Abstract
Objective Health care generated data have become an important source for clinical and genomic research. Often, investigators create and iteratively refine phenotype algorithms to achieve high positive predictive values (PPVs) or sensitivity, thereby identifying valid cases and controls. These algorithms achieve the greatest utility when validated and shared by multiple health care systems.
Materials and Methods We report the current status and impact of the Phenotype KnowledgeBase (PheKB, http://phekb.org), an online environment supporting the workflow of building, sharing, and validating electronic phenotype algorithms. We analyze the most frequent components used in algorithms and their performance at authoring institutions and secondary implementation sites.
Results As of June 2015, PheKB contained 30 finalized phenotype algorithms and 62 algorithms in development spanning a range of traits and diseases. Phenotypes have had over 3500 unique views in a 6-month period and have been reused by other institutions. International Classification of Disease codes were the most frequently used component, followed by medications and natural language processing. Among algorithms with published performance data, the median PPV was nearly identical when evaluated at the authoring institutions (n = 44; case 96.0%, control 100%) compared to implementation sites (n = 40; case 97.5%, control 100%).
Discussion These results demonstrate that a broad range of algorithms to mine electronic health record data from different health systems can be developed with high PPV, and algorithms developed at one site are generally transportable to others.
Conclusion By providing a central repository, PheKB enables improved development, transportability, and validity of algorithms for research-grade phenotypes using health care generated data.
Keywords: electronic health records, electronic phenotyping, natural language processing, genomic research, clinical research
Objective
The Electronic MEdical Records and GEnomics (eMERGE) Network,1 as well as other efforts,2–7 have demonstrated that electronic health record (EHR) data can be used to identify research-grade disease phenotypes with sufficient positive and negative predictive values for use in identifying traits and diseases for biomedical research and clinical care,6–11 recruitment for clinical trials,3,4 quality improvement studies,12 population-based health outcomes research,9 disease or drug safety surveillance,13,14 and genetic research.15–21 These studies have required accurate, sharable, and quickly adaptable phenotype algorithms implemented in varying clinical data repositories.22 Moreover, research has shown that once created, these algorithms can be ported from one institution to another, often with comparable performance.15,23–26 The Phenotype Knowledgebase (PheKB; available at http://phekb.org) was created as a workflow management system and learning center supporting the creation, validation, and dissemination of computable algorithms. PheKB has built-in tools specifically designed to enhance knowledge sharing and collaboration across sites. With feedback mechanisms and structured performance measures for implementations, PheKB facilitates the transportability of algorithms across different institutions, health care systems, and clinical data repositories and into multiple research applications.
Background and Significance
eMERGE is a national network organized and funded by the National Human Genome Research Institute that combines DNA biorepositories with EHRs for large-scale, high-throughput genetic research in support of implementing genomic medicine. During Phases I and II (2007–2015), the network developed and deployed more than 47 electronic phenotype algorithms across more than 55 000 subjects with dense genomic data. The eMERGE network found that phenotype development is both highly iterative and time consuming,16,22 often taking up to 6–8 months to develop and validate a single algorithm and requiring coordination and collaboration among informaticians and clinical experts. Phenotype algorithms typically use heuristic or machine learning algorithms, combining multiple data sources to achieve high positive predictive values (PPVs) in identifying cases and controls for phenotypes of interest.27 Critically, sharing phenotypic and genotypic data across sites allowed increased sample size and thus power for discovery and has been able to advance genomic discoveries based on this data.15,19,20,28,29 For example, in the study of ventricular conduction in normal hearts, eMERGE leveraged existing genotype data from multiple sites to increase the statistical power of the analysis, resulting in a significant result where single site results had been nonsignificant.24
The initial approach adopted in eMERGE was to store versions of phenotype algorithms on shared wiki pages. Although these postings provided a way to share the final pseudocode product, the approach lacked critical historical information collected, as each site worked through implementation within the specific data available in their clinical data repository. Access to versions with an understanding of the changes became critical for efficient workflow. Communicating changes in algorithms as they occur along with associated work documents, included flow charts, site adaptations, implementation notes, and validation tools enables validating sites to efficiently implement the lead site’s algorithm. PheKB was created to meet this need by offering a collaborative environment to support all stages of development, validation, implementation, data sharing, and dissemination. More recent collaborations with the Patient-Centered Outcomes Research Network (PCORnet), the NIH Collaboratory, and the Pharmacogenomics Research Network have yielded a broader engaged community providing a greater number of use cases, resulting in important evolution of the site. While PheKB typically supports rule-based phenotype algorithms, emerging statistical approaches to phenotyping5,30 are yielding data-driven approaches to phenotyping; one such example is currently available on PheKB.31 Lessons learned in multisite collaboration for phenotype algorithm development include early assessment of feasibility, appropriate versioning, standardizing data elements, data quality and validation checks, and methods for disseminating the results.22
Materials and Methods
PheKB was developed iteratively within the eMERGE Network starting from 2012 and is continuously enhanced as needs are identified and the field advances. The site leverages the Drupal content management system32 and includes custom-developed extensions to facilitate aspects of the phenotype algorithm workflow (see Results). Drupal was chosen largely due to local familiarity, its flexibility to support custom content types with arbitrary metadata tags, the ability to integrate custom code with off-the-shelf modules, and its longevity, though many other content management systems would support the needed functionality. PheKB required the ability to collect custom metadata for phenotypes and their implementations and support different views and searching based on these fields, implemented with standard Drupal modules. Nonstandard features including customized access controls, phenotype workflow, and integrated external code for data validation features, described in more detail below, were implemented with custom programming using Drupal’s standard application programming interface. Over the years, we have migrated websites across major revisions from Drupal 5 to Drupal 7 with relative ease and with each upgrade had major improvements.
The goal of PheKB is to enable a workflow with purposefully integrated tools and standards that guide the user in efficiently navigating each of these stages from initial development to public sharing and reuse. As the eMERGE Network Coordinating Center, Vanderbilt University led development of PheKB and solicited feedback from eMERGE members during face-to-face and online conference meetings. On-boarding new users has led to additional suggestions to improve the usability of the site. For additional support, the eMERGE Coordinating Center has provided “How To” documentation and held one-on-one help sessions to assist new users, less than 5 sessions for the over 250 registered users. The design objectives for PheKB were to: (1) be a reference site enabling asynchronous communication between collaborators, (2) manage levels of sharing through the iterative stages of development and implementation, (3) facilitate sharing results, (4) provide an archive of published algorithms and implementation results (with customizations) that can serve as a resource for others to reuse the algorithms with confidence, and (5) provide efficient search and navigation mechanisms.
PheKB accomplishes these functions by (1) extending user permissions with a user group system to apply the custom access rules for viewing, creation, and data access; (2) having user profiles and authentication with institution as a metadata field; (3) defining content types to collect phenotype metadata and files as well as supporting commenting on discussion boards around phenotypes; (4) creating a metadata element that flags phenotype status to varied levels of visibility; and (5) allowing search by keywords as well as metadata. The concept of metadata-driven application development is not new and is well established;33 a key component in development was to create a simple workflow methodology and lightweight interface to allow research teams to autonomously develop and share phenotype algorithms efficiently.
A Data Dictionary/Data Validation tool written as a Ruby web service was integrated via a custom Drupal module to identify errors and warnings in data dictionaries and data files associated with a given phenotype. As files are uploaded, PheKB is able to display errors and warnings about the structure and content of the files. Users will upload data dictionaries for a phenotype, and data files containing implementation results can be associated to one of those dictionaries. In this way, the system ensures that the data structure and values fit the rules established in the data dictionary. A key feature in the data validation tool and sharing is the role-based security to restrict viewing of datasets to only specific groups. While most commonly this is the access group created for the phenotype algorithm authoring institution, it is flexible enough to include other groups (such as a secondary analysis site). Other groups can be given read-only access and can view the phenotype artifacts and uploaded data, but only the algorithm owners and the data owners can download data. In this way, PheKB becomes a secure environment to centralize asynchronous data collection and validation.
Results
The current version of PheKB supports the following workflow and features.
Computable phenotype creation, validation, and implementation
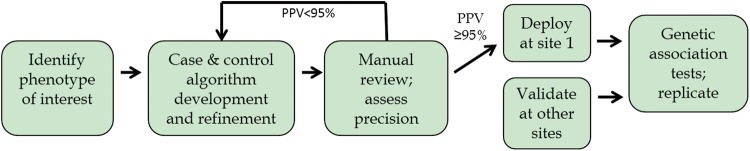
To facilitate electronic algorithm creation within the eMERGE Network, a site typically leads the study by assessing the feasibility, then develops a phenotype algorithm that is implemented and tested within their system. The algorithm is shared on PheKB for iterative feedback and revision as it is implemented (Figure 1). In addition to posting associated documents, researchers are encouraged to catalog their uploaded phenotype with metadata labels based on multidimensional representations, such as the methods and modalities used in the phenotype criteria (e.g., International Classification of Disease [ICD] codes, medications, natural language processing [NLP]), age, network, or institution affiliation. Uploaded documents typically include full descriptions of the computable algorithms including data types used, execution logic and variable dependencies, data definitions, and flow charts or other descriptive graphics. Although most files currently in PheKB are descriptive documents, such as human-readable portable document format files, users can also upload executable logic such as Konstanz Information Miner (KNIME)34 files and customized modules (e.g., for NLP tools). KNIME is a graphical workbench that includes extract, transform, and load functions and provides a framework to embed a logical workflow of steps for a phenotype algorithm. Sites within the eMERGE Network have used KNIME to create transportable phenotypes, and at least 7 of these have been shared on PheKB by at least 2 different sites, and have been implemented by at least 4 sites.25,35–37
Figure 1:
Approach to phenotyping.
Validation of the algorithm’s performance within different clinical data repositories is critical for establishing transportability to a large number of collaborating sites. The author of a phenotype and subsequent sites that utilize PheKB’s implementation pages can add site-specific implementation results in table format that capture data to calculate sensitivity, specificity, PPV, and negative predictive value, as well as total subjects identified.38 A commenting feature encourages discussion of poor or unexpected results, unique qualities of the data or the clinical repository supporting the implementation, and any changes or adaptations made to the phenotype algorithm to allow it to work within that site’s EHR system. These features assist future users in applying the algorithm to their unique repository as well as determining the algorithm’s suitability to the specific study or use case, e.g., identification of specific, narrowly defined disease cohort vs determining potential participants for broad-based recruitment.
The social networking framework for the website allows the user to control and define sharing of algorithm information. An algorithm is not publicly viewable until it is designated as “final” by the author. Earlier stages of sharing foster collaboration, allowing users to post and archive multiple iterations of the documents, comment and discuss issues in a whiteboard-type fashion, notify collaborators of changes, and provide implementation details. There are 3 stages of pre-final development that are viewable only by the selected “owner” and “view” groups: in-development, testing, and validated. Users join collaborative groups by sending a “membership request” to the appointed group administrator. For each phenotype, the author can designate an “owner group” and a “view group.” Collaborative owner groups allow member researchers to collectively revise a phenotype algorithm before making it available publicly. Phenotypes can be privately shared within view groups so these users can discuss, provide feedback, and upload implementation data for the phenotype. Users can also post their own implementation results through “implementation records,” which allow structured capture of a site-specific validation of a phenotype algorithm and any changes they may have made to adapt the phenotype to their environment. The site’s streamlined workflow requires minimal additional training for users, both for producers of phenotypes and consumers.
Data sharing tools
The Data Dictionary/Data Validation Tool validates covariate data definitions and associated data and is an embedded resource for registered users. The user uploads to the page associated to the phenotype, and the tool verifies the data dictionary file for adherence to standards and best practices. A predefined set of rules identifies deviations from the standard (“errors,” such as listing a variable twice) or recommendations for best practices (“warnings,” such as including an additional column). These checks are similar to (and inspired by) the checks run by the National Center for Biotechnology Information’s database of Genotypes and Phenotypes for phenotype submission files.39,40 Its integration in the phenotyping development workflow within PheKB ensures consistent formatting and coding of shared data sets. A data management function provides tracking tools for users to easily determine what data has been shared, what algorithm it is linked to, and by whom it was shared. The Data Dictionary/Data Validation Tool encourages data standardization and early stage quality control to efficiently share data for the merging of study sets. This minimizes reprocessing that may be otherwise needed by slight deviations from a published data dictionary.
Algorithm dissemination
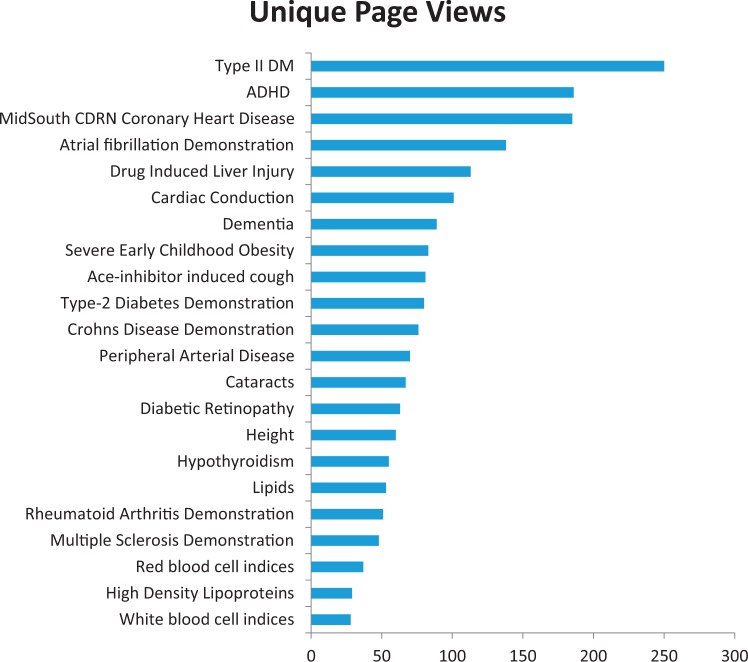
Finally, phenotype algorithms can be published on the site to allow for public sharing of definitions and results for reuse. Within PheKB, one can publicly share algorithms as well as multiple implementation results with recorded PPV, sensitivity, and site-specific notations. Performance data in the form of baseline implementation comparisons allows reuse of the algorithms with confidence. As seen in Figure 2, between December 1, 2014, and May 31, 2015, almost all of the public phenotypes have been uniquely viewed more than 50 times, with the Type 2 Diabetes algorithm having over 900 views since its publication. While many public algorithms have been developed by the eMERGE, the Mid-South Clinical Data Research Network Coronary Heart Disease algorithm is the third-most viewed algorithm, and was developed as part of PCORNet.4 Of the top 20 locations that have viewed this algorithm, 15 are institutions that are part of some Clinical Data Research Network sites and 4 are locations outside the United States.
Figure 2:
Views of Phenotypes on PheKB.
Having a well-defined corpus of developed phenotypes has been used to study algorithm portability,26 define recurring design elements in phenotype algorithms,41 and compare performance of different algorithms identifying the same disease.42 Leveraging past phenotype work, users can clone, edit, and link related phenotypes together (e.g., “diabetic retinopathy” is related to “type 2 diabetes,” as the former embeds the latter).
Using metadata, an algorithm can be searched on the inclusion or exclusion of classes of data elements (such as diagnoses or medications), authoring institution, author, network affiliation, or keyword. This functionality assists users in finding algorithms suited to their study as well as the capability of their clinical data repository. Electronic phenotype development efforts in eMERGE demonstrated that although algorithms showed considerable heterogeneity of specific features used, there was a strong degree of homogeneity in logic and classes of data elements.41
Use of PheKB
Currently, PheKB has 414 users from 52 institutions. Table 1 shows an overview of posted algorithms and contributing networks; the median number of algorithms per institution is 4 and the maximum number is 30. There are 30 public algorithms with 66 implementations currently available and 62 nonfinal algorithms with 83 associated implementations in various stages of development.
Table 1:
Phenotypes and Implementation Results on PheKB
| Institutions with users | 52 |
| Total phenotype algorithms | 92 |
| Public | 30 |
| Restricted/in development | 62 |
| With single site validationa | 51 |
| With external validationsa | 42 |
| Final (total) phenotype algorithm groups | |
| eMERGEa | 20 (43) |
| Pharmacogenomics Research Network/PGPopa | 0 (11) |
| Local/Othersa | 10 (8) |
| Total implementation resultsa | 149 (for 53 algorithms) range: 0–8/phenotypes |
| Total comments posted on shared algorithmsa | 384 (for 35 phenotypes) |
aIncludes phenotypes not yet public.
The most common data modality used for the algorithms were ICD codes, followed by medication data and NLP (Table 2).
Table 2:
Types of health care-related data modalities used in phenotype algorithms posted on PheKB
| Data modalities or methods | Number of phenotypes utilizing these features | ||
|---|---|---|---|
| Public (N = 30) | Non-publica (N = 62) | Percent of total (%) | |
| ICD-9 codes | 27 | 37 | 70 |
| Medications | 25 | 32 | 62 |
| Natural language processing | 21 | 21 | 46 |
| CPT codes | 14 | 24 | 41 |
| Laboratory test results | 14 | 21 | 38 |
Counts are out of 92 total algorithms at the time of this study.
aIncludes the all non-public statuses: “In development,” “Testing,” and “Validated.”
Of the 92 public and nonfinal algorithms, 51 have validation data from at least 1 site. Forty-three of the algorithms have multisite evaluations, with a median of 3 (range 1–8) external validations per algorithm. Median case PPV and case sensitivity were 96.5 and 100% with an interquartile range (IQR) of 9% and 8%, respectively. Median control PPV and control sensitivity were 100% and 99.0% with an IQR of 4% and 2%, respectively. The lower PPVs (see Figure 3) were associated with algorithms for rare phenotypes, which were tuned for sensitivity rather than PPV, such as drug-induced liver injury (PPV = 32%).31,32 Lower sensitivities included an algorithm to identify dementia (sensitivity = 37.1%), which utilized only ICD codes.
Figure 3:
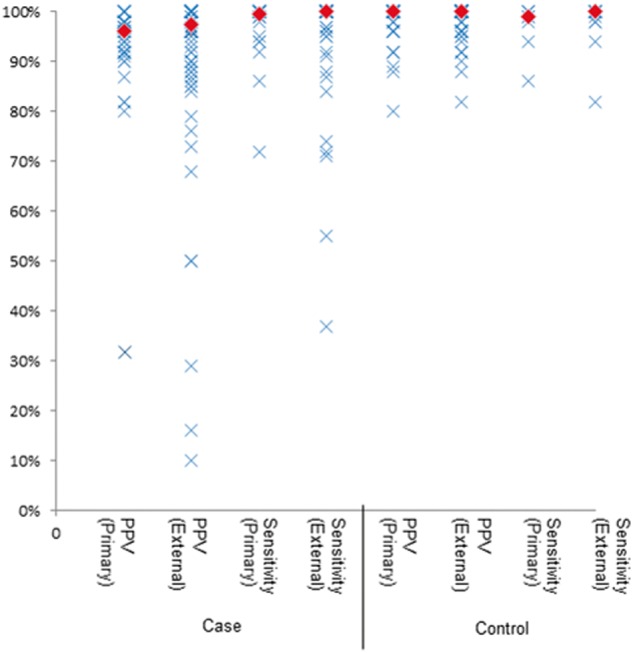
Primary and external site implementation distribution of results. Primary site refers to the algorithm’s performance at the authoring institution; external site refers to the results seen at sites other than the authoring institution. The diamonds represent the median results.
Performances on case and control algorithms for development-site (host) evaluations were similar to performance by external-site evaluations. Median case PPV and sensitivity were 96.0% and 99.5% for host evaluations, with IQRs of 6% and 5%, respectively. Median external site case PPV and sensitivity were 97.5% and 100% with variance of 1% and 9%, respectively. Median host control PPV and sensitivity were 100% and 99.0% with IQRs of 4% and 3%, whereas median external site control PPV and sensitivity were both 100% with IQRs of 4% and 2%.
Case study: multisite interactions during phenotype development in eMERGE
Among eMERGE network phenotyping workgroup use of PheKB for phenotype development and implementations, 384 comments have been made on 35 eMERGE phenotypes, with 1–31 comments per algorithm. Comments have occurred on algorithms at all stages of development, with the majority occurring on “validated” algorithms (when algorithms are typically being executed across eMERGE sites).
As an example, we characterize the development “life cycle” of a particular completed phenotype for Clostridium difficile colitis. There were 21 comments from 6 institutions, with 20 of those comments coming over a period 27 days in the active network-implementation phase. Eight exchanges clarified issues within the algorithm necessary for implementation at other sites. As the algorithm was developed in an adult population, other comments addressed the application in a pediatric population. Many users also identified areas of clarification that were necessary to collect required analysis covariates, as this collection is often as complex as implementation of the original phenotype algorithm. The comments and validations led to 4 revisions of the C. difficile colitis phenotype algorithm. Similar development life cycles occur with other phenotypes.
Discussion
PheKB enables streamlined development, sharing, and collaboration of algorithms designed to abstract research-quality phenotypes from health care data by design. PheKB also seeks to facilitate collaborative “next-generation phenotyping” as users investigate additional and more detailed phenotypes21,43 as well as varied use cases, such as recruitment and surveillance.
Analysis of current implementation results on PheKB show that the posted algorithms produce phenotypes with high precision and are transportable across different health care systems with generally similar high performance. In the setting of performing research on a given condition, PPV has generally been considered a more important metric than sensitivity,22,44,45 assuming sufficient sample size and that exclusions of cases are typically based on lack of available health care data instead of asserting a systematic bias toward a disease subset. As expected, more phenotypes had PPV evaluations than sensitivity evaluations. Algorithms with poorer PPV were often rare disease phenotypes in which manual review is feasible. While these results are subject to a reporting bias (authors may not post poorly performing algorithms), the breadth and diversity of implementations and phenotypes on PheKB suggest a wide variety of phenotypes can effectively be extracted using health care data and that these algorithms are transportable. The range of diseases investigated by the phenotypes posted spans circulatory, neurodevelopment, metabolic, and respiratory diseases and traits. This data adds importantly to specific phenotype evaluations,15–17,24,25,29 many of which are posted on PheKB.
Across institutions, the differences in EHRs require local customizations and workarounds of algorithms,22 which PheKB facilitates with the discussion boards and other features. By sharing phenotypes and their implementations broadly, researchers have also been able to identify commonalities between these phenotypes.41 Without an interface to capture data and system descriptions along with workflow, the algorithms risks increasing ambiguity and decreasing standardization. We hope that use of increasingly standardized definitions and pre-processing algorithms will reduce the need for local customization in the future. It is important to note that, because of the inevitable variation in workflow and data collection, standardization should provide context for site-specific needs that may be missing from the supporting publication using an algorithm. Communication about workflow and EHR differences can also improve transportability of phenotype algorithms through documentation of customizations fitted to specific EHRs, data repositories, or data types. PheKB makes research on phenotype algorithms easier by providing a knowledge base of what others have done, their implementation results, and details on what and why customizations have been made. We have seen that researchers have used PheKB as a resource to evaluate phenotype algorithms for commonalities41,42 and to compare results using different methods.8,46,47 PheKB algorithms have also proven a source of high-performing benchmarks against which novel approaches of statistical or knowledge resource mined approaches to automated phenotyping may be held.5,31,48
Currently, it is not clear why some algorithms may not perform as well at a given site as another, and the only method to identify poorer-performing algorithms is validation. Increasingly standardized algorithms and use of PheKB as a data aggregation tool could enable simple data profiling approaches to spot some inconsistencies (e.g., pregnant males, populations varying dramatically in covariate distributions, case/control count ratios, etc.). Future research into algorithms could identify factors associated with superior performance across different sites.
Conclusion
A key measure of long-term success for PheKB will be the growing library of phenotypes from a variety of networks and contributors, with subsequent reuse. It is difficult to directly track implementations of PheKB algorithms outside of those who have posted implementation data. However, based on a citation review, at least 40 non-eMERGE groups have deployed and then cited the Type 2 Diabetes algorithm (see Appendix).
Currently, most of the electronic phenotype algorithms on PheKB are in human-readable formats, which require human translation into executable code within local clinical repositories. However, several phenotype algorithms also include computable representations, such as in KNIME, or use statistical methods and approaches. As these methods evolve, PheKB will need to grow and adapt to fit the needs of these representations. Additionally, a key improvement for adoption of electronic phenotype algorithms will be creation of structured representations that enable rapid and error-free implementation and execution. One related approach has been the Quality Data Model49 and approaches built on common data models, such as Observational Health Data Sciences and Informatics42 and PCORnet. The format-agnostic approach of PheKB would allow such codified approaches, as well as the current data formats. Creating programmatic interfaces between PheKB and some of the networks to allow easy interchange of executable phenotype algorithms may be advisable.
Research using the EMR can be time consuming and difficult.27,43,44,50 The outgrowth of work in this area by the eMERGE Network is the workflow and tools built within PheKB. PheKB benefits from a consortium of academic institutions, and we encourage others to participate in sharing and disseminating research results. Next steps include collaborating with other consortia and institutions to develop improved tools that can further facilitate reuse, including algorithm comparisons by research topic and implementing data quality metrics.
Funding
This study was supported by the National Human Genomic Research Institute, grant numbers: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Harvard), U01HG006830 (Children’s Hospital of Philadelphia), U01HG006389 (Essentia Institute of Rural Health), U01HG006382 (Geisinger Clinic), U01HG006375 (Group Health Cooperative), U01HG06379 (Mayo Clinic), U01HG006380 (Mount Sinai School of Medicine), U01HG006388 (Northwestern University), U01HG006378 (Vanderbilt University), and U01HG006385 (Vanderbilt University serving as the Coordinating Center). Additionally, this study was supported by the National Institute of General Medical Sciences, grant numbers R01GM105688 and R01GM103859.
Competing Interests
We have read and understood the policy on declaration of interests and declare that we have no competing interests.
Acknowledgments
We gratefully acknowledge Stephanie Pretel (NIH/NLM/NCBI) for assistance and discussion for the development of the Data Dictionary/Data Validation.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med Off J Am Coll Med Genet. 2013;15:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chute CG, Pathak J, Savova GK, et al. The SHARPn project on secondary use of electronic medical record data: progress, plans, and possibilities. AMIA Annu Symp Proc. 2011;2011:248–256. [PMC free article] [PubMed] [Google Scholar]
- 3.Richesson RL, Hammond WE, Nahm M, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013;20:e226–e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleurence RL, Curtis LH, Califf RM, et al. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources [published online ahead of print April 29, 2015]. J Am Med Inform Assoc. doi:10.1093/jamia/ocv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah NH. Mining the ultimate phenome repository. Nat Biotechnol. 2013;31:1095–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boland MR, Tatonetti NP, Hripcsak G. Development and validation of a classification approach for extracting severity automatically from electronic health records. J Biomed Semant. 2015;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richesson RL, Rusincovitch SA, Wixted D, et al. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inform Assoc. 2013;20:e319–e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levison J, Triant V, Losina E, et al. Development and validation of a computer-based algorithm to identify foreign-born patients with HIV infection from the electronic medical record. Appl Clin Inform. 2014;5:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenman M, He J, Martin J, et al. Database queries for hospitalizations for acute congestive heart failure: flexible methods and validation based on set theory. J Am Med Inform Assoc. 2014;21:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah NH, LePendu P, Bauer-Mehren A, et al. Proton pump inhibitor usage and the risk of myocardial infarction in the general population. PLoS One. 2015;10:e0124653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Melton K, Lingren T, et al. Phenotyping for patient safety: algorithm development for electronic health record based automated adverse event and medical error detection in neonatal intensive care. J Am Med Inform Assoc. 2014;21:776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubberke ER, Nyazee HA, Yokoe DS, et al. Implementing automated surveillance for tracking clostridium difficile infection at multiple healthcare facilities. Infect Control Hosp Epidemiol. 2012;33:305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorberbaum T, Nasir M, Keiser MJ, et al. Systems pharmacology augments drug safety surveillance. Clin Pharmacol Ther. 2015;97:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peissig PL, Rasmussen LV, Berg RL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. 2012;19:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen-Torvik LJ, Pacheco JA, Wilke RA, et al. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin Transl Sci. 2012;5:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namjou B, Keddache M, Marsolo K, et al. EMR-linked GWAS study: investigation of variation landscape of loci for body mass index in children. Front Genet. 2013;4:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W-Q, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawatkar A, Chu L-H, Iyer R, et al. Development and validation of algorithms to identify acute diverticulitis [published online ahead of print September 25, 2014]. Pharmacoepidemiol Drug Saf. doi:10.1002/pds.3708. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie MD, Denny JC, Zuvich RL, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012;19:e162–e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc. 2011;2011:274–283. [PMC free article] [PubMed] [Google Scholar]
- 28.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie MD, Verma SS, Hall MA, et al. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol Vis. 2014;20:1281–1295. [PMC free article] [PubMed] [Google Scholar]
- 30.Yoni Halpern YC. Using anchors to estimate clinical state without labeled data. AMIA Annu Symp Proc 2014;2014:606–615. [PMC free article] [PubMed] [Google Scholar]
- 31.Savova GK, Fan J, Ye Z, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc. 2010;2010:722–726. [PMC free article] [PubMed] [Google Scholar]
- 32.Drupal. Drupal.https://www.drupal.org/. Accessed April 24 2015.
- 33.Fraternali P, Paolini P. Model-driven development of web applications: the AutoWeb system. ACM Trans Inf Syst. 2000;18:323–382. [Google Scholar]
- 34.KNIME. www.knime.org. Accessed December 28, 2015.
- 35.Muthalagu A, Pacheco JA, Aufox S, et al. A rigorous algorithm to detect and clean inaccurate adult height records within EHR systems. Appl Clin Inform. 2014;5:118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gawron AJ, Thompson WK, Keswani RN, et al. Anatomic and advanced adenoma detection rates as quality metrics determined via natural language processing. Am J Gastroenterol. 2014;109:1844–1849. [DOI] [PubMed] [Google Scholar]
- 37.Tromp G, Borthwick KM, Smelser DT, et al. Ephenotyping for abdominal aortic aneurysm in the electronic medical records and genomics (emerge) network: algorithm development and Konstanz Information Miner Workflow. Int J Biomed Data Min. 2015;4:113 http://www.omicsonline.com/open-access/ephenotyping-for-abdominal-aortic-aneurysm-in-the-electronic-medical-records-and-genomics-emerge-network-algorithm-development-andkonstanz-information-miner-workflow-2090-4924-1000113.php?aid=55667. Accessed July 31, 2015. [PMC free article] [PubMed] [Google Scholar]
- 38.Parikh R, Mathai A, Parikh S, et al. Understanding and using sensitivity, specificity and predictive values. Indian J Ophthalmol. 2008;56:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tryka KA, Hao L, Sturcke A, et al. NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen LV, Thompson WK, Pacheco JA, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms [published online ahead of print June 21, 2014]. J Biomed Inform. doi:10.1016/j.jbi.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Archer KR, Coronado RA, Haug CM, et al. A comparative effectiveness trial of postoperative management for lumbar spine surgery: changing behavior through physical therapy (CBPT) study protocol. BMC Musculoskelet Disord. 2014;15:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denny JC. Chapter 13: mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8:e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liaw S-T, Taggart J, Yu H, et al. Integrating electronic health record information to support integrated care: Practical application of ontologies to improve the accuracy of diabetes disease registers. J Biomed Inform. 2014;52:364–372. [DOI] [PubMed] [Google Scholar]
- 47.Roden DM, Xu H, Denny JC, et al. Electronic medical records as a tool in clinical pharmacology: opportunities and challenges. Clin Pharmacol Ther. 2012;91:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peissig PL, Santos Costa V, Caldwell MD, et al. Relational machine learning for electronic health record-driven phenotyping. J Biomed Inform. 2014;52:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. AMIA Annu Symp Proc. 2012;2012:911–920. [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar VD, Tipney HJ, editors. Mining the Electronic Health Record for Disease Knowledge - Springer. New York: Springer; 2014. http://link.springer.com/protocol/10.1007/978-1-4939-0709-0_15#page-1. Accessed June 5, 2015. [Google Scholar]