Abstract
Objective Traditionally, patient groups with a phenotype are selected through rule-based definitions whose creation and validation are time-consuming. Machine learning approaches to electronic phenotyping are limited by the paucity of labeled training datasets. We demonstrate the feasibility of utilizing semi-automatically labeled training sets to create phenotype models via machine learning, using a comprehensive representation of the patient medical record.
Methods We use a list of keywords specific to the phenotype of interest to generate noisy labeled training data. We train L1 penalized logistic regression models for a chronic and an acute disease and evaluate the performance of the models against a gold standard.
Results Our models for Type 2 diabetes mellitus and myocardial infarction achieve precision and accuracy of 0.90, 0.89, and 0.86, 0.89, respectively. Local implementations of the previously validated rule-based definitions for Type 2 diabetes mellitus and myocardial infarction achieve precision and accuracy of 0.96, 0.92 and 0.84, 0.87, respectively.
We have demonstrated feasibility of learning phenotype models using imperfectly labeled data for a chronic and acute phenotype. Further research in feature engineering and in specification of the keyword list can improve the performance of the models and the scalability of the approach.
Conclusions Our method provides an alternative to manual labeling for creating training sets for statistical models of phenotypes. Such an approach can accelerate research with large observational healthcare datasets and may also be used to create local phenotype models.
Keywords: Electronic health record, phenotyping, noisy labels, machine learning, high throughput
Introduction
Electronic health records (EHRs) have the potential to catalyze clinical research. 1–4 One of the first steps in using EHR data for research is to reliably identify a cohort of patients that have a condition of interest or a phenotype. Algorithms to search an EHR database for phenotypes can accelerate research, 5–7 and lead to new clinical discoveries. 8–11 Typically, methods for identifying patients with a given phenotype have relied on rule-based definitions, 12,13 which is time consuming, 14,15 Given the heterogeneity of the data models in use, missing data values, 16 and differences in standardization, 17 in commercial EHR systems such rule-based definitions are difficult to port across different EHR systems and institutions. Clinical phenotype descriptions that work across clinical data warehouses represent one of the key bottlenecks in clinical research. 18,19
Recently, statistical learning approaches have been employed for electronic phenotyping. Carrell et al. 20 have demonstrated the effectiveness of natural language processing based approaches in automatically reviewing the charts of breast cancer patients with a high degree of accuracy. 20 Carroll et al. 21 have demonstrated that rheumatoid arthritis regression models trained on labeled data can describe the phenotype and achieve high classifier performance with an area under the curve of 92–97%. 21 Liao et al. 19 have demonstrated a regression model for rheumatoid arthritis with a positive predictive value (PPV) of 94%. 19 Several studies, 19,20,22,23 advance the view that phenotype models using a diverse feature set, perform better than ones based on a single type of feature (such as diagnosis codes or medications). 24,25 Chen et al. 26 have shown that active learning techniques for rheumatoid arthritis, colorectal cancer, and venous thromboembolism outperform passive learning methods and achieve good generalizability. 26 As shown by Sinnott et al., 27 probabilistic modeling of phenotypes improves the statistical power of the genotype-phenotype association in genetic studies that utilize the EHR. There is general agreement that the rate-limiting step in the compilation of cohorts for clinical research is the generation of clinical phenotype descriptions, 3 and that manual creation of training sets for machine learning approaches is time intensive. 26,28
We demonstrate that by using semi-automatically assigned and possibly noisy labels in training data, we can build phenotype models that perform comparably to the rule-based phenotype definitions, and can be developed faster. In this context, noisy labels refer class labels that are wrong with a small probability, characterized by a labeling error rate, due to an imperfect labeling process. The assumption behind our work is that the large volume of training data which can be collected using an automated labeling process, can compensate for the inaccuracy in the labels. The basis of our assumption lies in the theory of noise-tolerant learning. 29,30 wherein by imposing a bound on the labeling error, and by using a sufficient number of training samples, models trained from very large data sets with noisy labels can be as good as those trained from data sets with clean labels. If successful, the use of such noise-tolerant learning may allow statistical phenotyping approaches to scale to hundreds of phenotypes.
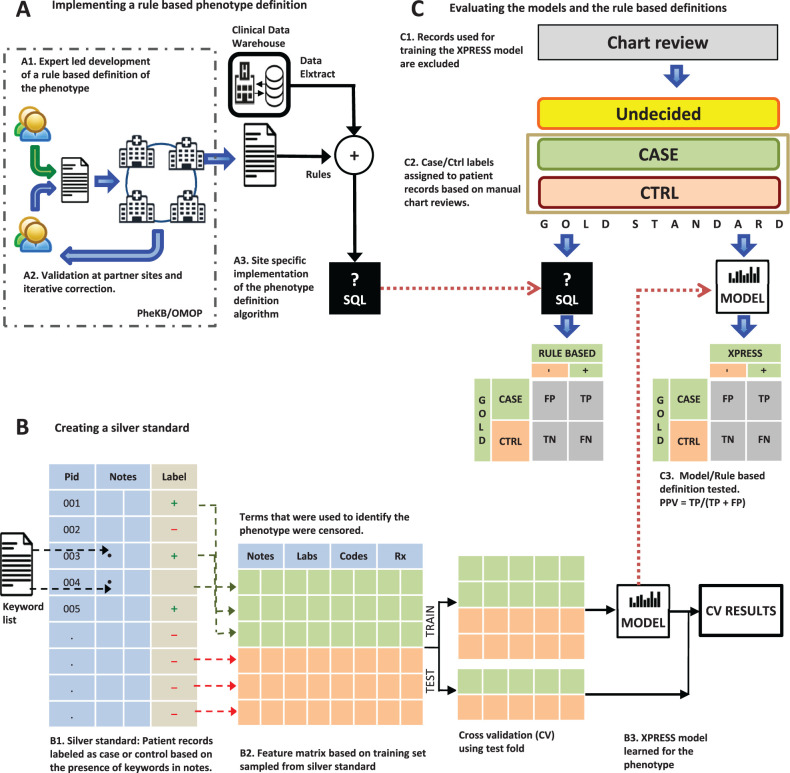
Using a machine learning technique capable of handling large feature sets, and semi-automatically assigned phenotype labels, we demonstrate the feasibility of rapidly learning phenotype definitions. We evaluate the statistically learned phenotype models of two diseases using a “gold standard” of manually reviewed patient charts. We discuss the relative importance of two key steps – label assignment and feature engineering – of our method. We also discuss the importance of the performance-effort tradeoff in alternative approaches for EHR-based phenotyping and the limitations of our work. Figure 1 illustrates our approach and overall workflow.
Figure 1:
Evaluating the performance of statistical models learned from semi-automatically labeled data with noisy labels ( A ) Existing rule-based phenotype definitions for the phenotypes are implemented using SQL. ( B ) Using a list of phenotype specific keywords, patient records are labeled has having or not having the phenotype; thus creating a noisy labeled training dataset. Features are constructed based on terms in notes, diagnostic codes, prescription, and lab orders. Keywords used in the noisy labeling are excluded. The data matrix is split into training and test sets for training a statistical model and for carrying out 5-fold cross-validation. ( C ) A manually reviewed gold standard set of patient records is created (excluding those used for training the model) and is used to evaluate both the rule-based definition and the statistical model for each phenotype.
Methods
Data
Phenotypes studied
We selected phenotypes for which rule-based definitions have been published by the Electronic Medical Records and Genomics (eMERGE), 14 and the Observational Medical Outcomes Partnership (OMOP) initiatives. 31 In eMERGE, a rule-based definition is developed by one of the partner institutions and is provided in the form of pseudo-code for site-specific implementation. After initial validation by the developers of the rule-based definition, other partner institutions implement and validate it at their respective sites via a manual chart review. The final definition is produced through iterative revisions and validations. The OMOP initiative defines phenotypes or health outcomes of interest (HOI), by systematically reviewing published literature on diagnostic criteria, coding guidelines, operational definitions and validation studies for phenotypes. Then they implement queries for applying the HOI definitions to an observational database, validate their results, and publish best practices for the HOI definition. From the 30 definitions published by eMERGE and the 34 definitions published by OMOP ( supplementary information S2 ), we chose Type 2 Diabetes Mellitus (T2DM) from eMERGE and Myocardial Infarction (MI) from the OMOP set, as examples of chronic and acute conditions, respectively.
Patient data
The patient dataset was extracted from the Stanford clinical data warehouseSCDW, which integrates data from Stanford Children’s Health and Stanford Health Care. The extract comprises 1.2 million patients, with 20.7 million encounters, 35 million coded diagnoses and procedures, 130 million laboratory tests, 14 million medication orders as well as pathology, radiology, and transcription reports totaling over 20 million clinical notes.
Our extract of patient data from January 1994 through June 2013 from Stanford Children’s Health and Stanford Health Care is stored in a structured and indexed form within a MySQL relational database. The pre-processing steps as well as details of the schema for the clinical data elements in the extracted data are shown in supplementary information S3 .
Methods
Implementing rule-based definitions
To compare the performance of the published rule-based definitions with that of the corresponding machine learned models, as illustrated in Figure 1 A, we implemented the rule-based definitions for T2DM and MI on our extract of the patient data. Implementing the rule-based definitions requires (a) mapping the definition variables to the respective clinical data elements and (b) writing the corresponding SQL queries.
Creation of a noisy labeled training set (silver standard) for statistical phenotype models
We labeled patients with and without the phenotype using semi-automatic labeling based on the presence or absence of highly specific phrases for the respective phenotypes. The choice of these phrases is the same as picking the “anchor” terms as described by Halpern et al. 32 The assumption is that if a patient exhibits the phenotype of interest then a doctor is likely to mention it in their notes, and that if a highly specific phrase is found the patient is likely to have the phenotype. For example, a phrase such as T2DM, in patient notes without a flag for negation or family history is taken to be a noisy label for the T2DM phenotype. An absence of T2DM specific keywords anywhere in the record is taken to indicate a “control.”
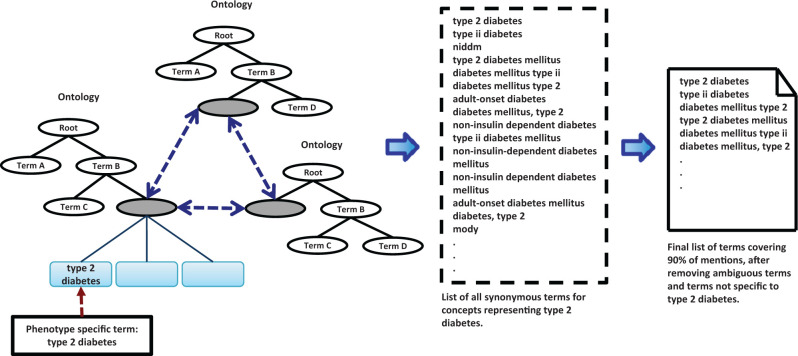
The goal of selecting keywords (shown in Figure 2 ) to assign phenotype labels, is to use a highly specific phrase for a given phenotype and identify its synonymous strings using 22 clinically relevant ontologies from the Unified Medical Language System (UMLS) and BioPortal. 34 To reduce the labeling error rate, the list of keywords is reviewed to identify high-frequency terms that are ambiguous. We sort the list of keywords by the number of patients in whose records they appear. Terms that collectively comprise 90% of the total number of patient counts are reviewed to remove ambiguous terms. For example, synonyms of “T2DM” include terms such as “MODY.” However, current medical practice makes a distinction between m ature o nset d iabetes of the y oung (MODY) and T2DM. Therefore, removing “MODY” from the keyword list is necessary for achieving search specificity.
Figure 2:
Construction of the list of keywords used to assign noisy labels. First, a list of synonymous terms for concepts representing the descriptive phrase for the phenotype is generated. The list is sorted by frequency of mentions and the terms covering 90% of the mentions are inspected to remove terms that are ambiguous or not specific to the phenotype of interest.
All patient notes in our data extract are pre-indexed with terms from the relevant ontologies in UMLS and BioPortal used in our previously published text-processing workflow (see Supplementary information S3 for details). 33 This keyword-search, which can be done in milliseconds, is cognizant of negation and family history contexts to avoid misattributions due to a term being mentioned in the family history or a rule-out diagnoses.
Labeling of the training dataset is semi-automatic because the keyword-list preparation requires manual intervention and some clinical expertise. Once the keyword list is prepared, the labeling of patients ( Figure 1 B) is done automatically. With this approach, we identified 32 581 possible cases for T2DM and 36 858 possible cases for MI; from which, we randomly sampled 750 patient records for each phenotype. We refer to these records as a “silver standard” set, having a “noisy” label for the phenotype of interest. For each phenotype, we constructed a random sample of 750 controls taken from all other patients in our extract disjoint with possible cases. We used this set of 1500 patient records to train a statistical model – referred to as a XPRESS model, for e X traction of P henotypes from R ecords using S ilver S tandards. 34
Creation of a clinician-reviewed evaluation set (gold standard)
We conducted an IRB-approved review of patient charts with the help of five clinicians to create an evaluation set for each phenotype consisting of clinician-labeled cases (patients with the phenotype) and controls (patients without the phenotype); see Figure 1 C. Each record was reviewed by two clinicians and ties were resolved on the basis of a majority vote after review by a third clinician. The clinicians reviewing the charts could label a record as “undecided” if, based on the chart contents, they were unsure about assigning a case or control label (see supplementary information S4 for protocol details). Patient records in the training data for the XPRESS models are disjoint from this evaluation set, which contains an equal number of cases and controls.
Building XPRESS models
Feature engineering
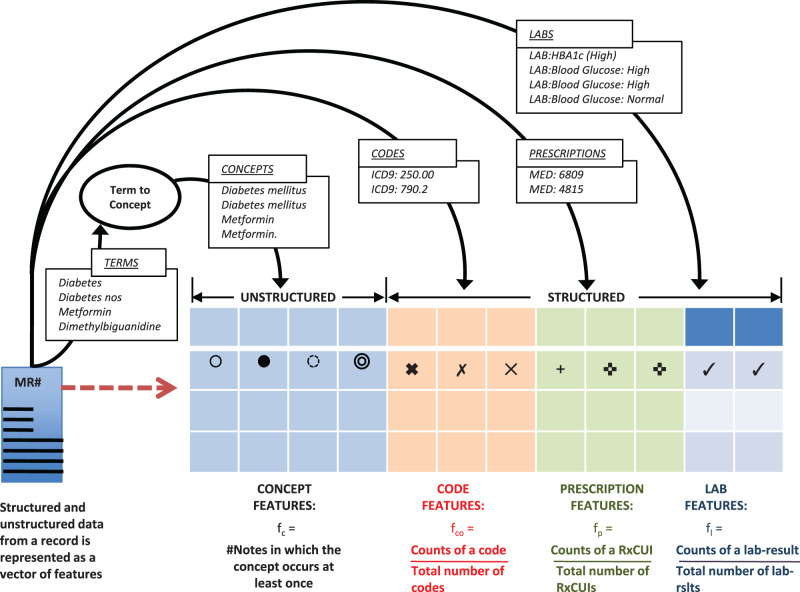
As shown in Figure 3 , we represented the structured and unstructured data from a patient record as features from four categories – terms (or concepts), prescriptions, laboratory test results, and diagnosis codes. Prescriptions, laboratory test results, and diagnosis codes were taken from the structured record whereas terms were extracted from free text (the section “Steps in processing clinical text” in supplementary information S3 , provides a description of the extraction method). We normalize terms into concepts in the same manner as in our earlier studies involving text mining on clinical notes – essentially using UMLS term-to-concept maps with suppression rules to weed out ambiguous mappings as described in Jung et al. 33 (see supplementary information S3 ). Such mapping reduces the total number of features as well as reduces the number of correlated features since synonyms get mapped to the same concept. For a concept, we used the number of distinct notes in which the concept occurs at least once (note frequency) as the feature representation. For prescriptions and diagnostic codes, we used the normalized counts of the active ingredient for each medication (RxNorm concept unique identifier) and the normalized counts of each International Classification of Diseases, revision 9 code as the respective features. For laboratory test results, we utilized the categorical result status for each ordered test (high/ normal/low or normal/abnormal as recorded in the EHR) and calculated a feature based on normalized counts for each test-result instance in the record. The number of features obtained was 23 717 (MI), and 25 045 (T2DM).
Figure 3:
Engineering features from structured and unstructured data elements in a patient record.
Learning statistical models from noisy labeled data
We defined the timestamp of the first note in the record that contains one of the keywords for a given phenotype as “time zero” and only extracted features from the record after that time. Our reason for using only the part of the record that follows time zero is that features occurring after the first mention of the phenotype related keywords are likely to characterize that phenotype. In contrast, if we wanted to create a model for predicting the onset of the phenotype, the portion of the record preceding time zero would be used.
Given the large number of features, we use a shrinkage method for learning from a sparse data set. By enforcing a penalty for the feature coefficients to be non-zero (i.e., “shrinking” them towards zero), such methods provide built in feature-selection. The feature coefficients indicate the relative importance of the features. We trained a L1 penalized logistic regression model for each phenotype using 5-fold cross-validation (CV). Under a constraint on the L1 norm of the coefficient vector, minimizing the negative log likelihood function of the model coefficients results in shrinking many of the coefficients to zero. 35 Standard implementations of L1 penalized logistic regression, 36,37 provide functions for selecting the optimal penalty. Computations were done on a system with 16 cores, 170 GB RAM. Training a XPRESS model for each of phenotype took 2–3 h.
Evaluation against a clinician-reviewed evaluation set
We applied the XPRESS model to each patient record in our evaluation data set to assign a case or control label and computed the precision and accuracy by comparing with the clinician assigned label. We also estimated the error rate of the noisy labeling process itself (i.e., the query used to define the “silver standard”) as illustrated in Figure 1 C. Finally, for reference, we assessed the performance of the rule-based phenotype definitions implemented on our data extract. The research was done under protocol numbers 24 883 (Expedited review, category 5) and 30 891 (Chart review), which were approved by Stanford University’s Institutional Review Board.
Results
Model performance
We first evaluated the models using 5-fold CV and examined the relative importance of the predictor variables ascertained by their respective coefficients. The mean precision and accuracy in 5-fold CV for T2DM were 0.86 and 0.84 and for MI—0.88 and 0.87, respectively. The top 10 features for the phenotype models are shown in Table 1 .
Table 1:
The top 10 features identified by the models for MI and T2DM
| MI | T2DM | ||
|---|---|---|---|
| Feature | Weight | Feature | Weight |
| cid:infarction | 0.0208 | cid: diabetes mellitus | 0.1031 |
| cid:onset of illness | 0.0101 | cid: metformin | 0.0455 |
| cid:cardiac arrhythmia | 0.0098 | Lab: GLUCOSE BY METER: high | 0.0332 |
| cid:coronary artery bypass surgery | 0.0085 | Lab: GLUCOSE, SER/PLAS: high | 0.0262 |
| cid:lasix | 0.0079 | cid: absence of sensation | 0.0184 |
| cid:bypass | 0.0067 | cid: edema | 0.0015 |
| cid:cerebrovascular accident | 0.0061 | cid: history of previous events | 0.0011 |
| cid:lupus erythematosus | 0.0058 | cid: mass of body structure | 0.0010 |
| lab:CK, MB (MASS):normal | 0.0051 | cid: skin appearance (normal) | 0.0008 |
| code:414.01(Coronary atherosclerosis of native coronary artery) | 0.0049 | cid: follow-up status | 0.0007 |
Performance assessment using a clinician-reviewed evaluation set
The results in Table 2 show the accuracy and PPV of the XPRESS models, using the clinician-reviewed evaluation set. We also present the accuracy and PPV of the rule-based definitions as well as the noisy labeling processes on the same gold standard. Each rule-based definition (T2DM and MI) took approximately one day to implement on our patient data extract. In the case of T2DM, our estimate of the performance of the rule-based definition is in agreement with the result published on PheKB, 18 by the eMERGE initiative, 14 which gives us confidence in the correctness of our local implementation of the rule-based definitions.
Table 2:
Performance assessed using a manually reviewed evaluation set for T2DM and MI
| T2DM | Cases | Ctrls | Acc | PPV |
|---|---|---|---|---|
| PheKB definition | 152 | 152 | 0.92 | 0.96 |
| Noisy labeling process | 152 | 152 | 0.89 | 0.81 |
| XPRESS | 152 | 152 | 0.89 | 0.90 |
| MI | Cases | Ctrls | Acc | PPV |
| OMOP definition | 94 | 94 | 0.87 | 0.84 |
| Noisy labeling process | 94 | 94 | 0.85 | 0.80 |
| XPRESS | 94 | 94 | 0.89 | 0.86 |
Numbers for “PheKB definition” show the performance of the rule-based definitions for identifying T2DM cases (authored by Vanderbilt University) applied to our database.
Numbers for “OMOP definition” show the performance of the rule-based definitions for identifying MI cases (broad definition with hospitalization) applied to our database.
Numbers in the “Noisy labeling process” row show the performance of the keywords based label assignment.
Numbers against “XPRESS” show the performance of the XPRESS models for identifying cases.
Learning good models with noisy labels
Noisy labels may be thought of as the output a procedure that returns a flipped (ie, wrong) label with a certain probability. Assuming a random classification noise model, the probability of label flipping is characterized by the classification error rate (τ). From the accuracy of the noisy labeling process reported in Table 2 , the classification error rate (τ) is estimated as 1 – Acc. In order for model-building using noisy labels to be feasible, we need the generalization error (defined as the probability that a model misclassifies a new observation drawn from the population) of an XPRESS model to approximate the generalization error of a model that best fits the data distribution. Simon, 29 and later Aslam et al., 30 formulate this guarantee as a sample complexity bound that is given as follows. If we define
D as the target data distribution consisting of observations and correct labels
D n as the data distribution consisting of observations and noisy labels
τ as the random classification error for D n
H as the class of learning algorithms to which our models belong
S as the set of m observations drawn from D n
as a model in H and trained on S
h * as a model in H that best fits the target distribution D
ε( ) as the generalization error of
ε( h *) as the generalization error of h *
Then for |ε( ) - ε( h *)| γ, with probability 1 - δ, it suffices that , where γ > 0 and 0
The case τ = 0 corresponds to observation data with clean labels and the case τ = 0.5 represents the case when the random flipping of labels makes learning impossible. For a given error bound γ, probability 1 − δ, and classification error rate τ, a learning algorithm can learn equally well from approximately m observations of noisy data what it can learn from m observations of clean data.
Assuming a 5% bound on the generalization error with a probability of at least 0.95, Table 3 shows the minimum sample size estimates m needed for our phenotype models.
Table 3:
Minimum sample size estimates for = 0.05 and = 0.05
| MI | T2DM | |||
|---|---|---|---|---|
| τ | m min | τ | m min | |
| XPRESS | 0.15 | 4135 | 0.11 | 3330 |
| Clean Labels | 0 | 2026 | 0 | 2026 |
At a 10% classification error rate in the noisy data, 50% more noisy observations are needed as compared to clean data and at 15% classification error rate, twice as many are required. As the error rate rises, the number of additional observations required increases and approaches infinity as τ goes to 0.5. Put another way, we can learn models with the same performance (PPV, Acc) from 2026 manually labeled, zero-error training samples, or from 4135 noisy labeled training samples. For XPRESS models, the cost of acquiring additional observations is negligible; creating 2026 manually labeled samples with zero error is difficult.
Discussion
Statistical phenotype models can provide high precision and recall but require the expert assignment of phenotype labels to patient records. 28 This expert-labeling requirement limits the use of such statistical learning approaches. Our work demonstrates the feasibility of learning phenotype models using noisy labeled training data for T2DM and MI, with performance evaluations based on clinician-reviewed patient records. Our noisy-labeling strategy achieves a labeling error rate of ≤15% for the two phenotypes. The resulting performance of our XPRESS models (PPV of 0.90 and 0.86 for T2DM and MI, respectively) compares favorably with the performance of the respective rule-based definitions (0.96 for T2DM and 0.84 for MI), evaluated using the same gold standard set of patients. The presence of well-known phenotype attributes in the top ranked features selected by the XPRESS models indicates that the performance is on account of learning generalizable features. We have also examined the trade-offs in a noisy labeling approach given the error-rate of the labeling process and the number of training instances required at a given error-rate. Understanding this trade-off is necessary for constructing phenotype models using noisy-labeled data. We also note the study by Yu et al., 38 who used manually selected concepts specific to a phenotype as their search criterion for labeling training instances. Their classifiers for coronary artery disease and for rheumatoid arthritis showed a PPV of 0.903 and 0.795 (false positive rate = 0.05). Their approach and results lend support to the argument that it is possible to learn good phenotype models from labeled datasets created via simple labeling techniques.
The development of a rule-based definition requires a rigorous validation, 14 and data fragmentation across multiple care facilities adversely impacts the performance of such phenotyping algorithms. 39 In the case of complex phenotypes, practices related to diagnoses, prescriptions, testing, and laboratory thresholds may vary across institutions. 15 Since XPRESS learns the phenotype definition in terms of a set of feature weights, agreeing on a shared feature space (such as the Systematized Nomenclature of Medicine, or Logical Observation Identifiers Names and Codes) across EHR systems provides a simple way to integrate patient and practice information across multiple source schemas. Thus, sharing an XPRESS workflow can be a potential alternative solution to the problem of porting a clinical phenotype description across different institutions and EHR systems.
Our results in Table 2 show that the models achieve nearly the same (T2DM) or better (MI) accuracy and precision compared to the rule-based definitions. As evident in the case of T2DM, a rule-based definition does provide some performance gain over XRESS, but would need significant development time. There are certainly situations that require higher precision. For such cases using a rule-based definition may be the only choice. However, for use cases that can tolerate a lower precision in phenotyping (eg, medical device surveillance, 40 and quality measurement, 41 ), the use of noisy labeled training data could work well.
XPRESS can enable rapid electronic phenotyping, 42 in multi-stakeholder research collaboratives such as the Observational Health Data Sciences and Informatics (OHDSI), 43 that covers over 600 million patients globally. An assumption of XPRESS (and of the noise tolerant learning approach) is that noisy labeled data is available in abundance. For rare phenotypes, this may not always be true. 44 The ability to aggregate training data from hundreds of millions of patients at OHDSI partner institutions, presents a unique opportunity to build phenotype models for conditions that may be under-represented in any single EHR database. It also provides an opportunity to extensively conduct cross-site validations on the XPRESS methodology as well as on specific phenotype models. In situations where the distribution of covariates in a target population may differ significantly from their global distribution, 45,46 the XPRESS method may be used to create local phenotype models.
Limitations
In the current work, we have only examined two phenotypes; making it difficult to generalize the findings. It is possible to conduct such an analysis for multiple phenotypes, if institutional mechanisms for sharing of manually curated evaluation sets can be worked out. We also acknowledge limitations in two additional areas, where improvements can reduce the knowledge engineering effort 47 namely, optimizing the labeling error rate and feature engineering. The first is in optimizing the labeling error rate. Given that negation and family history detection employs a set of regular expressions that can fail when the negation cue lies outside the token window. 48 Without tinkering with negation and history detection, one way to reduce the labeling error rate is by limiting the type of note in which the presence of the keywords is considered. For example, by searching only discharge notes, we could achieve a 15% labeling error rate in the MI training data, compared to an error rate of 34% when all note types are used. However, for T2DM, restricting to discharge notes resulted in just a 2% percent reduction in labeling error rate, which is not useful given the corresponding decrease in the number of training cases. For certain phenotypes it may be better to accept a higher labeling error rate in order to obtain a larger training data set. This trade-off between error rate and sample size, allows for a formal strategy for optimizing the labeling error rate. The second area of improvement is defining features that are informative across phenotypes, port well across different systems, and are tolerant to missing data. Given that the number of ways to compute feature representations for clinical concepts is large, an investigation of optimal feature engineering is likely to be worthwhile. 41 We acknowledge that better feature representations may be possible to further improve both the performance and the portability of the phenotype models.
Conclusion
We demonstrated the feasibility of using semi-automatically labeled, noisy training sets to create phenotype models from a comprehensive representation of the patient clinical record via machine learning. The XPRESS method provides an alternative to manual labeling for the creation of training sets to learn statistical models of phenotypes. The idea of using a specific phrase in patient notes as a positive predictor of the phenotype is not linked to the choice of the phenotype. As a result, the effort required to create a training dataset becomes negligible, making it feasible to apply the XPRESS method to multiple phenotypes. Adopting such an approach to electronic phenotyping could accelerate research studies carried out with large observational healthcare datasets.
Contributors
N.H.S., V.A., and T.P. envisioned the study. VA performed the experiments and compiled the results. T.P., who was also part of the Stanford clinical data warehouse team, implemented the PheKB and OMOP definitions at our local site, which included mapping of the clinical data elements in the patient data-extract to the respective variables in the PheKB and OMOP definitions. J.B. converted prototype code into the XPRESS software tools. V.G., T.I.L., E.P.M., T.E.S., and E.G. reviewed patient records to create a gold standard reference for phenotype labeled records that was used for evaluations. N.H.S., V.A., and T.P. participated in the editing of the manuscript. All authors approve of the final manuscript.
Funding
This work was supported in part by National Institutes of Health (NIH) grant U54 HG004028 for the National Center for Biomedical Ontology, National Library of Medicine (NLM) grant R01 LM011369, National Institute of General Medical Science (NIGMS) grant R01 GM101430 and an award from Janssen Research and Development for the 95 OHDSI ( www.ohdsi.org ) initiative.
Competing interests
None.
Supplementary Material
Acknowledgments
We thank the staff at the Stanford Center for Clinical Informatics for their help in the extraction of patient data from the Stanford clinical data warehouse. We would also like to thank Dr Harish Karnick and Dr Paea Le Pendu for their contributions to this study.
REFERENCES
- 1. Longhurst CA Harrington RA Shah NH . A ‘green button' for using aggregate patient data at the point of care .Health Aff (Millwood). 2014. ; 33 ( 7 ): 1229 – 1235 . [DOI] [PubMed] [Google Scholar]
- 2. Pathak J Kho AN Denny JC . Electronic health records-driven phenotyping: challenges, recent advances, and perspectives .J Am Med Inform Assoc. 2013. ; 20 ( e2 ): e206 – e211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shah NH . Mining the ultimate phenome repository .Nat Biotechnol. 2013. ; 31 ( 12 ): 1095 – 1097 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shivade C Raghavan P Fosler-Lussier E Embi PJ Elhadad N Johnson SB et al. . A review of approaches to identifying patient phenotype cohorts using electronic health records .J Am Med Inform Assoc. 2014. ; 21 ( 2 ): 221 – 230 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gottesman O Kuivaniemi H Tromp G Faucett WA Li R Manolio TA et al. . The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future .Genet Med. 2013. ; 15 ( 10 ): 761 – 771 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCarty CA Chisholm RL Chute CG Kullo IJ Jarvik GP Larson EB et al. . The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies .BMC Med Genomics. 2011. ; 4 : 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei WQ Denny JC . Extracting research-quality phenotypes from electronic health records to support precision medicine .Genome Med. 2015. ; 7 ( 1 ): 41 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denny JC Bastarache L Ritchie MD Carroll RJ Zink R Mosley JD et al. . Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data .Nat Biotechnol. 2013. ; 31 ( 12 ): 1102 – 1110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carroll RJ Eyler AE Denny JC . Intelligent use and clinical benefits of electronic health records in rheumatoid arthritis .Expert Rev Clin Immunol. 2015. ; 11 ( 3 ): 329 – 337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ritchie MD Verma SS Hall MA Goodloe RJ Berg RL Carrell DS et al. . Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci .Mol Vis. 2014. ; 20 : 1281 – 1295 . [PMC free article] [PubMed] [Google Scholar]
- 11. Lin C Karlson EW Dligach D Ramirez MP Miller TA Mo H et al. . Automatic identification of methotrexate-induced liver toxicity in patients with rheumatoid arthritis from the electronic medical record .J Am Med Inform Assoc. 2014. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang M Chen Y Liu M Rosenbloom ST Mani S Denny JC et al. . A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries .J Am Med Inform Assoc. 2011. ; 18 ( 5 ): 601 – 606 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wright A Pang J Feblowitz JC Maloney FL Wilcox AR Ramelson HZ et al. . A method and knowledge base for automated inference of patient problems from structured data in an electronic medical record . J Am Med Inform Assoc. 2011. ; 18 ( 6 ): 859 – 867 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newton KM Peissig PL Kho AN Bielinski SJ Berg RL Choudhary V et al. . Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network .J Am Med Inform Assoc. 2013. ; 20 ( e1 ): e147 – e154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Overby CL Pathak J Gottesman O Haerian K Perotte A Murphy S et al. . A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury .J Am Med Inform Assoc. 2013. ; 20 ( e2 ): e243 – e252 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayley KB Belnap T Savitz L Masica AL Shah N Fleming NS . Challenges in using electronic health record data for CER: experience of 4 learning organizations and solutions applied .Med Care. 2013. ; 51 ( 8 Suppl 3 ): S80 – S86 . [DOI] [PubMed] [Google Scholar]
- 17. Barlas S . Hospitals scramble to meet deadlines for adopting electronic health records: pharmacy systems will be updated slowly but surely .P T. 2011. ; 36 ( 1 ): 37 – 40 . [PMC free article] [PubMed] [Google Scholar]
- 18. PheKB . Phenotype KnowledgeBase . Available from: http://www.phekb.org . Accessed 12 February 2015. [Google Scholar]
- 19. Liao KP Cai T Gainer V Goryachev S Zeng-treitler Q Raychaudhuri S et al. . Electronic medical records for discovery research in rheumatoid arthritis .Arthritis Care Res (Hoboken). 2010. ; 62 ( 8 ): 1120 – 1127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carrell DS Halgrim S Tran DT Buist DS Chubak J Chapman WW et al. . Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence .Am J Epidemiol. 2014. ; 179 ( 6 ): 749 – 758 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carroll RJ Thompson WK Eyler AE Mandelin AM Cai T Zink RM et al. . Portability of an algorithm to identify rheumatoid arthritis in electronic health records .J Am Med Inform Assoc. 2012. ; 19 ( e1 ): e162 – e169 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boland MR Hripcsak G Shen Y Chung WK Weng C . Defining a comprehensive verotype using electronic health records for personalized medicine .J Am Med Inform Assoc. 2013. ; 20 ( e2 ): e232 – e238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tian TY Zlateva I Anderson DR . Using electronic health records data to identify patients with chronic pain in a primary care setting .J Am Med Inform Assoc. 2013. ; 20 ( e2 ): e275 – e280 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chute CG . Invited commentary: Observational research in the age of the electronic health record .Am J Epidemiol. 2014. ; 179 ( 6 ): 759 – 761 . [DOI] [PubMed] [Google Scholar]
- 25. Castro VM Apperson WK Gainer VS Ananthakrishnan AN Goodson AP Wang TD et al. . Evaluation of matched control algorithms in EHR-based phenotyping studies: a case study of inflammatory bowel disease comorbidities .J Biomed Inform. 2014. ; 52 : 105 – 111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y Carroll RJ Hinz ER Shah A Eyler AE Denny JC et al. . Applying active learning to high-throughput phenotyping algorithms for electronic health records data .J Am Med Inform Assoc. 2013. ; 20 ( e2 ): e253 – e259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sinnott JA Dai W Liao KP Shaw SY Ananthakrishnan AN Gainer VS et al. . Improving the power of genetic association tests with imperfect phenotype derived from electronic medical records .Hum Genet. 2014. ; 133 ( 11 ): 1369 – 1382 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peissig PL Santos Costa V Caldwell MD Rottscheit C Berg RL Mendonca EA et al. . Relational machine learning for electronic health record-driven phenotyping .J Biomed Inform. 2014. ; 52 : 260 – 270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simon HU . General bounds on the number of examples needed for learning probabilistic concepts .J Comput Syst Sci. 1996. ; 52 ( 2 ): 239 – 254 . [Google Scholar]
- 30. Aslam JA Decatur SE . On the sample complexity of noise-tolerant learning .Inform Process Lett. 1996. ; 57 ( 4 ): 189 – 195 . [Google Scholar]
- 31. Observational Medical Outcomes Partnership . Health Outcomes of Interest . Available from: http://omop.org/HOI Accessed 13 December 2014 . [Google Scholar]
- 32. Halpern Y Choi Y Horng S Sontag D . Electronic Medical Record Phenotyping using the Anchor & Learn Framework .J Am Med Inform Assoc. 2016. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung K LePendu P Iyer S Bauer-Mehren A Percha B Shah NH . Functional evaluation of out-of-the-box text-mining tools for data-mining tasks .J Am Med Inform Assoc. 2015. ; 22 ( 1 ): 121 – 131 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agarwal V Lependu P Podchiyska T Barber R Boland M Hripcsak G et al. . Using narratives as a source to automatically learn phenotype models 2014AMIA: in 1st Workshop on Data Mining for Medical Informatics. 2014; Annual Symposium: Washington DC . [Google Scholar]
- 35. Hastie T Tibshirani R Friedman J . The Elements of Statistical Learning . Vol. 2 . New York: : Springer; ; 2009. . [Google Scholar]
- 36. Friedman J Hastie T Tibshirani R . Regularization Paths for Generalized Linear Models via Coordinate Descent .J Stat Softw. 2010. ; 33 ( 1 ): 1 – 22 . [PMC free article] [PubMed] [Google Scholar]
- 37. Pedregosa F Varoquaux G Gramfort A Michel V Thirion B Grisel O et al. . Scikit-learn: Machine learning in Python .J Machine Learning Res. 2011. ; 12 : 2825 – 2830 . [Google Scholar]
- 38. Yu S Liao KP Shaw SY Gainer VS Churchill SE Szolovits P et al. . Toward high-throughput phenotyping: unbiased automated feature extraction and 90 selection from knowledge sources .J Am Med Inform Assoc. 2015. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei WQ Leibson CL Ransom JE Kho AN Caraballo PJ Chai HS et al. . Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus .J Am Med Inform Assoc. 2012. ; 19 ( 2 ): 219 – 224 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Platt R Carnahan RM Brown JS Chrischilles E Curtis LH Hennessy S et al. . The U.S. Food and Drug Administration's Mini-Sentinel program: status and direction .Pharmacoepidemiol Drug Saf. 2012. ; 21 Suppl 1 : 1 – 8 . [DOI] [PubMed] [Google Scholar]
- 41. Pathak J Bailey KR Beebe CE Bethard S Carrell DC Chen PJ et al. . Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium .J Am Med Inform Assoc. 2013. ; 20 ( e2 ): e341 – e348 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. OHDSI . Aphrodite . 2015. ; Available from: https://github.com/OHDSI/Aphrodite . Accessed 7 November 2015 . [Google Scholar]
- 43. OHDSI . Observational Health Data Sciences and Informatics . Available from: http://www.ohdsi.org . Accessed 7 November 2015 . [Google Scholar]
- 44. Denny JC . Chapter 13: Mining electronic health records in the genomics era .PLoS Comput Biol. 2012. ; 8 ( 12 ): e1002823 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ng K Sun J Hu J Wang F . Personalized Predictive Modeling and Risk Factor Identification using Patient Similarity .AMIA Jt Summits Transl Sci Proc. 2015. ; 2015 : 132 – 136 . [PMC free article] [PubMed] [Google Scholar]
- 46. Celi LAG Tang RJ Villarroel MC Davidzon GA Lester WT Chueh HC . A clinical database-driven approach to decision support: Predicting mortality among patients with acute kidney injury .J Healthcare Engineering. 2011. ; 2 ( 1 ): 97 – 110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hripcsak G Albers DJ . Next-generation phenotyping of electronic health records .J Am Med Inform Assoc. 2013. ; 20 ( 1 ): 117 – 121 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Agarwal S Yu H . Biomedical negation scope detection with conditional random fields .J Am Med Inform Assoc. 2010. ; 17 ( 6 ): 696 – 701 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.