Con-2, Con-3, Con-4), and clones of a single patient diagnosed with sporadic FTD (S-1, S-2). Red line represents relative gene expression equal to 1, representative of undifferentiated iPSCs. HSM represents adult human skeletal muscle, with each data point compared with each parent iPSC line, respectively. ∗, p < .05; ∗∗, p < .01. Abbreviations: FTD, frontotemporal dementia; HSM, human skeletal muscle; iPSC, induced pluripotent stem cell.
Serum-Free Medium Promotes Terminal Differentiation and Spontaneous Activity of Skeletal Myotubes
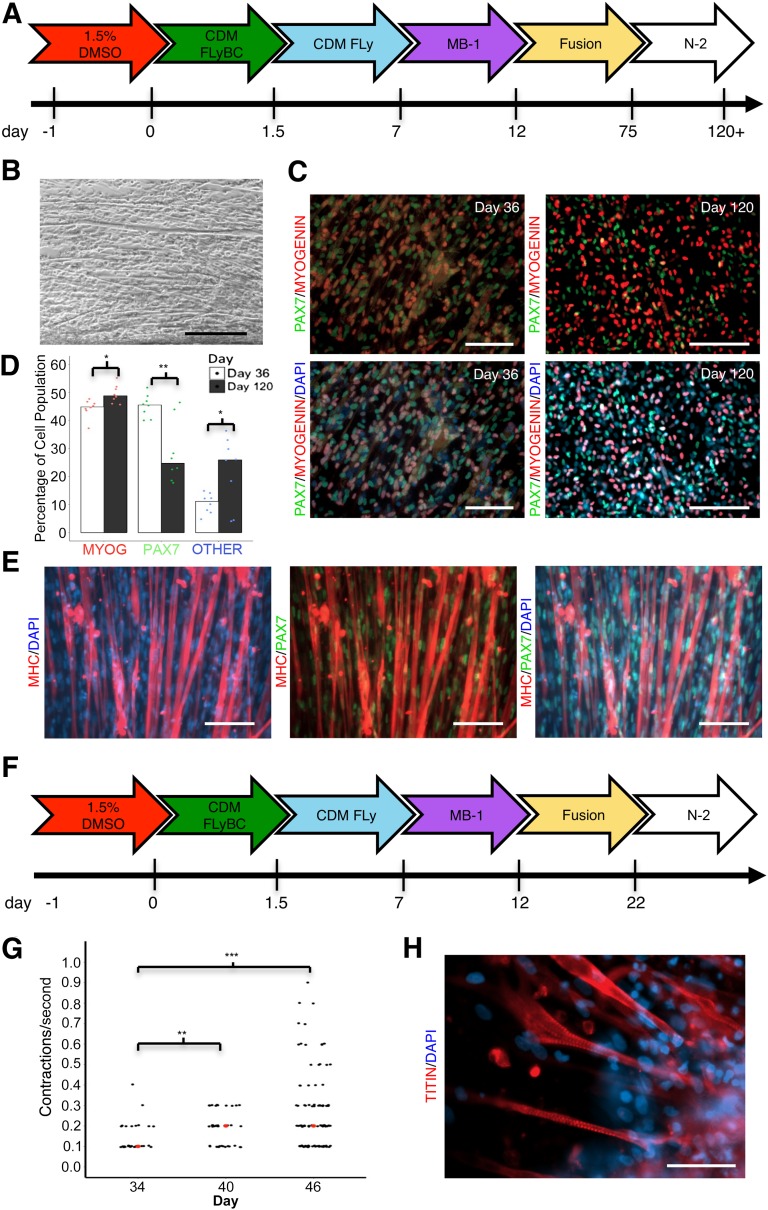
To our knowledge, four groups have reported spontaneous twitching of skeletal myotubes after differentiation from a pluripotent state without the use of transgene overexpression [4, 14, 17, 18]. We did not observe any spontaneous twitching in our cell cultures after 36 days in vitro and hypothesized that more time in culture was required to reach maturity (Fig. 4A). After 63 total days in fusion medium, we observed brief, spontaneous contractions in a small set of myotubes in addition to myotube thickening (Fig. 4B). Upon observation, we switched the medium of cells grown in tandem to a serum-free, N-2-based formulation, which promotes terminal differentiation [4, 14]. Seven to 10 days after the addition of N-2 medium, we observed robust spontaneous contractions throughout the cell cultures (supplemental online Movies 1 and 2). These cells continually contracted without exogenous stimulation for more than 3 weeks until fixation on day 120.
Figure 4.
The number of PAX7+ progenitor cells decreases over time in vitro. (A): Timeline schematic of medium formulations over a 120-day protocol. A small percentage of cells in fusion medium reach a spontaneously contractile state within 75 days. Switching to a serum-free N-2 medium promotes terminal differentiation coincident with spontaneous contractility within 7–10 days. Cells were grown on vitronectin-coated coverslips to prevent detachment of skeletal myotubes. (B): Phase contrast image of skeletal myotubes at day 70 show myotube thickening over time. Scale bar = 250 µm. (C): Immunostaining of myotubes for PAX7 and MYOG at days 36 and 120. Blue nuclei are stained with DAPI. Scale bar = 100 µm (left), 250 µm (right). (D): Quantification of myogenic cells on day 36 compared with day 120 shows a decrease in the percentage of PAX7+ cells over time (p = .0066) with coincident increases in MYOG+ cells (p = .023) and unidentified DAPI+ nuclei (p = .0446). ∗, p < .05; ∗∗, p < .01; n > 75 cells counted per data point). “Other” refers to unknown cell nuclei stained with DAPI. Bar plots represent the median. Data represents a single iPSC line (C9-2) differentiated in tandem plates in a single batch. (E): Immunostaining of myotubes at day 120 reveal PAX7+ cells intermingled with thickened MHC+ myotubes, reminiscent of a satellite cell niche. Scale bar = 100 µm. Line is C9-2. (F): Timeline schematic of the optimized protocol for obtaining spontaneously contractile myotubes in less than 36 days. (G): Quantification of single myotube contractions per second increased over a 12-day period. Myotube contractions per second were significantly higher on day 40 compared with day 34 (p = .0026) which continued in trend by day 46 (p = .0872). Red dots represent median values. iPSC line is C9-2. ∗∗, p < .01; ∗∗∗, p < .001. (H): Representative immunostaining of myotubes at day 36 reveal structural striations based on the presence of the sarcomeric protein TITIN. Scale bar = 50 µm. Abbreviations: CDM, chemically defined medium; DAPI, 4′,6-diamidino-2-phenylindole; iPSC, induced pluripotent stem cell; MHC, myosin heavy chain.
Following the protocols in Figures 1A and 4A, we quantified the ratio of PAX7+ or MYOG+ cells on day 36 compared with day 120 in a single-carrier iPSC line (Fig. 4C, 4D). At day 36, we observed up to 90% conversion into PAX7+ or MYOG+ myogenic cells. We also observed a 19.6% decrease in the amount of PAX7+ cells over time, suggesting that cells may continually become committed to terminal differentiation over time in vitro. Alternatively, differentiation efficiency may vary across trials, resulting in higher percentages of TUBB3+ neural crest-derived cells (supplemental online Fig. 7), which rely on similar signaling pathways during development [36, 37]. Nevertheless, a large amount of MHC+ myotubes surrounded by PAX7+ progenitor cells remained in culture even after 4 months, perhaps indicative of the satellite cell niche that is observed in vivo (Fig. 4E).
Because of the robust terminal differentiation phenotype observed after the addition of N-2 medium, we aimed to optimize the time frame of our protocol via the addition of N-2 medium after 10 days of fusion medium, or 22 days total from the start of differentiation (Fig. 4F). Using this strategy, we observed robust spontaneous myotube contractions as early as 34 days, which is the quickest time reported, to our knowledge. The median rate of spontaneous contractions increased over a 12-day period from 0.1 contractions/second to 0.3 (Fig. 4G, supplemental online Movie 3). Additionally, myotubes displayed striations as visualized by the sarcomeric protein TITIN (Fig. 4H). Importantly, cells were able to be passaged on day 23 or freeze-thawed on day 30 onto Matrigel-coated plates into N-2 medium with or without ROCK inhibitor, allowing for experimental scaling and long-term storage.
Skeletal Myotubes Derived From C9orf72 Repeat Expansion Carriers Contain RNA Foci but Do Not Exhibit TDP-43 Mislocalization
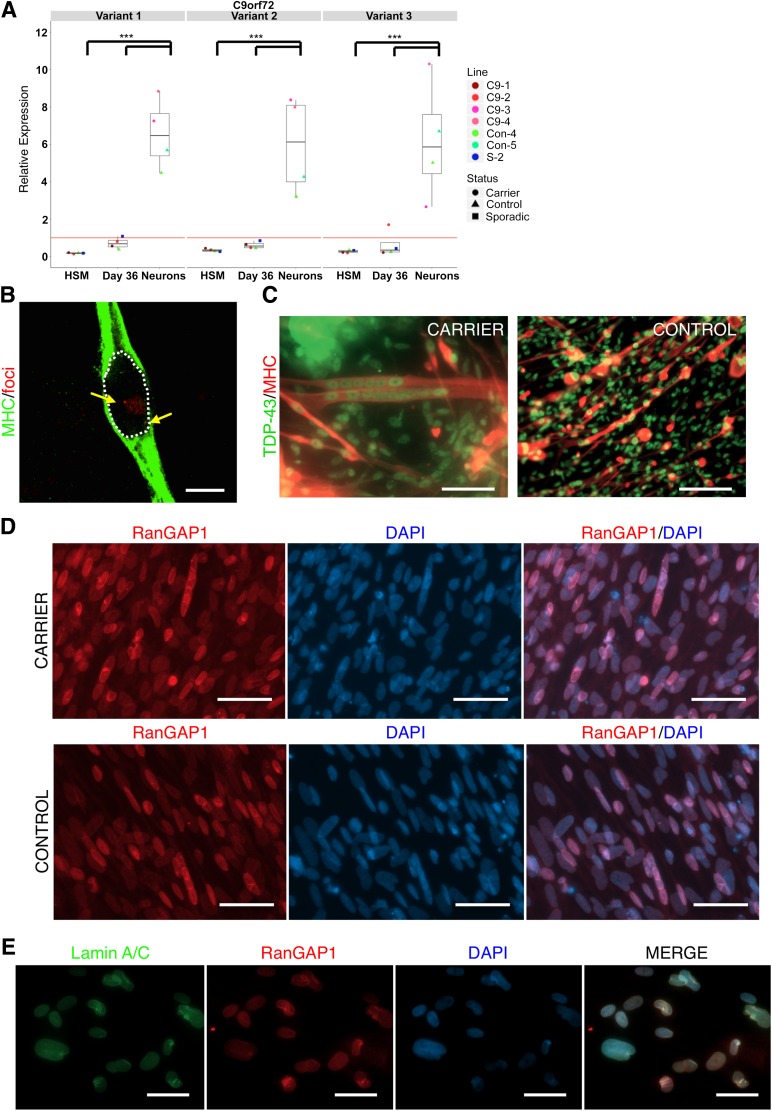
One of the three main hypotheses of how C9orf72 repeat expansions lead to ALS/FTD is the accumulation of RNA foci that may sequester important RNA-binding proteins, leading to splicing and transcriptome alterations [19, 38]. To determine the transcription levels of C9orf72 in skeletal muscle, we performed variant-specific qPCR on both iPSC-derived skeletal muscle at day 36 and iPSC-derived neurons. The results show that all variants of C9orf72 are expressed in both adult and iPSC-derived skeletal muscle, with iPSC-derived neurons showing the highest expression levels (supplemental online Fig. 8), as reported by other groups (Fig. 5A) [39]. We then performed RNA FISH on iPSC-derived skeletal muscle, which revealed the presence of RNA foci in MHC+ myofibers, indicating that RNA-related pathology in brain may also be present in muscle (Fig. 5B).
Figure 5.
Skeletal muscle derived from repeat expansion carriers contain RNA foci but do not exhibit mislocalized TDP-43. (A): qPCR detection of three separate transcript variants of C9orf72 in adult HSM, iPSC-derived muscle at day 36, and iPSC-derived cortical neurons (variant 1: NM_145005.6, variant 2: NM_018325.4, variant 3: NM_001256054.2). Data shown are pooled wells from single differentiation trials of four individual iPSC lines. Skeletal muscle lines were derived from clones of a single repeat expansion carrier diagnosed with FTD/ALS (C9-1, C9-2), one control individual (Con-4), and a single patient diagnosed with sporadic FTD (S-2). Neurons were derived from clones from two patients diagnosed with behavioral variant FTD (C9-3, C9-4). Red line represents relative gene expression equal to 1, representative of undifferentiated iPSCs. ∗∗∗, p < .001. (B): Confocal image shows an MHC+ iPSC-derived skeletal muscle cell containing two nuclear RNA foci. White dotted line indicates location of cell nucleus, yellow arrows indicate foci. Scale bar = 15 µm. (C): Immunostaining of TDP-43 reveals no instance of mislocalization in iPSC-derived skeletal muscle at day 120 in carrier or control lines. Scale bar = 50 µm. (D): RanGAP1 immunostaining shows typical perinuclear and nuclear localization in skeletal myotubes. Scale bar = 50 µm. (E): Nuclear folding evidenced by LMNA and RanGAP1 immunostaining is infrequently present in skeletal muscle derived from carriers and controls. Scale bar = 50 µm. Abbreviations: ALS, amyotrophic lateral sclerosis; DAPI, 4′,6-diamidino-2-phenylindole; FTD, frontotemporal dementia; HSM, human skeletal muscle; iPSC, induced pluripotent stem cell; LMNA, lamin A/C; MHC, myosin heavy chain; qPCR, quantitative polymerase chain reaction.
In IBMPFD, patients can exhibit identical TDP-43 and ubiquitin pathology in both brain and skeletal muscle [21]. Given the pathological overlap of IBMPFD with C9orf72-related motor neuron disease, we probed iPSC-derived skeletal myotubes at day 120 for TDP-43 mislocalization (Fig. 5C). We did not detect any instances of cytoplasmic TDP-43, indicating that C9orf72-related motor neuron disease does not display TDP-43 pathology or that these cells are too immature to properly model muscle pathology. Türk et al. have previously reported on a C9orf72 case study that displayed p62- and ubiquitin-positive inclusions in patient muscle biopsy [40]. We also probed iPSC-derived skeletal muscle for ubiquitin and p62 but did not observe any positive inclusions in our cells (data not shown).
More recent studies have shown that C9orf72-associated iPSC-derived neurons contain mislocalized RanGAP1 nuclear puncta and damaged nuclear envelope in Drosophila overexpressing expanded GGGGCC repeats, indicating impaired nucleocytoplasmic transport [41, 42]. Skeletal myotubes derived from expansion carriers and controls both exhibited typical RanGAP1 perinuclear staining with weaker distributions in the cytoplasm and nucleoplasm, with rare nuclear aggregates in both cases and controls (Fig. 5D) [41]. We infrequently observed aberrant nuclear membrane folding, as highlighted by LMNA staining and mirrored by RanGAP1 in skeletal muscle, but this phenomenon was present in both carriers and controls (Fig. 5E). Taken together, these results suggest that observed C9orf72-associated phenotypes may be restricted to neuronal subtypes, perhaps because of its high expression [39].
Discussion
Our study adds novel methodology to the growing body of protocols deriving skeletal myotubes from pluripotent stem cells for use in disease modeling, developmental studies, or drug discovery. There have been several studies thus far attempting a transgene-free approach, with various efficiencies [14, 15, 17, 18, 32, 43]. We likewise find that promotion of Wnt signaling via GSK3β inhibition strongly influences mesoderm commitment, and the combination of BMP4 and FGF2 signaling results in highly efficient mesoderm induction while recapitulating normal development as evidenced through a time-specific activation of early myogenic genes BRACHYURY T and PAX3, followed by PAX7 and late-stage markers such as MYOG, MYOD1, and MHC. Interestingly, we were still able to obtain myogenic cells with addition of BMP4 and FGF2 before adding CHIR99021 to our protocol, suggesting an important and nonindispensible role of BMP4 in myogenic induction, contrary to previous reports [18].
Our study differs from previously published studies that reported low myogenic induction efficiencies in the presence of serum [33, 43, 44]. In our protocol, we use MB-1 medium, which contains 15% FBS, followed by a fusion medium consisting of 2% horse serum, to achieve myogenic induction of up to 90% in 36 days. However, we likewise find that a serum-free N-2 medium necessitates myoblast fusion [4, 5, 7, 11, 14] enabling spontaneously contractile myotubes within 34 days. We believe this difference may be caused by the key additions of DMSO at the start of our protocol, which primes cells for differentiation, and PI3K inhibition, which has been shown to drive cells into a mesodermal lineage via activin A and nodal signaling [29–31]. Future optimization using serum-free medium formulations or cell sorting for markers such as NCAM or AchR [4, 15] may allow for use of iPSC-derived muscle in therapeutic regenerative medicine.
Success in maintaining a self-renewing satellite cell population in vitro would serve as an invaluable tool for future regenerative medicine purposes [45]. We observed that PAX7+ cells frequently lie adjacent to MHC+ myofibers, reminiscent of their in vivo niche between the myofiber plasma membrane and basal lamina (Fig. 4D). In tandem trials, we also observed a decreasing number of PAX7+ cells in vitro over time, indicating commitment to terminal differentiation (Fig. 4B). However, it is also possible that the 5% increase in MYOG+ cells and 19.6% decrease in PAX7+ cells are caused by initial overall decreased production of PAX7+ cells or increased production of neural crest-derived cell types. Further studies using a PAX7 reporter are needed to assess the percentage of PAX7+ cells that commit to terminal differentiation over time, what percentage of these cells coexpress the myogenic commitment factor MYF5, or whether addition of morphogens such as Wnt7a can maintain a symmetrically renewing population of progenitors indefinitely [46, 47].
The extracellular matrix (ECM) provides elasticity and stiffness qualities that exert strong influence over skeletal muscle proliferation and regenerative capacity [48–51]. The importance of the substrate in our studies is demonstrated by a failed recovery of day-5 mesodermal progenitors after FACS onto Matrigel, whereas the same Matrigel substrate can be used for passaging and expansion of cells later in differentiation on day 23. We believe this may be caused by Matrigel’s stiffness (Young’s modulus = 0.45 kPa [52]) being more similar to reported human adult muscle stiffness (0.5 kPa), allowing PAX7+ progenitors, which were not present at day 5, to proliferate on Matrigel [53]. Likewise, we occasionally observed detachment of skeletal myotubes along the edges of wells during terminal differentiation or because of strong contractile force (supplemental online Movie 1). Indeed, contractile cells remained attached for longer periods of time in dense areas or via growth on coverslips, the edges of which seemed to provide anchoring points and tension for contractile myotubes to attach, similarly to other methods using microfiber arrays or stainless-steel meshes [54, 55]. Using this strategy, we were able to observe contractile myotubes for >2 weeks. Further improvements may be made by more accurately mimicking in vivo properties and signaling cues of the ECM.
To demonstrate variability between iPSC lines and reproducibility of our protocol, we performed single differentiation trials among eight independent cell lines derived from six total individuals. As noted by others, we believe that the variability observed in gene expression between iPSC lines in our experiments may be largely influenced by imperfect initial seeding density [14, 15] but could also be influenced by interline variability based on genetic background of individual donors [56–58] which may be predictive of future differentiation potential [59]. For instance, lines Con-3 and Con-4 each had low levels of myogenic gene expression (Fig. 3), suggesting that they may be intrinsically skewed against mesodermal differentiation. Additional differentiation trials and iPSC lines are needed to accurately conclude which of these factors plays the governing role.
Evidence in animal models of ALS suggest that collapse of the neuromuscular junction and axonal retraction precede the death of the motor neuron, yet the role of skeletal muscle in the process remains largely unknown [60, 61]. We first determined that all variants of C9orf72 are expressed in iPSC-derived skeletal muscle, but at lower levels than iPSC-derived neurons. Interestingly, in the two lines we studied, repeat expansion carriers displayed a trend of higher levels of variants 1 and 2, contrary to most reports of mRNA haploinsufficiency in patient-derived tissues and cell lines (Fig. 5A) [24, 62], which warrants further investigation. We additionally probed for known pathological features associated with C9orf72-related ALS/FTD but found no evidence of TDP-43 mislocalization or ubiquitin/p62-positive inclusions, perhaps because of low levels of C9orf72 expression. In the vast majority of cases, RanGAP1 was properly localized to the perinuclear membrane (Fig. 5D), with few cases of aggregated RanGAP1 within the nucleus, seen in carriers and controls. In some instances, we observed a nuclear membrane phenotype via LMNA and RanGAP1 staining similar to that seen in patients with Hutchinson-Gilford progeria [63]. This phenotype, however, was observed in both carrier and control-derived cells (Fig. 5E), consistent with reported basal rates of altered nuclei of ∼10% [64]. Nevertheless, the localization of C9orf72 isoform b in the nuclear membrane and altered nuclear membrane proteins in patient postmortem tissues make a compelling case for the involvement of nuclear membrane breakdown in C9orf72-related disease [39, 65].
Conclusion
The data provided offer a robust method to obtain skeletal myotubes from pluripotent stem cells. This protocol can be adapted for obtaining skeletal myotubes from iPSCs from a variety of diseases for use in drug discovery and modeling of neuromuscular and musculoskeletal diseases. Further studies such as the tandem derivation of iPSC-derived motor neurons and skeletal muscle are needed to determine the contribution, if any, of the skeletal muscle and neuromuscular junction in C9orf72-related ALS and other motor neuron diseases.
Supplementary Material
Acknowledgments
This work was supported by CIRM Grant RL1-00650-1 (F.-B.G.) and National Institutes of Health Grants 5T32MH073526-08 (E.W.S.) and U54HD087101 (G.C.). G.C. was partially supported by the Tau Consortium and the John Douglas French Alzheimer’s Foundation. We thank Dr. Jacques Tremblay (supported by the Canadian Institute of Health Research) for supplying MB-1 medium, and the UCLA BSCRC Flow Cytometry Core, the UCLA CNSI ALMS microscopy core, Dr. Bob Farese (Harvard University), and members of Dr. April Pyle’s laboratory (UCLA) for helpful comments during the preparation of the manuscript.
Author Contributions
E.W.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.B., M.P., K.J.W., and S.A.: collection and/or assembly of data; A.K., F.-B.G., and B.L.M.: provision of study material or patients; G.C: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
G.C. has uncompensated research funding from the National Institutes of Health and private foundations. The other authors indicated no potential conflicts of interest.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Sakurai H, Okawa Y, Inami Y, et al. Paraxial mesodermal progenitors derived from mouse embryonic stem cells contribute to muscle regeneration via differentiation into muscle satellite cells. Stem Cells. 2008;26:1865–1873. doi: 10.1634/stemcells.2008-0173. [DOI] [PubMed] [Google Scholar]
- 4.Barberi T, Bradbury M, Dincer Z, et al. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat Med. 2007;13:642–648. doi: 10.1038/nm1533. [DOI] [PubMed] [Google Scholar]
- 5.Darabi R, Gehlbach K, Bachoo RM, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 6.Darabi R, Santos FNC, Filareto A, et al. Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells. 2011;29:777–790. doi: 10.1002/stem.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darabi R, Arpke RW, Irion S, et al. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka A, Woltjen K, Miyake K, et al. Efficient and reproducible myogenic differentiation from human iPS cells: Prospects for modeling Miyoshi myopathy in vitro. PLoS One. 2013;8:e61540. doi: 10.1371/journal.pone.0061540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albini S, Coutinho P, Malecova B, et al. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Reports. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goudenege S, Lebel C, Huot NB, et al. Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol Ther. 2012;20:2153–2167. doi: 10.1038/mt.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abujarour R, Bennett M, Valamehr B, et al. Myogenic differentiation of muscular dystrophy-specific induced pluripotent stem cells for use in drug discovery. Stem Cells Translational Medicine. 2014;3:149–160. doi: 10.5966/sctm.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maffioletti SM, Gerli MFM, Ragazzi M, et al. Efficient derivation and inducible differentiation of expandable skeletal myogenic cells from human ES and patient-specific iPS cells. Nat Protoc. 2015;10:941–958. doi: 10.1038/nprot.2015.057. [DOI] [PubMed] [Google Scholar]
- 13.Rao MS, Malik N. Assessing iPSC reprogramming methods for their suitability in translational medicine. J Cell Biochem. 2012;113:3061–3068. doi: 10.1002/jcb.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shelton M, Metz J, Liu J, et al. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Rep. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borchin B, Chen J, Barberi T. Derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Stem Cell Rep. 2013;1:620–631. doi: 10.1016/j.stemcr.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakurai H, Sakaguchi Y, Shoji E, et al. In vitro modeling of paraxial mesodermal progenitors derived from induced pluripotent stem cells. PLoS One. 2012;7:e47078. doi: 10.1371/journal.pone.0047078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosoyama T, McGivern JV, Van Dyke JM, et al. Derivation of myogenic progenitors directly from human pluripotent stem cells using a sphere-based culture. Stem Cells Translational Medicine. 2014;3:564–574. doi: 10.5966/sctm.2013-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chal J, Oginuma M, Al Tanoury Z, et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- 19.Ling S-C, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause S. Insights into muscle degeneration from heritable inclusion body myopathies. Front Aging Neurosci. 2015;7:13. doi: 10.3389/fnagi.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nalbandian A, Donkervoort S, Dec E, et al. The multiple faces of valosin-containing protein-associated diseases: Inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J Mol Neurosci. 2011;45:522–531. doi: 10.1007/s12031-011-9627-y. [DOI] [PubMed] [Google Scholar]
- 22.Müller F-J, Schuldt BM, Williams R, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sareen D, O’Rourke JG, Meera P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curran-Everett D. Explorations in statistics: The bootstrap. Adv Physiol Educ. 2009;33:286–292. doi: 10.1152/advan.00062.2009. [DOI] [PubMed] [Google Scholar]
- 26.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4:a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dosch R, Gawantka V, Delius H, et al. Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development. 1997;124:2325–2334. doi: 10.1242/dev.124.12.2325. [DOI] [PubMed] [Google Scholar]
- 28.Aulehla A, Pourquié O. Signaling gradients during paraxial mesoderm development. Cold Spring Harb Perspect Biol. 2010;2:a000869. doi: 10.1101/cshperspect.a000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean AB, D’Amour KA, Jones KL, et al. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 30.Chetty S, Pagliuca FW, Honore C, et al. A simple tool to improve pluripotent stem cell differentiation. Nat Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung C, Bernardo AS, Trotter MWB, et al. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat Biotechnol. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan JY, Sriram G, Rufaihah AJ, et al. Efficient derivation of lateral plate and paraxial mesoderm subtypes from human embryonic stem cells through GSKi-mediated differentiation. Stem Cells Dev. 2013;22:1893–1906. doi: 10.1089/scd.2012.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang Y, Suk S, Lin S, et al. Directed in vitro myogenesis of human embryonic stem cells and their in vivo engraftment. PLoS One. 2013;8:e72023. doi: 10.1371/journal.pone.0072023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olguín HC, Pisconti A. Marking the tempo for myogenesis: Pax7 and the regulation of muscle stem cell fate decisions. J Cell Mol Med. 2012;16:1013–1025. doi: 10.1111/j.1582-4934.2011.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- 37.Bhatt S, Diaz R, Trainor PA. Signals and switches in mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 2013;5:a008326. doi: 10.1101/cshperspect.a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prudencio M, Belzil VV, Batra R, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki N, Maroof AM, Merkle FT, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013;16:1725–1727. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Türk M, Haaker G, Winter L, et al. C9ORF72-ALS: P62- and ubiquitin-aggregation pathology in skeletal muscle. Muscle Nerve. 2014;50:454–455. doi: 10.1002/mus.24283. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, Donnelly CJ, Haeusler AR, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freibaum BD, Lu Y, Lopez-Gonzalez R, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan T, Liu J, Chu A, et al. Retinoic acid enhances skeletal myogenesis in human embryonic stem cells by expanding the premyogenic progenitor population. Stem Cell Rev. 2012;8:482–493. doi: 10.1007/s12015-011-9284-0. [DOI] [PubMed] [Google Scholar]
- 44.Awaya T, Kato T, Mizuno Y, et al. Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS One. 2012;7:e51638. doi: 10.1371/journal.pone.0051638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cosgrove BD, Sacco A, Gilbert PM, et al. A home away from home: Challenges and opportunities in engineering in vitro muscle satellite cell niches. Differentiation. 2009;78:185–194. doi: 10.1016/j.diff.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuang S, Kuroda K, Le Grand F, et al. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Grand F, Jones AE, Seale V, et al. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romanazzo S, Forte G, Ebara M, et al. Substrate stiffness affects skeletal myoblast differentiation in vitro. Sci Technol Adv Mater. 2012;13:064211. doi: 10.1088/1468-6996/13/6/064211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert PM, Havenstrite KL, Magnusson KEG, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosgrove BD, Gilbert PM, Porpiglia E, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trensz F, Lucien F, Couture V, et al. Increased microenvironment stiffness in damaged myofibers promotes myogenic progenitor cell proliferation. Skelet Muscle. 2015;5:5. doi: 10.1186/s13395-015-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soofi SS, Last JA, Liliensiek SJ, et al. The elastic modulus of Matrigel as determined by atomic force microscopy. J Struct Biol. 2009;167:216–219. doi: 10.1016/j.jsb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacraz G, Rouleau AJ, Couture V, et al. Increased stiffness in aged skeletal muscle impairs muscle progenitor cell proliferative activity. PLoS One. 2015;10:e0136217. doi: 10.1371/journal.pone.0136217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber A, Pickett A, Shakesheff KM. Reconstruction of spatially orientated myotubes in vitro using electrospun, parallel microfibre arrays. Eur Cell Mater. 2007;14:56–63. doi: 10.22203/ecm.v014a06. [DOI] [PubMed] [Google Scholar]
- 55.Vandenburgh HH, Karlisch P, Farr L. Maintenance of highly contractile tissue-cultured avian skeletal myotubes in collagen gel. In Vitro Cell Dev Biol. 1988;24:166–174. doi: 10.1007/BF02623542. [DOI] [PubMed] [Google Scholar]
- 56.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rouhani F, Kumasaka N, de Brito MC, et al. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doi A, Park I-H, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsankov AM, Akopian V, Pop R, et al. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupuis L, Loeffler J-P. Neuromuscular junction destruction during amyotrophic lateral sclerosis: insights from transgenic models. Curr Opin Pharmacol. 2009;9:341–346. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Fischer LR, Culver DG, Tennant P, et al. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Donnelly CJ, Zhang P-W, Pham JT, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang SH, Andres DA, Spielmann HP, et al. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J Clin Invest. 2008;118:3291–3300. doi: 10.1172/JCI35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao S, MacNair L, McGoldrick P, et al. Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann Neurol. 2015;78:568–583. doi: 10.1002/ana.24469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.