Abstract
Objective To help cancer registrars efficiently and accurately identify reportable cancer cases.
Material and Methods The Cancer Registry Control Panel (CRCP) was developed to detect mentions of reportable cancer cases using a pipeline built on the Unstructured Information Management Architecture – Asynchronous Scaleout (UIMA-AS) architecture containing the National Library of Medicine’s UIMA MetaMap annotator as well as a variety of rule-based UIMA annotators that primarily act to filter out concepts referring to nonreportable cancers. CRCP inspects pathology reports nightly to identify pathology records containing relevant cancer concepts and combines this with diagnosis codes from the Clinical Electronic Data Warehouse to identify candidate cancer patients using supervised machine learning. Cancer mentions are highlighted in all candidate clinical notes and then sorted in CRCP’s web interface for faster validation by cancer registrars.
Results CRCP achieved an accuracy of 0.872 and detected reportable cancer cases with a precision of 0.843 and a recall of 0.848. CRCP increases throughput by 22.6% over a baseline (manual review) pathology report inspection system while achieving a higher precision and recall. Depending on registrar time constraints, CRCP can increase recall to 0.939 at the expense of precision by incorporating a data source information feature.
Conclusion CRCP demonstrates accurate results when applying natural language processing features to the problem of detecting patients with cases of reportable cancer from clinical notes. We show that implementing only a portion of cancer reporting rules in the form of regular expressions is sufficient to increase the precision, recall, and speed of the detection of reportable cancer cases when combined with off-the-shelf information extraction software and machine learning.
Keywords: natural language processing, machine learning, information extraction, neoplasms, electronic health records, user-computer interface
Background and Significance
Most countries have government-mandated reporting requirements for a number of different diseases. In the United States, the National Cancer Registrars Association represents more than 5000 cancer registry professionals and Certified Tumor Registrars (CTRs),1 whose job is to identify patients with reportable cases of cancer and extract information from their medical records for government-mandated reporting. As one of 45 states participating in the National Program of Cancer Registries, Alabama collects cancer registrar reports on reportable cancer cases from all participating healthcare institutions and communicates them to the federal government. Teams of CTRs in each participating hospital discover and abstract the required data for each reportable cancer case, typically by manually reviewing clinical documentation. In aggregate, federally reported data on cancer cases are used to generate nation-wide cancer statistics, to educate the public, to better understand the epidemiology of cancer and cancer outcomes, as well as to inform public policy. Because of limited federal funding and an aging population with an increasing incidence of cancer, CTRs are increasingly hard-pressed to meet their reporting goals without clinical informatics tools to facilitate cancer case detection and reporting.
A recent review of text mining of cancer-related information2 revealed that the recognition of cancer entities in clinical data has focused on symbolic methods, primarily information extraction using dictionary-based lookup or regular expressions. The “overwhelming majority”2 of these approaches use MetaMap3,4 and the Unified Medical Language System (UMLS) to identify cancer entities in clinical data, and their results were dependent on the type of text and the cancer analyzed. However, most of this work did not use clinical text, and, when clinical text (including pathology reports) was used, it was for the purposes of information extraction,5–8 rather than for determining cancer cases’ status (reportable or nonreportable). There are a few exceptions, including studies of determining cancer status for pancreatic cancer9 and colorectal cancer10 from clinical text, but there are no examples in the literature of determining patients’ cancer status from clinical text for cancer as a general class to aid in government-mandated reporting. To meet this challenge and to increase the volume and accuracy of reported cancer cases at the University of Alabama at Birmingham (UAB), we developed the Cancer Registry Control Panel (CRCP). Our goals were to:
Automate cancer case detection by applying natural language processing (NLP) techniques and querying cancer-related International Classification of Diseases – 9 (ICD-9) codes (in a system version called “CRCP-DUAL”) and contrast the precision of this approach with that of manual review of pathology reports by CTRs.
Facilitate CTRs’ review of clinical documentation by highlighting cancer-related text as defined by multiple standardized terminologies.
Develop a machine-learning algorithm that can predict patients’ reportable cancer status based on ICD-9 codes and NLP-extracted information.
MATERIALS AND METHODS
Setting
The reported initiative was the result of the collaboration of the UAB Center for Clinical and Translational Science and the UAB Hospital CTR team within the Health Information Management program. The UAB Institutional Review Board (IRB) reviewed and approved this investigation (IRB protocol X121114001).
Architecture
CRCP is web application with a JavaScript/HTML front end and a middle tier based on Ruby on Rails with a MySQL database. The NLP infrastructure is based on Unstructured Information Management Architecture – Asynchronous Scaleout11 and uses the UMLS12 2013AB data, North American Association of Central Cancer Registries (NAACCR) search criteria,13 and Facility Ontology Registry Data Standards terms14 to identify reportable cancer cases and/or to highlight text of interest to the CTRs. Herein, we describe three iterations of CRCP:
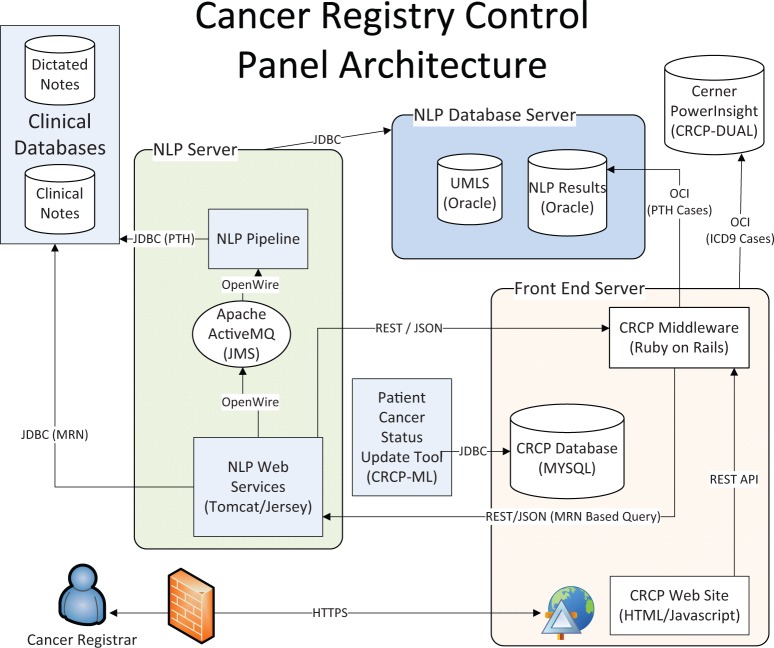
CRCP-ML: The current CRCP system, which utilizes machine learning through the Patient Cancer Status Update Tool to determine patients’ cancer status. Its architecture diagram is shown in Figure 1.
CRCP-DUAL (RANKED): This system predates CRCP-ML and lacks the Patient Cancer Status Update Tool. Data generated from the use of this system was used to train CRCP-ML. CRCP-DUAL identified suspect reportable cancer cases for a user-defined reporting period using ICD-9 billing codes or positive mentions of NLP cancer-related UMLS Concept Unique Identifiers (CUIs). Suspect cancer cases detected by both NLP and ICD-9 codes were first shown to the CTRs, then NLP-only cases, and finally ICD-9 code-only cases. An early pilot of CRCP-DUAL (UNRANKED) did not include this ranking and instead presented cases from oldest to newest. CRCP-DUAL (RANKED) is hereafter referred to as CRCP-DUAL.
CRCP-NLP: This was the original prototype of CRCP, which detected reportable cancer cases based on the presence of non-negated NLP cancer-related CUIs. It was used for a 2-week period, and its results were then compared with the pre-CRCP process in which pathology reports were manually reviewed. In addition, CRCP-NLP lacked the components to query Cerner PowerInsight™ for ICD-9 codes.
Figure 1:
CRCP architecture. CRCP-NLP and CRCP-DUAL lack the Patient Cancer Status Update Tool. CRCP-NLP also lacks the ability to query Cerner PowerInsight™ for ICD-9 codes.
Workflow
CTRs at UAB identify reportable cancer cases in two phases. In the first phase, they inspect all pathology reports produced since the previous workday for evidence of reportable cancer cases. Once a putative cancer case is found, the CTRs access UAB’s electronic medical record to validate the case and abstract reportable details to enter in Metriq™, a commercial product used for cancer case tracking and reporting. This product is distinct and downstream from our NLP pipeline, and its use does not affect the generalizability of the results described herein.
This clinical document-based manual review process was supplanted by CRCP, which allows CTRs to validate predicted reportable cancer patients. Predicted reportable cancer patients are identified nightly as those patients for whom either a cancer-related CUI (all CRCP versions) was identified in a pathology report or for whom a reportable cancer-associated ICD-9 code (CRCP-DUAL and CRCP-ML) was identified in any clinical document. Each night, a batch process sends the union of all ICD-9 code- and NLP-associated potential cancer cases (identified by medical record numbers) to the NLP infrastructure. All clinical documents for each input medical record number are marked with reportable cancer case annotations using the same NLP process that is used for pathology reports, to facilitate the CTR’s subsequent review of said documentation.
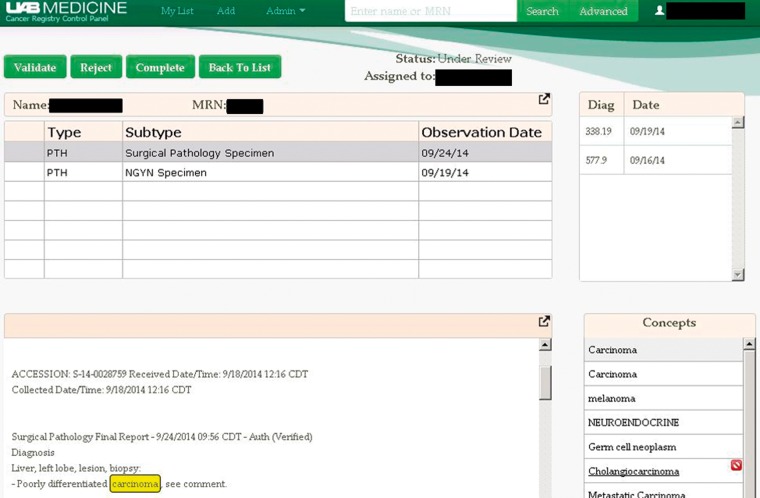
Putative cancer cases identified by the system and those validated by CTRs can be retrieved as needed (see Supplementary Figure 1) and then reviewed in the case review screen (shown in Figure 2). This screen is where the CTR can validate a putative cancer case, reject the case, or notify CRCP that the case has been completed, once it has been abstracted and entered into Metriq. This workflow is the same in all versions of CRCP.
Figure 2:
Screenshot of the CRCP worklist. Cancer-related concepts for the selected document are highlighted. All ICD-9 codes for the selected patient are displayed.
NLP Processing Pipeline
The initial identification of cancer concepts is done using NLM’s MetaMap Unstructured Information Management Architecture annotator and NAACCR term exact-matching, followed by a variety of custom annotators applied to exclude (or re-include) concepts from the initial concept list. Table 1 provides a description and motivation for each filter, and Supplementary File 1 contains the specific details necessary for reimplementation.
Table 1:
CRCP Filtering Rules
| Filter Description | Rationale |
|---|---|
| Document segmentation filter | Exclude reportable cancer cases found in document sections that are expected to generate false positives. |
| UMLS semantic-type filter | Include only MetaMap concepts with UMLS semantic-type neoplasms to filter out non-neoplastic cancers. |
| Positive patient assertion filter | Restrict concept to non-negated mentions that refer to the patient, not a family history of cancer. |
| CUI-specific MetaMap abbreviation filter | Exclude known MetaMap false positive cancer cases based on incorrect abbreviation mapping. |
| CUI-based reporting rule filter | Exclude nonreportable cancer cases that map to a list of 899 CUIs, which we developed. |
| Non-CUI reporting rule filter | Remove nonreportable cancer cases that cannot be filtered out using a pure CUI-based approach. For example, mentions of reportable cases containing “benign” or CUI concept text are filtered out, but are subsequently re-included if they are associated with the central nervous system, because such neoplasms are reportable. |
CUI, Concept Unique Identifier; UMLS, Unified Medical Language System.
The current CRCP processing pipeline does not distinguish between cancer cases that are reportable for UAB or reportable for another institution (but which have interacted with the UAB health system), and only identifies that the case is reportable. Finally, the number of rules applied in the CRCP system has steadily increased based on CTR feedback as the system has evolved. Consequently, CRCP-NLP had a much less extensive rule set than CRCP-DUAL, which underwent a number of revisions before becoming stable in October 2014. Since that point, the rules for CRCP-DUAL have remained unchanged and are identical to the rules for CRCP-ML.
CRCP-ML Machine Learning Algorithm Features and Implementation
The CRCP-ML algorithm was developed using the Mallet toolkit. In the algorithm, identifying cancer status (as reportable or nonreportable) was formulated as a multiclass patient classification problem, with each patient characterized by one or more of five types of features.
CUI name: The names of the concepts identified by our NLP pipeline for detecting mentions of reportable cancer cases.
ICD-9 codes (cancer): All reportable cancer case ICD-9 codes found in the patient data. The cancer-associated ICD-9 codes for this feature are identified by a regular expression based on NAACCR guidelines.
ICD-9 codes (any): Any ICD-9 code, whether cancer-related or not, in addition to the “ICD-9 codes (cancer)” reportable cancer-associated ICD-9 billing codes.
Prefix feature: Same as the “CUI name” feature, but prefixed by the document type and document subtype abbreviation; for example, “PTH: Surgical_Pathology_Specimen”.
TonsillarCarcinoma.” This feature was motivated by the idea that a mention of cancer appearing in one type of document, such as an outpatient clinic note, may be less indicative than such a mention appearing in another type of document, eg, a pathology report for a surgical pathology specimen.
Source: Specifies how the putative reportable cancer case was identified: by ICD-9 code, by NLP, or by both. This feature was the primary way that CRCP presented cases to the CTRs prior to the inclusion of machine learning; cases detected by both methods were first shown to the CTRs, then cases detected by NLP, and, finally, cases detected by ICD-9 codes.
We tested Mallet’s maximum entropy, naïve Bayes, and decision tree algorithms for classification, using an 80/20 training/test division, with parameters as shown in Supplementary File 2. We selected the maximum entropy model because it consistently gave the best precision, recall, and F-measure for detecting cancer.
Machine Learning Data
A gold standard that used a consistent (unchanging) set of rules was derived from CRCP-DUAL data on 2014 cancer cases, from October 2014 to May 2015. The cancer status of all the patients included in the gold standard was vetted by CTRs and represents the same data that UAB reports to the State of Alabama. The gold standard consists of 3087 cancer cases, categorized as follows: 292 detected only from NLP-identified CUIs in pathology reports, 185 detected only from ICD-9 codes, and 2610 detected from both NLP-identified CUIs and ICD-9 codes. This data, including noncancer cases and totals, is plotted on the primary Y-axis of Figure 3.
Figure 3:
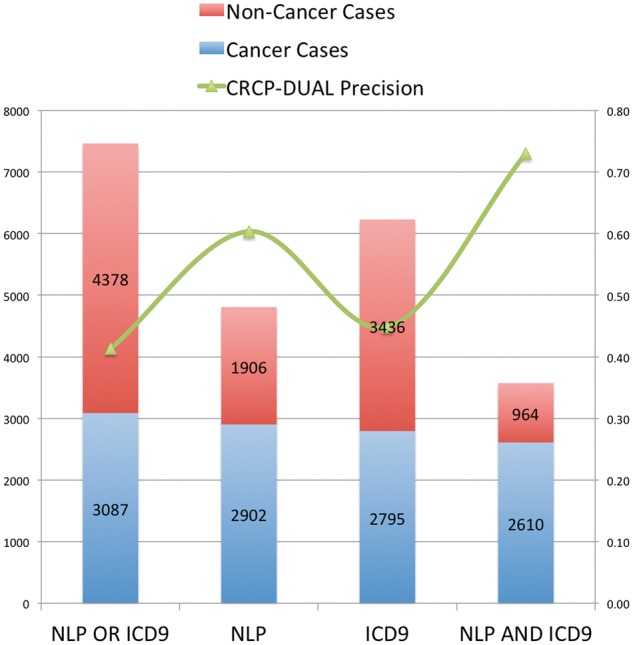
CRCP-DUAL dataset and precision results. The X-axis indicates the system type. Cancer case counts are plotted on the primary Y-axis, and the secondary Y-axis indicates the precision of CRCP-DUAL on that class of cases.
Analyses
We performed three sets of analyses.
Analysis 1. We prototyped the CRCP-NLP system to test the hypothesis that the first phase of the process of identifying reportable cancer cases (identifying potentially reportable cases) could be automated using off-the-shelf NLP components to identify mentions of cancer in clinical documents. We compared this system (which utilizes only NLP-processed pathology reports to detect reportable cancer cases) against the manual process of inspecting pathology reports that UAB used at the time. We divided CTRs into two groups: one that manually processed all cancer cases for a 2-week period, and one that processed the same cases using CRCP-NLP. Tests were performed to discover if there were differences in precision and recall between the manual review process and the CRCP-NLP process. In cases in which both proportions were based on the same patients, McNemar’s test for paired data was used. In cases in which the two proportions were based on some, but not all, of the same patients, a hybrid paired and unpaired approach was used.15
Analysis 2. We tested the hypothesis that the throughput of cancer CTRs (measured as the number of cases completed per month) could be increased by CRCP-DUAL when both NLP and ICD-9 codes are used for cancer case identification. CRCP-DUAL (not CRCP-ML) was selected as the system for the throughput comparison, because it was the only stable system in place long enough to generate sufficient data for such a comparison.
Analysis 3. Finally, we tested the hypothesis that the accuracy and F-measure of reportable cancer patient identification could be increased by including machine learning in the process and analyzed the errors of a system that included machine learning to reveal other potential avenues for performance increases.
The statistical analyses were performed using SAS software version 9.3 (SAS Institute Inc., Cary, NC, USA).
RESULTS
Analysis 1 – Comparison of Manual Review Process with CRCP-NLP Process
Table 2 shows the comparison of the previously used manual pathology report review process and the CRCP-NLP reportable cancer case detection process.
Table 2:
Manual Review of Pathology Reports vs CRCP-NLP Reportable Cancer Case Detection
| System | Patients entering process | CTR-reviewed patients | CTR-validated patients | CTR-rejected patients | Precisiona | Recallb |
|---|---|---|---|---|---|---|
| Manual review | 2125 | 2125 | 344 | 1781 | 0.162 | 0.586 |
| CRCP-NLP | 2382 | 1864 | 445 | 1419* | 0.239 | 0.621 |
CRCP-NLP, Cancer Registry Control Panel – natural language processing; CTR, Certified Tumor Registrar. *The CRCP-NLP process rejected an additional 518 patients (2382-1864) without CTR review, because those patients had no NLP-identified cancer mentions in any of their documents. aP < .0001. bP = .2342.
Table 2 shows that cancer cases presented to CTRs through the CRCP-NLP system vs the manual process were significantly (P < .0001) more likely to be regarded as reportable by the CTRs. CRCP-NLP also found more reportable cases than the manual review process (0.621 recall vs 0.586 recall) considering only cases that were visible to that system, a result that was not statistically significant (P = .2342). However, CRCP-NLP identified and reviewed 14% more pathology reports (corresponding to 12% more patients) than were reviewed in the manual process, because the manual review process eliminated some classes of pathology reports from the CTR review (they were never printed out), whereas CRCP-NLP processed all pathology reports. If this improved coverage provided by CRCP-NLP is considered, recall is instead 0.619 for CRCP and 0.522 for the manual review process and is statistically significant (P < .001).
CRCP-NLP also accurately eliminated patients whose records did not require human review. Of the 518 patients identified by CRCP-NLP as not requiring human review, 444 of these were also reviewed by the manual process. All of these cases were rejected, with the exception of two cases, one of which was a January 2012 case that had been missed by the manual review process earlier and another patient whose cancer was judged to be nonreportable at the time but who went on to develop a reportable cancer.
Another result of the CRCP-NLP system is that it eliminates duplicate work, including duplicate patient review and duplicate document review. In the manual review process, we observed that 20.2% of the CTR reviews were of duplicate patients, and 10.5% of CTR-reviewed reports were duplicate documents. The CRCP-NLP system eliminated duplicate patient reviews completely by orienting the review process at the patient level. The document-level focus of the manual review process vs the patient-level focus of CRCP makes calculating and comparing interannotator agreement problematic, but we did compute both a raw agreement rate of 0.894 at the patient level and a κ-score of 0.7055 between CTRs using CRCP-NLP and CTRs using the manual review process.
Analysis 2 – Precision and Case Throughput with CRCP-DUAL
Figure 3 plots reportable cancer case precision (the proportion of cases presented to the CTRs that were determined to be reportable cancers) for CRCP-DUAL for the dataset described in the Materials and Methods section on the secondary Y-axis. In CRCP-DUAL, cancer cases are sorted and presented to registrars such that cases with both ICD-9 code and NLP evidence (“dual” cases) are presented first, followed by those with only NLP evidence, and finally those cases with only ICD-9 code evidence. In practice, the cancer registry would use CRCP-DUAL to examine all “dual-detected” cases (precision: 73.03%), some or all of the NLP-only detected cases (precision: 23.66%), and, rarely, ICD-9 code-only cases (precision: 6.96%), depending on the registry team’s workload or the current case completion rate. This huge drop in precision for cases that lack both ICD-9 code- and NLP-based evidence indicated that the vast majority of reportable cancer cases (we estimate over 93%, based on the results for the “source only” algorithm, presented in Table 3) were detected from both ICD-9 code- and NLP-based evidence.
Table 3:
Reportable Cancer Case Detection Algorithm Test Results
| Algorithm | Number of features | Accuracy (%) | Cancer precision (%) | Cancer recall (%) | Cancer F-measure (%) | Training test skew (%) |
|---|---|---|---|---|---|---|
| Source only (∼CRCP-DUAL) | 3 | 79.92 | 68.96 | 93.87 | 79.51 | 0.86 |
| Prefix feature only | 1847 | 77.38 | 78.31 | 62.90 | 69.77 | 7.26 |
| CUI name only | 1307 | 82.87 | 87.60 | 68.39 | 76.81 | 3.02 |
| ICD-9 codes (cancer) only | 541 | 83.80 | 80.39 | 80.65 | 80.52 | −10.15 |
| ICD-9 codes (all) only | 4921 | 78.18 | 72.48 | 76.45 | 74.41 | 7.89 |
| CUI name and prefix feature | 3153 | 83.94 | 86.82 | 72.26 | 78.87 | 5.67 |
| CUI name and ICD-9 codes (cancer) | 1847 | 87.15 | 84.30 | 84.84 | 84.57 | 1.30 |
| CUI name and ICD-9 codes (all) | 6227 | 82.73 | 79.29 | 79.03 | 79.16 | 10.35 |
| Source, CUI name, and ICD-9 codes (cancer) | 1849 | 86.61 | 84.09 | 83.55 | 83.82 | 2.82 |
| Prefix feature, CUI name, and ICD-9 codes (cancer) | 3693 | 87.15 | 84.08 | 85.16 | 84.62 | 4.04 |
| Source, prefix feature, and ICD-9 codes (cancer) | 2389 | 85.41 | 82.32 | 82.58 | 82.45 | 3.86 |
| Source, prefix feature, CUI name, and ICD-9 codes (cancer) | 3695 | 87.02 | 85.86 | 82.26 | 84.02 | 4.63 |
| Source, prefix feature, CUI name, and ICD-9 codes (all) | 8075 | 83.27 | 82.01 | 76.45 | 79.13 | 11.70 |
CUI, Concept Unit Identifier; ICD-9, International Classification of Disease – 9. Bolded text indicates the feature set that was ultimately selected for implementation.
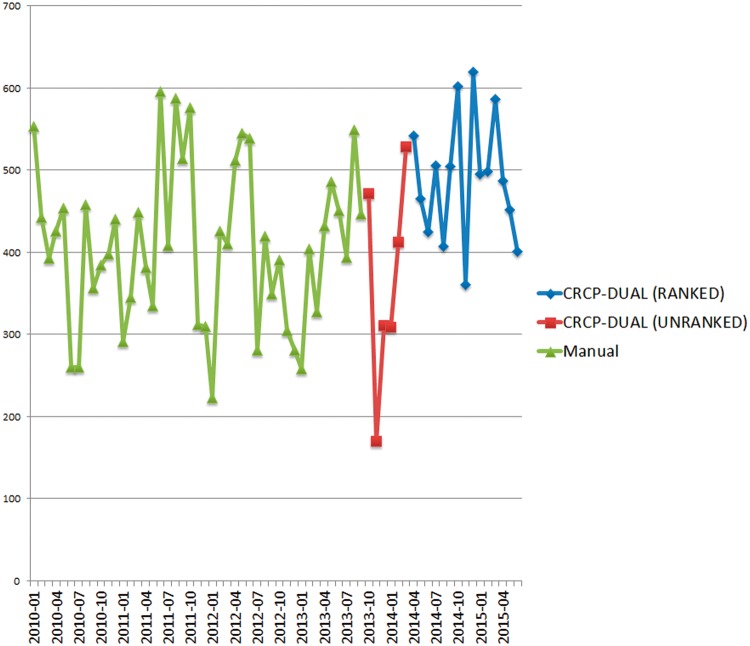
Figure 4 shows monthly reportable cancer case completion rates at UAB since 2010, based on Metriq data. Case completion rates when using CRCP-DUAL are 23.6% higher than those when using the manual review process, and even higher if the reduction in CTR staff that occurred during our study period (loss of 1 CTR) is taken into account. One source of this throughput increase is that 51.2% of incoming patients (from pathology reports) are triaged as “unreviewable,” because the NLP process has failed to identify any cancer concepts in the clinical documentation, thus significantly reducing the CTRs review burden.
Figure 4:
Monthly UAB cancer registry case completion rate. The Y-axis indicates the number of cases completed. and the X-axis shows the month of the cases’ completion. Each line represents a different yearly reporting period. CRCP-DUAL has been in use since April 2014.
Analysis 3 – Patient Classification Results with CRCP-ML
Table 3 shows the results for the test set data. The precision, recall, and F-measure are shown only for the cancer label, because the purpose of CRCP is to detect cancer cases. The “training test skew” was computed as the difference between the accuracy of the training set (data not shown) and the accuracy of the test set, and gives some indication of the relative stability and consistency of the algorithm. Generally, the results for the training set were superior, resulting in a positive skew, but some features yielded similar (source) or superior (ICD-9 codes) results for the test set. Ultimately, we selected the algorithm that used only cancer ICD-9 codes and CUI names as features as our new classifier for CRCP-ML. because of its higher precision and stability. CRCP-DUAL’s performance can be approximated from the “source only” results, because this classifier predicts reportable cancer cases when both sources of data (ICD-9 codes and NLP) are present.
Error Analysis
A total of 40 misclassifications (20 false positives and 20 false negatives) were randomly selected for further analysis. About half of the false positives were actually reportable cancer cases, but were not reportable by UAB and, thus, were still considered false positives. This is because CRCP (all versions) makes no effort to determine which institution a cancer is reportable for, only that it is reportable. The other false positives were generally the results of poor or no regular expression coverage of cancer disease attributes, as shown in Table 4.
Table 4:
Error Analysis
| Error class | Error type | Count |
|---|---|---|
| Patient history of cancer | False positive | 3 |
| Family history of cancer | False positive | 1 |
| Cancer not reportable by UAB – ICD-9 codes | False positive | 4 |
| Cancer not reportable by UAB – NLP + ICD-9 codes | False positive | 6 |
| Cancer at this body location not reportable | False positive | 2 |
| Uncertain language means not reportable | False positive | 2 |
| Negation of cancer mention not detected | False positive | 1 |
| Secondary tumor is not reportable | False positive | 1 |
| No NLP CUIs and coarse ICD-9 codes | False negative | 5 |
| Coarse NLP CUIs and no ICD-9 codes | False negative | 6 |
| Coarse NLP CUIs and coarse ICD-9 codes | False negative | 5 |
| NLP or machine learning failures | False negative | 4 |
CUI, Concept Unit Identifier; ICD-9, International Classification of Diseases – 9; NLP, natural language processing; UAB, University of Alabama at Birmingham.
The false negative errors generally fall into two categories. About half of the false negatives lacked either an ICD-9 billing code or an NLP hit. The remaining false negatives contained very few data points: 20% had only one distinct CUI and one distinct ICD-9 code, 15% had only three codes of any type, and the remaining 15% had more than three codes of any type, but they tend to be highly repetitive. This lack of information makes it difficult for the classifier to reach a decision for these more unreliable codes. A common theme for missed cancer cases are coarse concepts (whether CUIs or ICD-9 codes) that are sometimes, but not always, associated with a reportable cancer, such as adenocarcinoma, for which reporting is dependent on body location or with commonly misdiagnosed conditions, including a number of blood-related cancers. Another source of false negatives are patients whose clinical documentation includes CUIs corresponding to reportable cancer codes that were classified as “negative” by the cancer status algorithm. These codes were often associated with false positives, including codes for “cancer” or “carcinoma,” which can appear in many different contexts, or abbreviations such as “HCC” (for hepatocellular carcinoma).
Discussion
With CRCP-NLP, we show that extracting cancer-related CUIs from pathology reports helps CTRs improve both the precision and recall of identifying reportable cancer cases. Additionally, we show that a sufficiently useful set of reportable cancer codes can be defined by a combination of UMLS semantic types, specific CUIs, and a small set of filtering rules, as outlined in Table 1.
Our results from using CRCP-DUAL show that we can increase the precision and throughput of the process of identifying reportable cancer cases by including ICD-9 codes for reportable cancers in the process, provided that the list of codes is suitably refined. Although we originally tried to follow the Alabama State Cancer Registry guidelines for detecting cancer cases, we found that including ICD-9 codes such as 042 (for HIV) and similar general codes that pull in broadly cancer-related conditions resulted in too many false positives. Furthermore, including nonreportable cancer-specific ICD-9 codes in our machine-learning algorithm did not lead to a performance gain, as shown in Table 3 in the “ICD-9 codes (all)” columns.
One lesson learned from our deployment of CRCP-DUAL is that the system must match the CTRs’ workflow. The deployment of our prototype version of CRCP-DUAL (UNRANKED) resulted in the registrars being overwhelmed with old putative cancer cases, which subsequently resulted in a spike of cases from 2012 and earlier being submitted. Additionally, because the UNRANKED version of CRCP-DUAL was optimized for recall rather than precision, the registrars were frustrated by having to examine so many false positive cases, which resulted in a severe drop-off in tool usage that was not fixed until the deployment of the RANKED version of CRCP-DUAL. Based on this, we believe that optimizing the F-measure is more appropriate than optimizing recall. Working closely with CTRs, we were able to refine the system and achieve improved throughout despite a reduction in our hospital’s CTR workforce during the study period. Anecdotally, the CTRs were very pleased with the highlighting of cancer concepts in all clinical documentation. This was done to focus their review, targeting their extraction of information for federally mandated reporting of management and outcomes data for each case within Metriq. As a whole, the quantifiable impact of CRCP-DUAL on efficiency and the enthusiastic adoption of this approach by CTRs highlights its potential to improve federally mandated cancer case reporting using NLP.
With CRCP-ML, we show that including machine learning in the process of identifying reportable cancer cases could increase the accuracy of case detection as well as improve the F-measure and precision of cancer case identification. However, our simple “source only” feature gave a recall of 93.87%, indicating that, currently, no machine learning is needed in the CRCP process if the goal is to maximize cancer case recall. With CRCP-ML, we are currently experimenting with Mallet confidence scores to discover and/or order cancer cases for registration consumption. Supplementary Figure 2 shows the distribution of Mallet confidence scores for CRCP-ML; at a confidence of 50, about half of the cases are expected to be reportable cancer cases. Currently, these scores determine the order of the presentation of new cases for a given monthly window in CRCP-ML, with cases with the highest confidence level presented first.
Limitations
We only directly compared CRCP-NLP to the original manual review process, due to resource constraints. However, because we know that CRCP-DUAL and CRCP-ML outperform CRCP-NLP in metrics such as precision and recall, it is reasonable to expect that both CRCP-DUAL and CRCP-ML will perform as well as or significantly better than the manual review process. We rely on proxy data, such as monthly state case submissions, to provide an estimate. Unfortunately, yearly changes in coding practices as well as multiple changes in registrar personnel and leadership make comparisons difficult. However, because overall registry staff levels have generally been reduced in the time that CRCP-DUAL has been in use vs the comparative period when the manual review process was used, our actual throughput increase could be as high as 43%.
Because CRCP was deployed in the United States, we use ICD-9 codes for billing data, potentially making CRCP less useful internationally. However, we have already showed that CRCP-NLP alone can outperform a manual pathology report-based review process, and replacing ICD-9 codes with ICD-10 codes should be feasible, because registrars submit cases 4–6 months after they become reportable, leaving sufficient time to generate training data for ICD-10 codes or other coding systems used internationally as needed.
The calculation of interannotator agreement is also problematic for our tool, because the manual review process validates cancer cases at the document level, whereas CRCP validates cancer cases at the patient level. Thus, the manual review process may flag different documents from the same patient as indicating reportable or nonreportable cancers. This does not necessarily reflect errors in annotator agreement, just that only a portion of a patient’s documents may indicate a reportable cancer. Our reported agreement rate and κ-score are therefore estimated from the reportable cancer status of the first document (ie, whether or not that document indicates a reportable cancer case) reviewed for the patient; actual agreement is expected to be higher. Indeed, one of the strengths of our approach is the ability of the tool to present and summarize reportable cancer cases at the patient level (rather than the document level).
Finally, CRCP uses rules for document segmentation generated solely from UAB training data. Thus, the reliability of our set of regular expressions is expected to be reduced at other institutions to the extent that provider and sectioning practices differ.
CONCLUSION
The key contribution of this manuscript is to take technology and approaches from one discipline and apply them to cancer detection, a manual, time consuming, and costly process that is a federally mandated requirement for all health systems in the United States. Our focus was not on developing new NLP technologies, but rather bringing the strengths of these technologies to bear on a real-world clinical problem. We feel that this is a significant contribution towards bringing NLP tools to common use cases that have been neglected to date.
We show herein that, despite the variety and complexity of cancer, it is possible to use NLP and machine learning to accurately detect patients’ reportable cancer status. This can be achieved despite the fact that not all the rules for reportable cancer cases are currently implemented in CRCP, indicating that a basic set of rules can cover the vast majority of cancer cases. Additionally, we show that an NLP analysis of pathology reports without ICD-9 billing codes is sufficient to help cancer registrars identify cancer cases, although the tool’s performance is improved with the addition of both ICD-9 codes and machine learning. It is important to note that although it seems intuitive that a reportable cancer detection system should focus on recall, the reality is that cancer registrars are under pressure to find cases quickly, and architects designing such systems should aim to strike a balance between precision and recall if they want to maximize reportable caner case throughput.
CONTRIBUTORS
All authors contributed significant draft revisions and analyzed results. G.G. took the lead in paper conception, J.W. in data interpretation, M.W. in registrar data acquisition, A.O.W. in statistical analysis, and S.B. in machine learning. J.D.O. wrote the initial draft of the paper, implemented the NLP and machine learning portions of CRCP, and analyzed all the data. We acknowledge Randy Paries for the front-end development of CRCP, Raghu Mannam for the development of the Ruby on Rails middleware component, and all UAB cancer registrars for providing critical feedback.
FUNDING
The research reported in this paper was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR00165. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
COMPETING INTERESTS
None of the authors has a financial interest in CRCP, but we have discussed commercialization and licensing possibilities.
REFERENCES
- 1.National Cancer Registrars Association. Become a Cancer Registrar. 2014. http://www.ncra-usa.org/files/public/BecomeaCancerRegistrar2014%29.pdf. Accessed May 1, 2015.
- 2.Spasić I, Livsey J, Keane JA, et al. Text mining of cancer-related information: review of current status and future directions. Int J Med Inf. 2014;83:605–623. [DOI] [PubMed] [Google Scholar]
- 3.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Proceedings of the AMIA Symposium. American Medical Informatics Association 2001; 2001: 17–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a disease knowledge representation model. J Biomed Inform. 2009;42:937–949. [DOI] [PubMed] [Google Scholar]
- 6.Napolitano G, Fox C, Middleton R, et al. Pattern-based information extraction from pathology reports for cancer registration. Cancer Causes Control. 2010;21:1887–1894. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen AN, Lawley MJ, Hansen DP, et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc. 2010;17:440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Anderson K, Grann VR, et al. Facilitating cancer research using natural language processing of pathology reports. Medinfo. 2004;11:565–572. [PubMed] [Google Scholar]
- 9.Friedlin J, Overhage M, Al-Haddad MA, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. In: AMIA Annual Symposium Proceedings. 2010. American Medical Informatics Association; 2010: 237–241. [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. In: AMIA Annual Symposium Proceedings 2011. American Medical Informatics Association; 2011: 1564–1572. [PMC free article] [PubMed] [Google Scholar]
- 11.The Apache Software Foundation. Getting Started: Apache UIMA Asynchronous Scaleout. 2013. http://incubator.apache.org/uima/doc-uimaas-what.html. Accessed September 10, 2015.
- 12.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North American Association of Cancer Registries. Search Terms List for Screening Pathology Reports. 2010. http://www.naaccr.org/LinkClick.aspx?fileticket=3by-8n_JswA%3d&tabid=128&mid=468. Accessed September 10, 2015.
- 14.The American College of Surgeons. Facility Oncology Registry Data Standards. Secondary Facility Oncology Registry Data Standards. 2015. https://www.facs.org/∼/media/files/quality%20programs/cancer/coc/fords/fords%202015.ashx. Accessed September 10, 2015.
- 15.Bland JM. Comparing proportions in overlapping samples. Undated. https://www-users.york.ac.uk/∼mb55/overlap.pdf. Accessed November 15, 2015. [Google Scholar]