Abstract
Background: O-GlcNAcylation is a single sugar attachment of serine and/or threonine residues on intracellular proteins. Recent reports reveal that it can modify several secretory proteins; however, the underlying mechanisms are largely unexplored. Materials and Methods: To investigate whether extracellular vesicles (EVs) carry secretory O-GlcNAc-modified proteins that were isolated from colorectal cancer (CRC) cells, two-dimensional gel electrophoresis followed with O-GlcNAc immunoblotting and liquid chromatography-tandem mass spectrometry (LC-MS/MS) were applied. Results: It was revealed that the O-GlcNAc modification of many EV proteins was increased in metastatic cells. Among these, transitional endoplasmic reticulum ATPase (TER ATPase) and RuVB-like1 were successfully confirmed for the O-GlcNAc modification in which the levels were significantly higher in EVs of metastatic CRC cell line. Conclusion: These data, demonstrate that proteins carried by EVs are O-GlcNAc-modified. Importantly, elevated aberrant O-GlcNAcylation of EV proteins might serve as a potential biomarker of metastatic CRC.
Keywords: Colorectal cancer, extracellular vesicles, O-GlcNAcylation, metastasis, RuVB-like1, transitional endoplasmic reticulum ATPase
Title
Colorectal cancer (CRC) is one of the most common cancers worldwide. Based on the World Health Organization GLOBOCAN database in 2012, CRC is the third most common cancer (1.36 million cases) and ranks as the fourth cause of death (694,000 cases) from overall cancers (1). Cancer patients usually are not diagnosed until the disease has progressed to advanced stages. At the advanced-late stage, cancer cells invade from the primary sites and spread throughout the bloodstream in the body to form secondary tumors at distal sites, in a process called metastasis. Distant metastases are the major leading causes of death of patients with cancers, including CRC (2). The mechanisms of CRC metastasis are under investigation (3). Seeking new biomarkers of cancer metastasis, therefore, will improve understanding of cancer biology, as well as stimulate the development of effective drugs against metastasis.
Extracellular vesicles (EVs) is a term currently used for vesicles that are enclosed by the phospholipid bilayer of the cell membrane, filled with cytosolic machineries and secreted to the outer environment by various cell types (4,5). Two major EVs that have been widely studied are “exosomes” and “microvesicles”. Exosomes originate from the endolysosomal pathway involving the fusion of multivesicular bodies (MVBs) with plasma membrane to release vesicles to the outer milieu (6). Microvesicles, also called ectosomes, are released by budding of plasma membrane (5). EVs contain several biological molecular cargos, including proteins, nucleic acids and lipids, which mediate proximal and distal cell communication (4). Growing evidence indicates that EVs are involved in various disorders. EVs found in biological samples can represent the original cells. Thus, in cancer, the component of EVs derived from cancer cells and body fluids from cancer patients are distinct from those of normal donors (7,8). Tumor-derived EV proteins and RNAs are known for their critical roles in malignancy, angiogenesis, invasiveness, metastasis and drug resistance (9). In addition, EVs derived from tumor cells can alter the tumor microenvironment by changing the composition of extracellular matrix, inducing cancer-associated fibroblasts and mediating immune responses (10,11). Due to the fact that EVs are readily observed in various specimens, they are promising tools for predicting the early onset of cancer.
O-GlcNAcylation is a glycosylation that mainly modifies nuclear, cytosolic and mitochondrial proteins (12). Two key enzymes, O-GlcNAc transferase (13) and O-GlcNAcase (14), catalyze the transfer of O-GlcNAc moiety to the hydroxyl group of serine and/or threonine residues on target proteins and the removal of O-GlcNAc from O-GlcNAc-modified proteins, respectively (12). The enzymatic activity of O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) is sensitive to the concentration of its donor substrate, uridine diphospho-N-acetylglucosamine (UDP-GlcNAc) (15), a high-energy small molecule synthesized as the end product of the hexosamine biosynthetic pathway (HBP). The HBP has been considered as a nutrient sensor since changes in cellular levels of various nutrients, including glucose, fatty acids and amino acids, dramatically affect the level of UDP-GlcNAc synthesis (16). Higher energy consumption is commonly observed in most types of cancer (17,18). Therefore, an elevated nutrient flux directly affects the HBP and also O-GlcNAcylation. Notably, levels of O-GlcNAcylation and OGT expression are relatively higher in many types of cancer (19).
Recently, we reported that O-GlcNAcyation level of extracellular proteins was increased in breast cancer cell secretion compared to that of normal breast cell secretion (20). In the present study, we investigated whether (i) proteins secreted from colorectal cells were O-GlcNAc modified and (ii) O-GlcNAc-modified proteins can be released from cells by the secretory pathway via EVs. EVs were successfully isolated from non-metastatic (SW480) and metastatic (SW620) CRC cells. Gel-based proteomics of EV proteins followed by O-GlcNAc immunoblotting revealed the O-GlcNAc modification of many EV proteins for the first time. The possible roles of modified proteins, in the extracellular vesicle formation and associated with CRC metastasis, are discussed.
Materials and Methods
Cell culture. Human colorectal cancer cell lines, HT29, SW480 and SW620 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Normal colon epithelial cell, CCD 841 CoN (ATCC) was provided by Dr. Jutamaad Satayavivad, Chulabhorn Research Institute, Thailand. All cell culture medium and supplementary products were purchased from Invitrogen (Carlsbad, CA, USA). Approximately, 2×106 cells of CCD841 CoN, HT29, SW480 and SW620 were seeded in T175 flasks. CCD841 CoN cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 1% L-glutamine, while HT29, SW480 and SW620 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium, supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin and maintained at 37˚C in a humidified 5% CO2 incubator. At 70 % confluence, cells were washed three times with 15 ml of serum-free medium to remove any remaining FBS. CCD841 CoN cells were cultured in serum-free DMEM, while HT29, SW480 and SW620 cells were cultured in serum-free RPMI-1640 medium for 24 h prior to collecting conditioned medium (CM). To extract proteins from cells, culture cells were lysed with 1× RIPA supplemented with 1% protease inhibitor cocktail (Sigma-Aldrich, St.Louis, MO, USA) and 100 μM of Thiamet-G (Sigma-Aldrich), an O-GlcNAcase (OGA) inhibitor as described previously (20). Protein concentration was determined using Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA).
Secretory protein (secretome) and EV isolation. Secretory proteins, isolated as serum-free CM, and EVs were prepared with some modifications according to previous report (21). CM collected from CCD841 CoN, HT29, SW480 and SW620 cells was centrifuged at 480 × g for 5 min and 2,000 × g for 10 minutes to remove floating cells and cell debris, respectively. The supernatants were then filtered through 0.1 nm filter membrane using VacuCap® vacuum filtration devices (Pall Corporation, Washington, NY, USA) to remove vesicles with a size larger than 100 nm. The filtrate was concentrated to approximately 1 ml using 3K cut-off Amicon® Ultra-15 centrifugal filter devices (Millipore, Bedford, MA, USA). The concentrated filtrate was further centrifuged at 10,000 ×g for 30 minutes at 4˚C to pellet microvesicles. From this step, concentrated conditioned media (CCM) were collected for secretome analysis. For EV isolation, CCM was then overlaid on 30% sucrose in 20 mM HEPES, pH 7.4 and centrifuged at 184,000 × g for 18 hours at 4˚C. EV pellets were then washed once in 20 mM HEPES, pH 7.4 by centrifugation at 184,000 × g for 1 hour at 4˚C. The pellets were finally collected to further analysis.
OGA, peptide -N-Glycosidase F (PNGase F) digestion and on-blot β-Elimination. OGA (14) (R&D Systems, Minneapolis, MN, USA) was used to digest O-GlcNAc residues from proteins enriched from cell secretions and EV fractions at a ratio of 10 μg of secreted or EV proteins per 0.05 μg OGA according to a previously reported method (20). PNGase F (New England Biolabs, Beverly, MA, USA) was used to remove N-linked-glycan by following manufacturer’s protocol. On blot β-Elimination experiment was performed to remove O-Glycan according to a previous report (22).
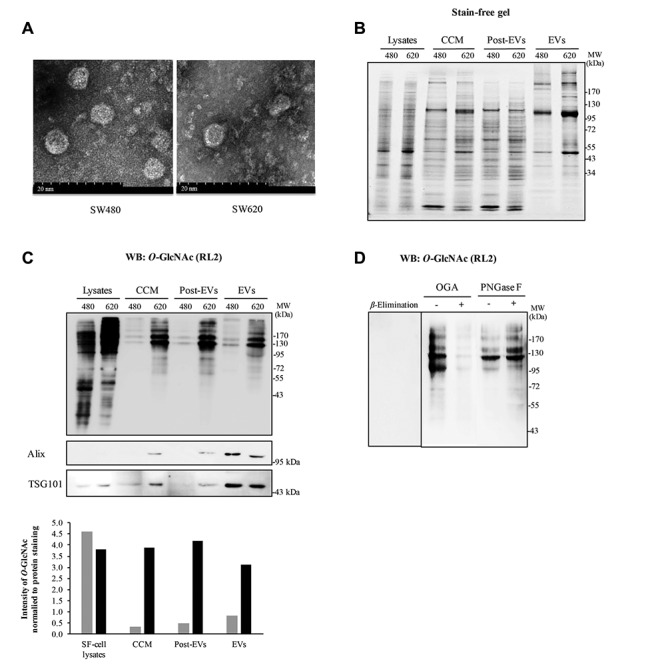
Electron microscopy of EVs. After isolation of fresh EVs, EV pellets were re-suspended and fixed in 1% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) in 0.2 M sodium cacodylate buffer at 4˚C overnight. On the next day, EVs were applied to glow-discharged carbon-coated copper grids (Electron Microscopy Sciences). After allowing the EVs to adsorb completely, the grids were rinsed with droplets of deionized water and stained with a mixture of 2% uranyl acetate (Electron Microscopy Sciences). Electron micrographs were recorded using a HT7700 transmission electron microscope (Hitachi, Krefeld, Germany) with an acceleration voltage of 80 kV at Kasetsart University Research and Development Institute (KURDI, Bangkok, Thailand).
Western blotting. Protein samples (10 μg) were separated in 10% sodium dodecyl suphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. Blots were stained with SYPRO Ruby (Molecular Probes, Eugene, OR, USA) in order to determine total protein loading before probing with specific antibodies. In some experiments, stain-free SDS-PAGE gels (Bio-Rad Laboratories) were used to determine total protein loading prior to transferring into PVDF membranes. Then, the stained blots were probed with antibodies specific to O-GlcNAc (RL2), Alix, TSG101, Calsynthenin-1, TER ATPase and RuvB-like1 (all antibodies were from Abcam, Cambridge, UK). Bands on immunoblots were detected using WesternBright ECL (Advansta, Menlo Park, CA, USA) and chemiluminescent signals images were captured using ImageQuant™ LAS 4000 digital imaging system (GE Healthcare, Buckinghamshire, UK).
Two-dimensional gel electrophoresis. Proteins harvested from CCM and EVs from SW480 and SW620 cells (100 μg/gel) were separated using two-dimensional gel electrophoresis (2DE). The 2DE was performed as previously described (23) after which proteins in gels were transferred to PVDF membranes. The membranes were stained by SYPRO Ruby (Molecular Probes) using the manufacturer’s recommendations and the fluorescent signals were scanned using Ettan DIGE Imager (GE Healthcare) prior to probing with an O-GlcNAc antibody, RL2 (Abcam).
In-gel digestion and protein identification by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In-gel trypsin digestion of EV protein spots with O-GlcNAcylation was performed as previously described (24). The digested peptides were then identified using Nanoflow liquid chromatography coupled with the amaZon speed ion trap mass spectrometry (Bruker, Billerica, MA, USA) as previously described (23). MASCOT search with NCBInr version 20130630 sequence databases (http://www.matrixscience.com) was performed in order to identify the protein spots. Search parameters were set following a previous study (23).
Confirmation of O-GlcNAc-modified EV proteins. Due to the limitation in amount of proteins recovered from the EV isolation steps (about 200 μg of EV proteins successfully harvested from 20 culture flasks of T175 flask), confirmation of O-GlcNAc-modified EV proteins could not be readily evidenced by performing standard immunoprecipitation. Therefore, in order to validate O-GlcNAcylated proteins from small amounts of EV proteins, double 2DE immunoblots were performed. Two identical gels of 2DE immunoblotting membranes of EV proteins from SW480 and SW620 (100 μg/gel) were used. Total proteins on immunoblots were stained with SYPRO Ruby (S1) prior to be detected using specific antibodies. The first membrane was immunoblotted with antibody specific to O-GlcNAc (RL2) (S2). The second membrane was first immunoblotted with Calsynthenin-1, secondly, reprobed with TER ATPase and lastly reprobed with RuvB-like1, respectively (S3). Intensity of O-GlcNAc-modified protein spots (probed with RL2) and target proteins (probed with Calsynthenin-1, TER ATPase and RuvB-like1) on blots were measured using ImageQuant™ LAS 4000 digital imaging system (GE Healthcare). Then, the immunoblots (S2, S3) were aligned to the SYPRO Ruby image (S1) to assign the location of O-GlcNAc-modified protein on each membrane. This validation confirmed if the targeted proteins were O-GlcNAc modified. Intensity of O-GlcNAcylation (S2) was normalized by intensity of protein staining of individual spot (S1).
Statistical analysis. The statistical analysis was performed using unpaired Student’s t-test for differences between the two groups. Statistical significance was defined at p<0.05.
Results
O-GlcNAcyation level of intracellular and secreted proteins from colorectal cells. O-GlcNAcylation has been widely found in the cells; however, reports suggest that some extracellular proteins are also modified by this glycosylation. We, therefore, hypothesized that some secreted proteins may be modified by O-GlcNAc in colorectal cells. In this study, the epithelial normal colon cells, CCD841 CoN and three CRC cell lines, including HT29, SW480 and SW620, were cultured in monolayer system. Serum-free medium was used to avoid interference by high-abundance proteins, such as serum albumin. Intracellular proteins and proteins’ conditioned medium from cultures deprived of serum for 24 hours were separated in 10% SDS-PAGE, immunoblotted with O-GlcNAc antibody (RL2). The total protein on the immunoblot was stained using SYPRO Ruby (Figure 1A). Immunoblots of O-GlcNAc demonstrated that RL2 antibody could detect the O-GlcNAcylation of secreted proteins in all cells with different intensity (Figure 1B). By measuring the extent of O-GlcNAc from immunoblots, the results showed a much lower O-GlcNAc on secreted proteins from normal colon cells, CCD841 CoN, in comparison to those of the other three CRC cell lines. In addition, the patterns of O-GlcNAc-modified proteins between serum-free medium cell lysates and secreted proteins differed. Furthermore, comparison of non-metastatic SW480 and metastatic SW620 cells derived from the same patient revealed a higher O-GlcNAcylation in SW620.
To test the specificity of RL2 antibody used to detect O-GlcNAc, we treated secreted proteins with OGA and PNGase F enzymes. While OGA cleaved O-linked-GlcNAc from serine and threonine residues, PNGase F removed virtually all N-linked glycan from all asparagine-linked complexes. The untreated and treated groups were separated on SDS-PAGE and immunoblotted with RL2 antibody. The results showed that OGA-treated proteins displayed much fainter O-GlcNAc bands detected by RL2 antibody compared to untreated secretory proteins (Figure 1C). β-Elimination of O-GlcNAc on immunoblot also demonstrated the specificity of RL2 antibody since there was no band detected by RL2 (Figure 1C). Moreover, no differences in O-GlcNAc band intensity was observed between PNGase F treated and untreated secretory proteins, thus confirming the specificity of RL2 antibody to O-linked-GlcNAc (Figure 1D).
O-GlcNAc modification of EV proteins secreted from CRC cell lines. Some evidence suggests the possible O-GlcNAcylation of secreted proteins, the mechanism of which is still unexplained. In this study, it was proposed that EVs might be a carrier of O-GlcNAcylated proteins for secretion. Enriched exosomes of CM collected from non-metastatic (SW480) and metastatic (SW620) CRC cells were prepared by ultra-centrifugation in 30% sucrose. Transmission electron microscopy revealed that enriched exosome of SW480 and SW620 had an average size between 40-80 nm (Figure 2A). In addition, samples from each step of EV isolation, including CCM, post-EV fraction (a sucrose cushion) and EV pellets, were collected. The pattern of total proteins was visualized by fluorescence of stain-free SDS-PAGE gel (Figure 2B). The results showed the distinct protein pattern observed in EVs when compared to those from the other fractions. Comparing between two CRC cell lines, SW620 cells showed a higher number of protein bands, as well as enhanced intensity of some specific bands (Figure 2B). Immunoblotting of O-GlcNAc clearly showed that certain secretory proteins from all fractions (CCM, post-EVs and EVs) were modified by O-GlcNAcylation and those were different from intracellular O-GlcNAc patterns (Figure 2C). Of interest, O-GlcNAcylation levels were dramatically enhanced in EV and post-EV fraction of metastatic SW620 cells, while much less content was found in those of SW480 cells. Importantly, two EV protein markers, Alix and TSG101, confirmed an enrichment of EVs in the final fraction from isolation step (Figure 2C). In addition, the glycosylation of EV proteins detected by RL2 antibody was confirmed to be O-GlcNAcylation using on-blot β-Elimination, treatments of OGA and PNGase F, respectively (Figure 2D).
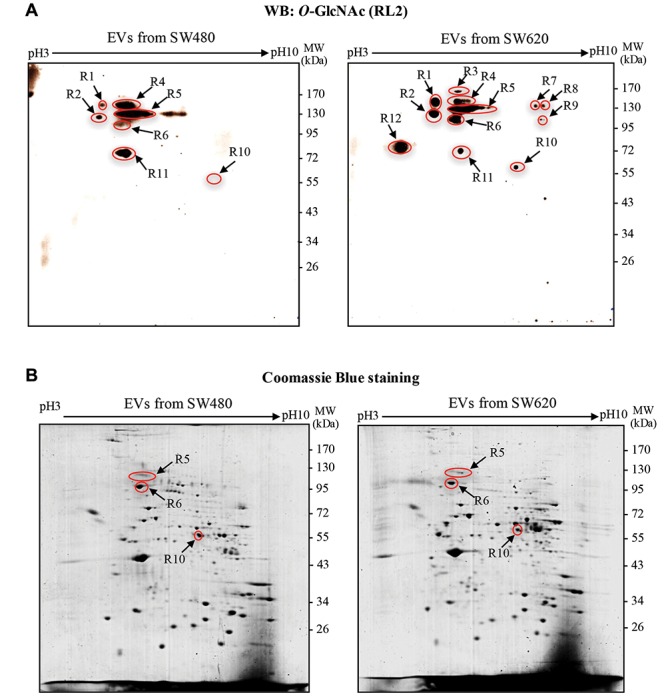
Identification of O-GlcNAcylation of EV proteins isolated from CM of SW480 and SW620 cells. 2DE followed by immunoblotting with antibody against O-GlcNAc (RL2) was performed to study O-GlcNAcylation of EV proteins from non-metastatic (SW480) and metastatic (SW620) CRC cells. Freshly prepared EVs isolated by ultracentrifugation were collected and lysed in 2D-lysis buffer. Immunoblotting of O-GlcNAc demonstrated that EVs secreted from both SW480 and SW620 cells contained O-GlcNAc-modified proteins (Figure 3A). Twelve O-GlcNAc spots were mapped and ten spots observed in EVs of SW620 cells demonstrated higher O-GlcNAc intensity than those of SW480 cells, while two spots (R4 and R11) showed the opposite. Spots of EV protein were visualized by SYPRO Ruby fluorescent dye staining prior to be probed with RL2 antibody. The O-GlcNAc spots were then aligned to the protein spots and only three O-GlcNAc spots (R5, R6 and R10) perfectly matched the protein spots, as indicated in Figure 3B. The matched protein spots on 2DE gels were then digested, trypsinized and identified by LC-MS/MS. The identified EV proteins included calsyntenin-1, TER ATPase and RuvB-like 1 (Table I). All of these identified proteins were previously reported as vesicle proteins, especially found in exosomes according to “ExoCarta” and “Vesiclepedia”, the databases for EVs (Table I).
Table I. List of identified O-GlcNAc-modified proteins from EVs secreted by SW480 and SW620 cells identified by LC-MS/MS.
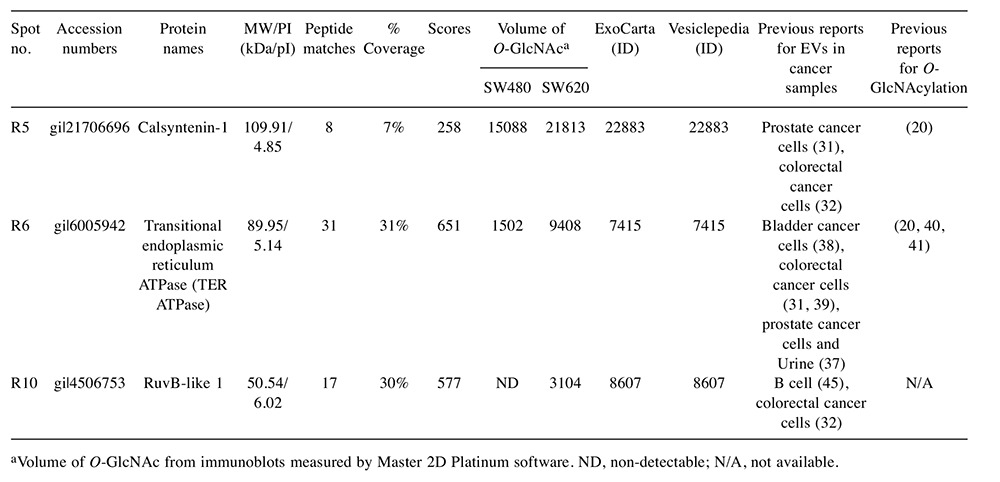
aVolume of O-GlcNAc from immunoblots measured by Master 2D Platinum software. ND, non-detectable; N/A, not available
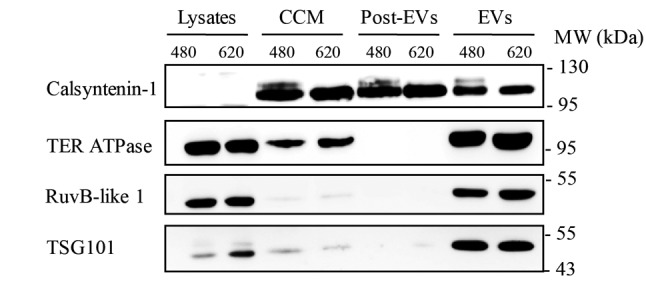
Validation of O-GlcNAc modification of EV proteins. To confirm whether the proteins identified by LC-MS/MS, including calsyntenin-1, TER ATPase and RuvB-like 1, were enriched in the EV fraction, samples from each isolation step were collected and separated by SDS-PAGE. Immunoblotting of candidate proteins, as well as TSG101, showed that all identified proteins were clearly enriched in EVs derived from both SW480 and SW620 cells (Figure 4). TER ATPase and RuvB-like1 were predominantly observed in the EV fractions of SW480 and SW620, while calsyntenin-1 was found in both post-EVs and EVs fractions (Figure 4).
Figure 4. Western blots of calsyntenin-1, TER ATPase and RuvB-like 1, as well as TSG101, in different fractions collected from EV isolation of SW480 and SW620 cells. CCM refers to concentrated conditioned medium and post-EVs refers to sucrose cushion fraction.
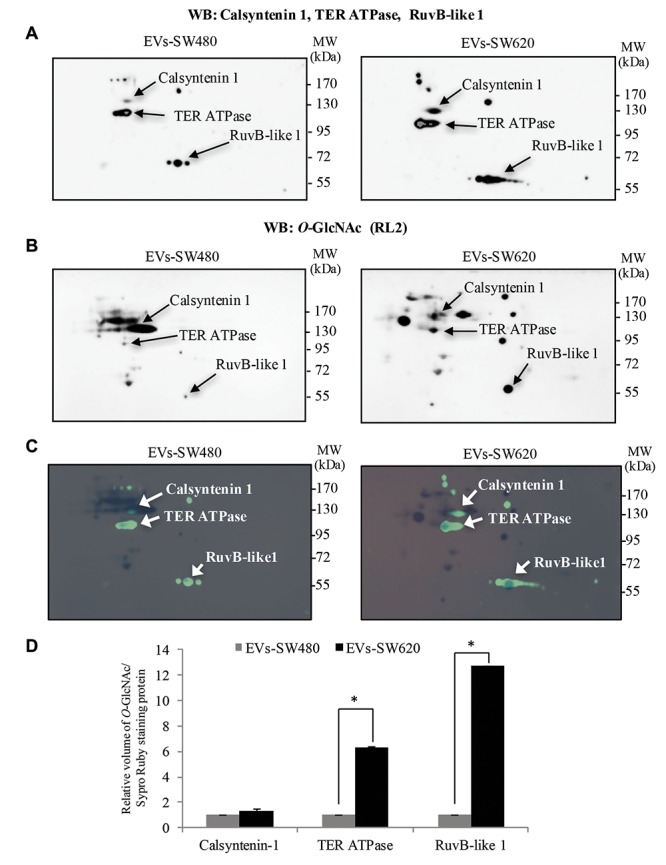
In addition, O-GlcNAc modification of these identified proteins was further verified using double 2DE immunoblots. EV proteins from SW480 and SW620 CRC cells were separated in 2DE and detected with (i) O-GlcNAc antibody (RL2) for the first immunoblot and (ii) antibodies specific to calsyntenin-1, TER ATPase and RuvB-like 1 for the second immunoblot. As shown in Figure 5A, calsyntenin-1, TER ATPase and RuvB-like 1 were enriched in EVs isolated from SW480 and SW620 cells. Immunoblots of O-GlcNAc were aligned to SYPRO Ruby spots as shown in Figure 5B. Overall, the results demonstrated that calsyntenin-1, TER ATPase and RuvB-like 1 spots were perfectly matched to the O-GlcNAc spots (Figure 5C). Thus, it was confirmed that EVs secreted by SW480 and SW620 cells carried calsyntenin-1, TER ATPase and RuvB-like 1 protein. Importantly, these vesicle proteins were modified by O-GlcNAcylation. The relative content of O-GlcNAc on individual EV proteins was measured and normalized to SYPRO Ruby staining. O-GlcNAc-calsyntenin-1 tended to increase but was not statistically different (p=0.14, EVs-SW480 vs. EVs-SW620). However, O-GlcNAcylation of TER ATPase and RuvB-like 1 in EVs from metastatic SW620 cells was significantly higher than from non-metastatic SW480 cells, respectively (Figure 5D).
Figure 5. Validation of O-GlcNAc-modified EV proteins released from SW480 and SW620 cells. Double 2DE immunoblots of EV proteins of interest. A: A representative of the first 2DE Western blot (WB) of calsyntenin-1, TER ATPase and RuvB-like and B: A representative of the second 2DE Western blot of O-GlcNAcylated proteins. C: Immunoblots of indicated EV-proteins (depicted in green) were overlaid onto immunoblots of O-GlcNAc (depicted in black). D: Bar graph showing the volume of O-GlcNAc of calsyntenin-1, TER ATPase and RuvB-like 1 normalized to SYPRO Ruby staining proteins measured by ImageMaster 2D Platinum version 7.0. An asterisk (*) represents statistically significant difference of O-GlcNAcylation levels between EV proteins from SW480 and from SW620 at p≤0.05. WB, Western blot.
Discussion
To date, O-GlcNAcylation has been identified as an exclusive glycosylation of numerous intracellular proteins enriched in cytosol, nucleus and mitochondria (12). This study, for the first time, demonstrates O-GlcNAcylation of vesicular proteins secreted by CRC cells. Here, we used a 2DE-gel approach combined to immunoblotting technique for detecting O-GlcNAc modification of EV proteins. Even though 2DE has been used as a sample separation method since the beginning of proteomic study, it currently remains a technique of choice for specific research area. The combinational method has been applied for studying O-GlcNAc-modified proteins in some reports like, for instance, secretory proteins in pig nasal mucus and the identification of membrane proteins of CRC cell lines that were recognized by lectin from Helix pomatia (HPA) (25,26). These examples showed the power of the resolution of 2DE that allows visualizing the modified-proteins on a 2D map for further identification by mass spectrometry.
Soluble secreted proteins were collected from CM of CCD841 Con (normal epithelial colon cells) and three CRC cell lines (HT29, SW480 and SW620). O-GlcNAc modification of secretory proteins was observed in all CRC cell types (Figure 1). Using OGA and PNGase F digestion, as well as on-blot β-Elimination, these extracellular proteins were confirmed to be O-GlcNAc modified (Figures 1 and (Figure 2). These findings are similar to our recent report of extracellular O-GlcNAcylation in the secreted products from breast cancer cells (20). Extracellular O-GlcNAc level was dramatically increased in secretions of both CRC and breast cancer cell lines in comparison to those from their normal cells. In addition, the patterns of O-GlcNAc-modified protein differed between intercellular and extracellular compartments. These findings indicate that not all intracellular O-GlcNAc-modified proteins are released or secreted extracellularly.
Figure 1. O-GlcNAcylation of proteins secreted from normal epithelial colon cell and three different CRC cells. A: A representative SYPRO Ruby stained protein gel and B: a representative Western blot of O-GlcNAcylated proteins from cell lysates cultured in serum free-medium (SF-cell lysates) and their secreted proteins collected from conditioned medium (CM). C and D: A representative western blot of O-GlcNAcylated proteins from the secreted proteins treated, or not, with O-GlcNCAase, on-blot β-Elimination and with PNGase F, respectively. WB, Western blot.
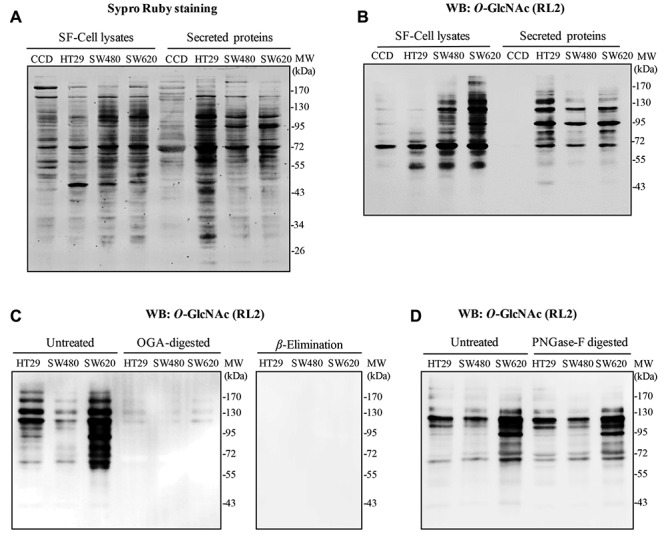
Figure 2. EVs isolated from conditioned medium of SW480 and SW620 cells containing O-GlcNAcylated proteins. A: Representative captured pictures using transmission electron microscopy revealing enriched EVs of SW480 and SW620 cells varied in size from 40-80 nm. Scale bar = 20 nm. B: Stain-free SDS-PAGE gel demonstrated the pattern of total proteins from cell lysates cultured in serum free-medium (SF-cell lysates) and different fractions of EV preparation steps (CCM, post-EVs and EVs). C: Detection of O-GlcNAcylated proteins from SF-cell lysates and different fractions of EV preparation steps by Western blotting of O-GlcNAc using RL2 antibody. Exosomal markers (Alix and TSG101) showed an enrichment of exosomes in the final fraction (EVs). D: Western blots of O-GlcNAc of EV proteins from SW620 cells treated with OGA, PNGase F and on-blot β- Elimination. Post-EVs refers to sucrose cushion fraction. 480 refers to SW480, while 620 refers to SW620.
Presently, few studies have been reported on the O-GlcNAc modification of secretory proteins. O-GlcNAcylation of extracellular proteins was first reported on the 20th epidermal growth factor (EGF) domain (EGF20) of Notch receptors in Drosophila (27). EGF domain-specific O-GlcNAc transferase (EOGT) was recently identified as a novel OGT catalyzing addition of O-GlcNAc molecule on extracellular proteins containing EGF domain (28). EOGT was localized in the lumen of the endoplasmic reticulum (ER) and transferred GlcNAc to target proteins at the conserved sequence of EGF domain containing six cysteine residues (14). Interestingly, the O-GlcNAc on EGF domain in HEK293T cells was further glycosylated by adding galactose (Gal) to form a new glycan structure, called O-LacNAc (O-linked N-acetyllactosamine) (29). Recently, the same glycan structure of O-GlcNAc and Gal was also observed in human umbilical cord blood-derived mesenchymal stromal cells (30). Another study revealed a potential O-GlcNAc-modification of olfactory binding proteins (OBPs) isolated from pig nasal mucus (26). The presence of the ortholog Eogt gene was shown in the pig genome by amplifying the Eotg gene from six different tissues of pigs. Together, it is possible that O-GlcNAcylation of soluble secreted proteins might be catalyzed by EOGT in ER after which the proteins are secreted to the external environment through the conventional ER-Golgi pathway.
Indeed, proteins may not only be secreted through the classical ER-Golgi system but also other intricate mechanisms, including exosomes of endosomal origin, plasma-budding microvesicles, etc. Thus, an alternative secretory pathway for O-GlcNAc-modified extracellular protein should not be ruled out. Here, EVs were enriched from CM of CRC cells by ultracentrifugation in 30% sucrose, with Alix and TSG101 being used as EV markers (Figures 2 and 4). O-GlcNAc-modified proteins in enriched EV fraction of SW480 and SW620 cells were successfully identified. Notably, most EV proteins from SW620 displayed remarkably higher O-GlcNAcylation than those of SW480 cells (Figure 3). Moreover, using the same 2DE proteomic approach, we found that many more O-GlcNAc-modified protein spots were found in CM (secretome) compared to EVs fractions (data not shown). This suggests that there are many secretory pathways for extracellular O-GlcNAc-modified proteins and extracellular vesicles represent one of these secretions.
Figure 3. O-GlcNAc modification of EV proteins from SW480 and SW620 cells. A: Representative 2DE immunoblots of O-GlcNAcylated EV proteins isolated from SW480 and SW620 cells. Arrows indicate O-GlcNAc-protein spots. B: Representative 2DE gels of EV proteins from SW480 and SW620 cells. Total protein spots were visualized by staining the gels with Coomassie Brilliant Blue R250. Arrows indicate protein spots that could be matched with O-GlcNAc immunoblots. WB, Western blot.
Proteomic study of exosomal proteins derived from the primary non-metastatic colorectal cancer cells SW480 and metastatic colorectal cancer cells SW620 revealed distinct protein contents between these two cell lines (21,31). SW480-exosomes contained proteins involved in cell adhesion, while SW620-exosomal proteins had a role in cancer progression and in the metastatic phenotypes of cancer, including multi-drug resistance (31). Other work has also demonstrated that exosomes isolated from SW620 contain major proteins of metastatic factors, signal transduction molecules, lipid raft and lipid raft associated components (21).
Differences in the levels of O-GlcNAcylation between EVs proteins from non-metastatic and metastatic CRC cells indicate the need for further study on an association between this glycosylation and tumorigenesis. Since calsyntenin-1, TER ATPase and RuvB-like 1 were present in EVs of SW480 and SW620 cells with different O-GlcNAc modification levels and have been categorized in the databases of exosomes and vesicles (ExoCarta and Vesiclepedia), the functions and characteristic features of these proteins are discussed here.
Calsyntenin-1 (also known as alcadein-α) is a transmembrane protein and a member of the cadherin superfamily of cell adhesion molecules (13). A major function of calsyntenin-1 is as a cargo-docking protein involved in axonal anterograde transport by mediating vesicles’ transport along axons on kinesin-1 motors (32). Calsyntenin-1 has been reported to be an exosomal protein purified from prostate cancer cells and CRC cells (33,34). Moreover, calsyntenin-1 was found in exosomes derived from prostate cancer cell lines but not from normal cells (33). Of note, calsyntenin-1 was post-transcriptionally modified with both high-mannose and complex N-glycans in HEK293 cells and adult mouse brain (35). The complex N-glycosylation facilitates the trafficking between calsyntenin-1 and kinesin-1, a microtubule motor protein involved in the intracellular vesicle transportation (34). Recently, our group found that O-GlcNAc-calsyntenin-1 level was increased in secretions from breast cancer cells compared to normal breast cells (20). Although the effect of this glycosylation was not investigated, increased O-GlcNAcylation of calsyntenin-1 in the extracellular compartment might be associated with malignancy.
TER ATPase, also known as vasolin-containing protein, is in the AAA+ (ATPases associated with diverse cellular activities) ATPase family (36). As a high-abundant protein in cells, it is associated in various cellular activities involved in regulation of the cell cycle, formation of the nuclear envelope, biogenesis of the Golgi and ubiquitin proteasome degradation (37). An important and well-studied role of TER ATPase is in mediating protein homeostasis by targeting ubiquitinylated proteins from ER to proteasome degradation (38). Interestingly, it is implicated in the degradation of various proteins relating to cancer. TER ATPase has also been found in exosomes derived from various cancer cells, including CRC, prostate cancer and bladder cancer cells (33,39,40). Moreover, TER ATPase was commonly found in exosomes derived from CRC cells, including HT29, SW480, SW620 and LIM1215 (31). The O-GlcNAcylation of TER ATPase was found in Xenopus laevis and Rattus norvegicus, although the O-GlcNAc sites have not determined yet (41,42). We also reported that O-GlcNAc-TER ATPase level was higher in breast cancer cell secretion than in normal breast cell secretion (20). How this glycosylation affects the functions of extracellular TER ATPase in CRC needs further elucidation.
RuvB-like1 (Pontin52 or TIP49) also belongs to AAA+ ATPase superfamily (43). The functions of RuvB-like1 involve both ATPase-dependent and ATPase-independent activities (44). It is associated with a wide-range of cellular mechanisms relevant to tumorigenesis, including transcription regulation, DNA repair, cell growth, cell invasion, etc. The well-documented roles of RuvB-like1 involve the interaction with various important transcription factors related to cancer malignancy, including β-catenin and c-myc (45). RuvB-like1was observed in exosomes derived from the CRC cell line, HT29 (34), and a B cell line as well (46). Since the O-GlcNAc glycosylation of RuvB-like1 has never been reported, its O-GlcNAc sites were computationally predicted using two available websites including YinOYang and O-GlcNAcScan (http://www.cbs.dtu.dk/services/YinOYang/ and http:// cbsb.lombardi.georgetown.edu/hulab/OGAP.html). YinOYang predicted four O-GlcNAc sites, Thr74, Ser321, Ser322 and Thr402, while O-GlcNAcScan predicted five sites, Thr9, Thr153, Thr163, Thr269 and Ser424.
In conclusion, we demonstrated that O-GlcNAc modification of some extracellular proteins in CRC secretion was increased in comparison to that of normal colorectal cell secretion. This is consistent with our recent report on breast cancer cell secretion. Increased O-GlcNAcylation of extracellular proteins may open up a new aspect of this glycosylation in malignant phenotypes. Moreover, this study, for the first time, demonstrated that O-GlcNAcylation was present in EV proteins. Interestingly, O-GlcNAcylation of EVs derived from metastatic cells (SW620) was higher than that from non-metastatic cells (SW480). Three O-GlcNAc-modified proteins, including calsythenin-1, TER ATPase and RuVB-like1, were extracted from EVs of CRC cells. Even though the functions of these proteins in tumorigenesis have not been investigated, increased O-GlcNAcylation in metastatic cells indicated possible roles of this glycosylation in cancer metastasis. Furthermore, the alteration in O-GlcNAc extent on EVs proteins of metastatic cells may be of interest as a predictive biomarker for CRC metastasis and progression.
Acknowledgements
This work was supported by the Chulabhorn Research Institute, Chulabhorn Graduate Institute and National Science and Technology Development Agency (Grant no. P-12 01487), Thailand.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Penna C, Nordlinger B. Colorectal metastasis (liver and lung) Surg Clin North Am. 2002;82:1075–1090. doi: 10.1016/s0039-6109(02)00051-8. x-xi. [DOI] [PubMed] [Google Scholar]
- 3.Ciombor KK, Wu C, Goldberg RM. Recent therapeutic advances in the treatment of colorectal cancer. Annu Rev Med. 2015;66:83–95. doi: 10.1146/annurev-med-051513-102539. [DOI] [PubMed] [Google Scholar]
- 4.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 5.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Masyuk AI, Masyuk TV, Larusso NF. Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J Hepato. 2013;59:621–625. doi: 10.1016/j.jhep.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PloS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7:2088–2096. doi: 10.1021/pr700775x. [DOI] [PubMed] [Google Scholar]
- 9.Gangoda L, Boukouris S, Liem M, Kalra H, Mathivanan S. Extracellular vesicles including exosomes are mediators of signal transduction: are they protective or pathogenic? Proteomics. 2015;15:260–271. doi: 10.1002/pmic.201400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 11.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PloS One. 2010;5:e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 13.Vogt L, Schrimpf SP, Meskenaite V, Frischknecht R, Kinter J, Leone DP, Ziegler U, Sonderegger P. Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol Cell Neurosci. 2001;17:151–166. doi: 10.1006/mcne.2000.0937. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa M, Sawaguchi S, Furukawa K, Okajima T. N-acetylglucosamine modification in the lumen of the endoplasmic reticulum. Biochim Biophys Acta. 2015;1850:1319–1324. doi: 10.1016/j.bbagen.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 16.Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010;584:2526–2538. doi: 10.1016/j.febslet.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 17.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 19.Chaiyawat P, Netsirisawan P, Svasti J, Champattanachai V. Aberrant O-GlcNAcylated Proteins: New Perspectives in Breast and Colorectal Cancer. Front Endocrinol (Lausanne) 2014;5:193. doi: 10.3389/fendo.2014.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Netsirisawan P, Chokchaichamnankit D, Srisomsap C, Svasti J, Champattanachai V. Proteomic Analysis Reveals Aberrant O-GlcNAcylation of Extracellular Proteins from Breast Cancer Cell Secretion. Cancer Genomics Proteomics. 2015;12:201–209. [PubMed] [Google Scholar]
- 21.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, Xue Y, Xu T, Zhu HJ, Simpson RJ. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 22.Zachara NE, Vosseller K, Hart GW. Detection and analysis of proteins modified by O-linked N-acetylglucosamine. Curr Protoc Mol Biol Chapter: Unit-7.6. 2011 doi: 10.1002/0471142727.mb1706s95. doi:10.1002/0471142727.mb1706s95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaiyawat P, Chokchaichamnankit D, Lirdprapamongkol K, Srisomsap C, Svasti J, Champattanachai V. Alteration of O-GlcNAcylation affects serine phosphorylation and regulates gene expression and activity of pyruvate kinase M2 in colorectal cancer cells. Oncol Rep. 2015;34:1933–1942. doi: 10.3892/or.2015.4178. [DOI] [PubMed] [Google Scholar]
- 24.Srisomsap C, Sawangareetrakul P, Subhasitanont P, Panichakul T, Keeratichamroen S, Lirdprapamongkol K, Chokchaichamnankit D, Sirisinha S, Svasti J. Proteomic analysis of cholangiocarcinoma cell line. Proteomics. 2004;4:1135–1144. doi: 10.1002/pmic.200300651. [DOI] [PubMed] [Google Scholar]
- 25.Saint-Guirons J, Zeqiraj E, Schumacher U, Greenwell P, Dwek M. Proteome analysis of metastatic colorectal cancer cells recognized by the lectin Helix pomatia agglutinin (HPA) Proteomics. 2007;7:4082–4089. doi: 10.1002/pmic.200700434. [DOI] [PubMed] [Google Scholar]
- 26.Nagnan-Le Meillour P, Vercoutter-Edouart AS, Hilliou F, Le Danvic C, Levy F. Proteomic Analysis of Pig (Sus scrofa) Olfactory Soluble Proteome Reveals O-Linked-N-Acetylgluco-saminylation of Secreted Odorant-Binding Proteins. Front Endocrinol (Lausanne) 2014;5:202. doi: 10.3389/fendo.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 28.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat Commun. 2011;2:583. doi: 10.1038/ncomms1591. [DOI] [PubMed] [Google Scholar]
- 29.Sakaidani Y, Ichiyanagi N, Saito C, Nomura T, Ito M, Nishio Y, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem Biophys Res Commun. 2012;419:14–19. doi: 10.1016/j.bbrc.2012.01.098. [DOI] [PubMed] [Google Scholar]
- 30.Suila H, Hirvonen T, Ritamo I, Natunen S, Tuimala J, Laitinen S, Anderson H, Nystedt J, Rabina J, Valmu L. Extracellular o-linked N-acetylglucosamine is enriched in stem cells derived from human umbilical cord blood. Biores Open Access. 2014;3:39–44. doi: 10.1089/biores.2013.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, Kim YK, Kim KP, Gho YS. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles. 2012;1:10.3402/jev.v1i0.18704. doi: 10.3402/jev.v1i0.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagnoni A, Perkinton MS, Gray EH, Francis PT, Noble W, Miller CC. Calsyntenin-1 mediates axonal transport of the amyloid precursor protein and regulates Abeta production. Hum Mol Genet. 2012;21:2845–2854. doi: 10.1093/hmg/dds109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosseini-Beheshti E, Pham S, Adomat H, Li N, Tomlinson Guns ES. Exosomes as biomarker enriched microvesicles: characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes. Mol Cell Proteomics. 2012;11:863–885. doi: 10.1074/mcp.M111.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, Gho YS. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 35.Araki Y, Kawano T, Taru H, Saito Y, Wada S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, Matsuno K, Kinjo M, Suzuki T. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26:1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure – diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 37.Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnaghi P, D'Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O'Brien T, Jackson PK, Isacchi A. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 39.Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol Cell Proteomics. 2013;12:587–598. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dehennaut V, Slomianny MC, Page A, Vercoutter-Edouart AS, Jessus C, Michalski JC, Vilain JP, Bodart JF, Lefebvre T. Identification of structural and functional O-linked N-acetylglucosamine-bearing proteins in Xenopus laevis oocyte. Mol Cell Proteomics. 2008;7:2229–2245. doi: 10.1074/mcp.M700494-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Park J, Kwon H, Kang Y, Kim Y. Proteomic analysis of O-GlcNAc modifications derived from streptozotocin and glucosamine induced beta-cell apoptosis. J Biochem Mol Biol. 2007;40:1058–1068. doi: 10.5483/bmbrep.2007.40.6.1058. [DOI] [PubMed] [Google Scholar]
- 43.Huen J, Kakihara Y, Ugwu F, Cheung KL, Ortega J, Houry WA. Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem Cell Biol. 2010;88:29–40. doi: 10.1139/o09-122. [DOI] [PubMed] [Google Scholar]
- 44.Grigoletto A, Lestienne P, Rosenbaum J. The multifaceted proteins Reptin and Pontin as major players in cancer. Biochim Biophys Acta. 2011;1815:147–157. doi: 10.1016/j.bbcan.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17:187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Buschow SI, van Balkom BW, Aalberts M, Heck AJ, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol Cell Biol. 2010;88:851–856. doi: 10.1038/icb.2010.64. [DOI] [PubMed] [Google Scholar]


