Abstract
The prevalence and antibiotic susceptibility of intestinal carriage of Gram-negative bacteria among preterm infants admitted to the neonatal intensive care unit (NICU) in a tertiary teaching hospital in Malaysia were determined. A total of 34 stool specimens were obtained from preterm infants upon admission and once weekly up to two weeks during hospitalization. The presumptive colonies of Escherichia coli and Klebsiella pneumoniae were selected for identification, antibiotic susceptibility testing, and subtyping by using pulsed-field gel electrophoresis (PFGE). Out of 76 Gram-negative isolates, highest resistance was detected for amoxicillin/clavulanate (30.8%, n = 16), ceftriaxone (42.3%, n = 22), ceftazidime (28.8%, n = 15), cefoxitin (28.8%, n = 15), aztreonam (36.5%, n = 19), and polymyxin B (23.1%, n = 12). Three colistin resistant K. pneumoniae have also been detected based on E-test analysis. Thirty-nine isolates of K. pneumoniae and 20 isolates of E. coli were resistant to more than three antimicrobial classes and were categorized as multidrug resistant (MDR). PFGE analysis revealed a higher diversity in pulsotypes for K. pneumoniae (18 pulsotypes) in comparison to E. coli (four pulsotypes). In addition, a total of fifteen pulsotypes was observed from 39 MDR K. pneumoniae. The risk factors for antibiotic resistance were assessed using random forest analysis. Gender was found to be the most important predictor for colistin resistant while length, OFC, and delivery mode were showing greater predictive power in the polymyxin B resistance. This study revealed worrying prevalence rates of intestinal carriage of multidrug-resistant K. pneumoniae and E. coli of hospitalized preterm infants in Malaysia, particularly high resistance to polymyxins.
Keywords: Preterm infants, Meconium, Multidrug resistant, Gram-negative enteric, PFGE
Introduction
Infants admitted to neonatal intensive care units (NICUs) are at high risk for developing healthcare-associated infection due to the severity of their illness and exposure to invasive medical devices. Healthcare-associated neonatal infection caused by multidrug-resistant Gram-negative (MDRGN) bacilli has emerged as a substantial health problem in developing countries.1,2 Several studies have been carried out to characterize the epidemiology of nosocomial MDRGN by identifying risk factors for their acquisition and to trace horizontal transmission between newborns.3−6 However, the studies have yielded quite different findings because the transmission of MDRGN is mainly dependent on the interaction between human and environmental ecology of NICUs.5
The infections with Gram-negative bacilli that are resistant to multiple antibacterial drugs have been increasingly reported in NICUs.2,7 This is mainly due to empiric treatment with antimicrobials in the NICUs. Repeated courses of antibiotic treatment and long-term exposure to antibiotics have resulted in an increase in antibiotic-resistant organisms.8,9 In addition, antibiotics are also administered to preterm infants who are at risk of infections soon after birth, especially those with very low birth weight.10
The frequent use of broad-spectrum antibiotics in hospital has given rise to the selection of resistant strains.11 It has also been recognized that antibiotic resistance in the nosocomial setting is an important factor that influences the neonatal outcomes. For instance, high fecal carriage rates have been identified as a significant risk factor associated with ESBL K. pneumoniae infections.12 Nevertheless, few studies have elucidated such risk in neonatal ESBL Klebsiella spp. colonization.13 In Malaysia, MDRGN infection among infants remains a major threat. Previously, Ariffin et al.7 have defined the pattern of antibiotic resistance amongst Gram-negative bacilli in NICU and PICU, where all isolates showed low susceptibility rates to the third generation cephalosporins (47–62%) and more than 50% of K. pneumoniae and E. coli isolates were extended-spectrum beta-lactamase (ESBL) producing strains.7 K. pneumoniae is among the commonest Gram-negative bacteria causing late-onset bacteremia in the NICU of a tertiary teaching hospital located in northeastern Malaysia.14 This pathogen was associated with nosocomial infection and high infection rates among ventilated infants.15–17
The present study was conducted in a tertiary level III NICU in Malaysia. The objectives were to determine the antimicrobial susceptibility patterns and pulsotypes of K. pneumoniae and E. coli isolated from the NICU where broad-spectrum antibiotics are frequently used. The outcome of the antibiotics currently used for empirical therapy was also recorded and associated with the demographic and clinical characteristics of the infants.
Materials and methods
Patient and study design
Setting
Preterm infants (gestational age of less than 37 weeks) admitted to the NICU of a tertiary hospital in Kuala Lumpur, Malaysia, were prospectively enrolled and sampled. No exclusion criterion was applied to the preterm infants group. This study did not involve any intervention apart from the routine medical procedures, but the major demographic and clinical data for each participant were extracted from the hospital records (Table 1). Informed consents were obtained from parents and the study protocol was approved by the institutional ethics committee (MEC ID: 201310-0267). Confidentiality and anonymity of the subjects were maintained, and sample collection was carried out according to institutional ethics guidelines.
Table 1.
Demographic and clinical information of the infant cohort
| Infant | Gestational age (weeks) | Race | Gender | Birth weight (g) | Birth length (cm) | Birth head of circumference (cm) | Maternal GPA | Delivery mode | Sample collection (day of life) | Isolation of K. pneumoniae | Isolation of E. coli |
|---|---|---|---|---|---|---|---|---|---|---|---|
| B1 | 33 | Chinese | Female | 1570 | 40 | 28 | G1P0A0 | Cesarean section | 0, 7, 14 | Positive | Positive |
| B2 | 33 | Chinese | Male | 1700 | 39 | 28 | G4P2A1 | Cesarean section | 0, 7 | Positive | Positive |
| B4 | 32 | Malay | Male | 1505 | 38 | 28 | G1P0A0 | Vaginal | 0, 7, 14 | Negative | Negative |
| B6 | 30 | Chinese | Female | 1390 | 37 | 28 | G1P0A0 | Cesarean section | 0, 7, 14 | Positive | Negative |
| B7 | 30 | Chinese | Female | 490 | 28 | 21 | G1P0A0 | Cesarean section | 0, 7, 14 | Negative | Negative |
| B8 | 30 | Malay | Male | 1295 | 37 | 26.5 | G1P0A0 | Vaginal | 0, 7, 14 | Positive | Positive |
| B9 | 35 | Indian | Female | 1455 | 40 | 28 | G2P1A0 | Cesarean section | 0, 7, 14 | Positive | Positive |
| B11 | 36 | Malay | Female | 1555 | 37 | 29.2 | G1P0A0 | Cesarean section | 0, 14 | Positive | Positive |
| B12 | 34 | Chinese | Female | 2845 | 45 | 33 | G3P2A0 | Cesarean section | 0 | Negative | Negative |
| B13 | 32 | Malay | Female | 1420 | 41 | 26.5 | G3P0A2 | Cesarean section | 0, 7, 14 | Positive | Positive |
| B14 | 32 | Malay | Female | 785 | 32.5 | 25.5 | G3P0A2 | Cesarean section | 0, 7, 14 | Positive | Positive |
| B15 | 32 | Malay | Female | 1540 | 38 | 28 | G1P0A0 | Cesarean section | 0 | Negative | Negative |
| B16 | 30 | Malay | Female | 902 | 34 | 25 | G1P0A0 | Cesarean section | 0 | Negative | Negative |
| B17 | 30 | Malay | Female | 1240 | 39 | 26.5 | G1P0A0 | Cesarean section | 0 | Positive | Negative |
| B18 | 30 | Malay | Male | 1220 | 37.5 | 26.3 | G1P0A0 | Cesarean section | 0 | Positive | Negative |
| B19 | 33 | Indian | Male | 1905 | 41 | 28 | G1P0A0 | Vaginal | 0 | Negative | Negative |
Collection of specimens
Stool specimens were obtained within 48 h of admission, and on the 7th day and 14th day of life during their stay in the NICU. All the samples were stored at −30 °C until analysis.
Microbiological methods
Isolation of Gram-negative bacteria
Stool specimens were diluted 1:10 in phosphate-buffered saline (PBS) and inoculated onto CHROMagar Orientation plates (CHROMagar™ Orientation; Becton Dickenson, Heidelberg, Germany). The plates were incubated aerobically at 37 °C. After 20 h, the plates were examined for growth and colony characteristics. E. coli presented pink colonies while K. pneumoniae exhibited metallic blue colonies on the CHROMagar Orientation plates. Three to five colonies for each distinct color group were randomly selected and cultured on a non-selective agar.
Identification
Overnight culture of the isolates was suspended and washed in PBS. Cells were centrifuged at 13,000 rpm for two minutes to pellet the cells. A volume of 200 μL of ultrapure water was added to each tube. The cells were then subjected to heating at 99 °C for 10 min and snapped cool on ice for 5 min. The PCR specific identification of E. coli was performed using the primers phoA1 (5′GTG ACA AAA GCC CGG ACA CCA TAA AT 3′) and phoA2 (5′TAC ACT GTC ATT ACG TTG CGG ATT TGG 3′)18 while identification of K. pneumoniae was performed using the primer pair mdh1 (5′CCC AAC TCG CTT CAG GTT CAG 3′) and mdh2 (5′CCG TTT TTC CCC AGC AGC AG 3′).19 PCRs for the identification of E. coli and K. pneumoniae were performed in Veriti® 96-Well Fast Thermal Cycler (Applied Biosystems, Califonia, USA) as previously described by Kong et al.18 and Diancourt et al.19, respectively. Gel electrophoresis was carried out by applying 2 μL of PCR products to 1% agarose gel. Gels were run for 30 min at 100 V in 0.5x TBE (tris-borate-EDTA) buffer. DNA molecular weight marker 100 bp DNA ladder (Promega, Wisconsin, USA) was used as size standard. Gels were visualized by UV illumination after staining with SYBR® Safe DNA Gel Stain (Life Technologies, CA, USA).
Determination of susceptibility
Susceptibility testing was performed by disk diffusion test according to the CLSI guidelines.20 The 0.5 McFarland (1.0 for mucoid phenotype) of test organism was uniformly inoculated over the Mueller-Hinton agar (MHA). Susceptibilities of the isolates toward 14 antibiotics such as amoxicillin (10 μg), amoxicillin/ clavulanic acid (20/10 μg), ceftriaxone (30 μg), ceftazidime (30 μg), cefoxitin (30 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg), gentamicin (10 μg), tobramycin (10 μg), ciprofloxacin (5 μg), polymyxin B (300unit), colistin sulfate (polymyxin E; 10 μg), and chloramphenicol (30 μg) were determined. E.coli ATCC 25922, P. aeruginosa ATCC 27853, and K. pneumoniae ATCC 700603 were used as quality control strains for antibiotic susceptibility testing. Isolates showing intermediate antimicrobial susceptibility were considered to be resistant. Isolates were assigned as multiple drug resistance (MDR) when they are resistant to at least one member from three different antibiotic classes.21 In addition, based on the CLSI guidelines, putative ESBL producers were determined.
E-test for colistin-resistant strains
The colistin E-test (AB Biodisk; Biomérieux, Sweden) was performed on MHA according to the manufacturer’s procedures guidelines. The MIC was determined at the intersection of the inhibition zone with the strip. Resistant strains were defined as isolates with MICs for colistin > 2 mg/L using the CLSI breakpoint for Pseudomonas aeruginosa.
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed according to established protocol from PulseNet USA.22 Genomic DNA of E. coli and K. pneumoniae was digested with XbaI restriction enzyme (12 U per plug) (Promega, Wincousin, USA). XbaI-digested Salmonella Braenderup (H9812) was used as the DNA size marker. Pulsotypes were analyzed using BioNumerics v6.0 software (Applied Maths NV, Sint-Martens-Latem, Belgium). The variability of the strains was determined on the basis of the Dice coefficient of similarity (F) and the unweighted pair group method with arithmetic averages (UPGMA) clustering analysis at 1.5% position tolerance. Interpretive clustering of the PFGE patterns was based on the banding patterns and the variability of the isolates as previously described by Barrett et al.23
Risk assessment
Eleven demographic variables, including gestational age, gender, race, birth weight, length, head circumferences, delivery mode, maternal gravida/para/abortus (GPA), and breastfeeding were tested as independent variables in the classification of the antibiotic resistance profiles such as resistant to at least three beta-lactam antibiotics, resistant to polymyxin B, resistant to colistin (based on E-test), and resistant to gentamicin. Random forest was carried out using Rattle R program package to identify and validate predictors with respect to the risk factors for antibiotic resistance.24 This recursive partitioning method is selected as it has been shown to perform well under the small sample size and relatively larger categorical predictors problems.25
Results
Microbiological methods
In total, 34 stool specimens from 16 infants were collected. Most of the meconium samples were unculturable using selective agar. K. pneumoniae and E. coli were isolated from ten (29.4%) and seven (20.6%) specimens, respectively. Demographic and clinical information of the infant cohort were also extracted and tabulated in Table 1. In total, 24 E. coli and 52 K. pneumoniae were isolated, identified and tested for antimicrobial resistance. Disk diffusion test was carried out on the K. pneumoniae and E. coli isolates (Table 2). For K. pneumoniae isolates, high resistance rates were detected for amoxicillin/ clavulanate (30.8%, n = 16), ceftriaxone (42.3%, n = 22), ceftazidime (28.8%, n = 15), cefoxitin (28.8%, n = 15), aztreonam (36.5%, n = 19), and polymyxin B (23.1%, n = 12). Compared to K. pneumoniae, E. coli isolates were overall more resistant to the tested third generation cephalosporins such as ceftriaxone (83.3%, n = 20) and ceftazidime (70.8%, n = 17). High resistance rates were also observed for amoxicillin (91.7%, n = 22) and aztreonam (83.3%, n = 20). None of the isolates was resistant to gentamicin, tobramycin, ciprofloxacin, colistin, and chloramphenicol. Eight K. pneumoniae and three E. coli isolates were resistant to either imipenem or meropenem. Seven K. pneumoniae isolates showed resistance to colistin based on the disk diffusion method and were further confirmed by using E-test. Out of the seven colistin-resistant isolates tested using E-test, three isolates were confirmed as colistin-resistant with the MIC more than 256 μg/mL. Based on the CLSI initial screening test for Extended Spectrum Beta-Lactamase (ESBLs), 24 isolates of K. pneumoniae and 21 isolates of E. coli were identified as putative ESBL-producers, subject to further confirmatory tests. Twenty isolates of K. pneumoniae and six isolates of E. coli were resistant to more than three antimicrobial classes, hence categorized as MDR strains.
Table 2.
Antimicrobial activity of 16 antibiotics tested against K. pneumoniae and E. coli by disk diffusion test
| No. of K. pneumoniae isolates (n = 52) |
No. of E. coli isolates (n = 24) |
|||||
|---|---|---|---|---|---|---|
| Antibiotics | Susceptible | Resistant | Resistance rate (%) | Susceptible | Resistant | Resistance rate (%) |
| Amoxicillin (AML) | 0 | 52 | 100 | 2 | 22 | 91.7 |
| Amoxicillin/clavulanic acid (AMC) | 36 | 16 | 30.8 | 23 | 1 | 4.2 |
| Ceftriaxone (CRO) | 30 | 22 | 42.3 | 4 | 20 | 83.3 |
| Ceftazidime (CAZ) | 37 | 15 | 28.8 | 7 | 17 | 70.8 |
| Cefoxitin (FOX) | 37 | 15 | 28.8 | 23 | 1 | 4.2 |
| Aztreonam (ATM) | 33 | 19 | 36.5 | 4 | 20 | 83.3 |
| Imipenem (IMP) | 47 | 5 | 9.6 | 22 | 2 | 8.3 |
| Meropenem (MEM) | 49 | 3 | 5.8 | 23 | 1 | 4.2 |
| Gentamicin (CN) | 43 | 9 | 17.3 | 24 | 0 | 0 |
| Tobramycin (TOB) | 45 | 7 | 13.5 | 24 | 0 | 0 |
| Ciprofloxacin (CIP) | 48 | 4 | 7.7 | 24 | 0 | 0 |
| Polymyxin B (PB) | 40 | 12 | 23.1 | 23 | 1 | 4.2 |
| Colistin sulfate (CT) | 45 | 7 | 13.5 | 24 | 0 | 0 |
| Chloramphenicol (C) | 0 | 52 | 100 | 24 | 0 | 0 |
Pulsed-field gel electrophoresis
For isolates from the same specimen, only those showing distinct antibiogram were subjected to PFGE. Isolates from the same specimen with identical antibiotic profile were omitted in this experiment. Among the 37 selected isolates, K. pneumoniae yielded only 18 different pulsotypes (KPXbal 1–18; Fig. 1) and one isolate from B2 was untypable. Isolates from specimens of infant B9 (K11, K14, K16, K44, K46, K47, and K48) yielded the highest number of different pulsotypes (three pulsotypes) while isolates from infant B6 (K37, K39, and K41), B17 (K30, K57, and K58), and B18 (K33 and K35) gave variable MDR profiles despite displaying identical pulsotype. At the similarity of 80%, three major clusters were observed. Cluster A was comprised of isolates from infant B6 (day 7 and day 14) and B9 (day 14). Cluster B was the largest cluster, which mainly composed of isolates from B8 (day 7 and day 14), B13 (day 7 and day 14), and B14 (day 7 and day 14). Finally, cluster C was comprised of isolates from B8 (day 7 and day 14) and B9 (day 7 and day 14). Interestingly, the isolates from B13 and B14 although displayed different antibiotic susceptibility profiles, were sharing similar pulsotypes (KPXbaI 8, 10, and 15). The 37 MDR isolates were distributed among 15 distinct pulsotypes, indicating low levels of clonality among the MDR strains. E. coli isolates yielded only five different pulsotypes (ECXbaI 1–4; Fig. 2). Isolates from day 7 and day 14 collection of infant B1 showed same pulsotype but different antibiograms. For instance, isolate E1 was multidrug resistant (MDR) while E3 was only resistant to imipenem. ECXbaI 1 was the most common pulsotype which shared by isolates from B8 (day 14), B9 (day 14), B11 (day 14), B13 (day 7 and day 14), B14 (day 7 and day 14). Similar to K. pneumoniae, isolates from B13 and B14 shown high similarity and were up of the same pulsotype regardless of collection date and antibiotic susceptibility profiles.
Figure 1.
PFGE profiles of the non-repeated Klebsiella pneumonaie strains of different antibiotic susceptibility profiles that isolated from preterm infants during hospitalization.
Notes: Stool samples were collected in 3 timepoints, namely D1: day 1 (meconium); D7: day 7, and D14: day 14. Three major clusters were observed based on the similarity of 80%. Putative ESBL producers were indicated with*. CLSI recommended breakpoints for screening of E.coli and K. pneumoniae by disk diffusion for ESBL producers confirmatory test are lower than the CLSI published disk diffusion breakpoints for resistance. Thus, many isolates that have been categorized as susceptible may be considered as putative ESBL producers.
Figure 2.
PFGE profiles of the non-repeated Escherichia coli strains of different antibiotic susceptibility profiles that isolated from preterm infants during hospitalization.
Notes: Stool samples were collected in 3 timepoints, namely D1: day 1 (meconium); D7: day 7, and D14: day 14. However, E. coli was not isolated from the meconium samples. Majority of the strains were of pulsotype ECXbal I. Putative ESBL producers were indicated with *. CLSI recommended breakpoints for screening of E.coli and K. pneumoniae by disk diffusion for ESBL producers confirmatory test are lower than the CLSI published disk diffusion breakpoints for resistance. Thus, many isolates that have been categorized as susceptible may be considered as putative ESBL producers.
Risk assessment
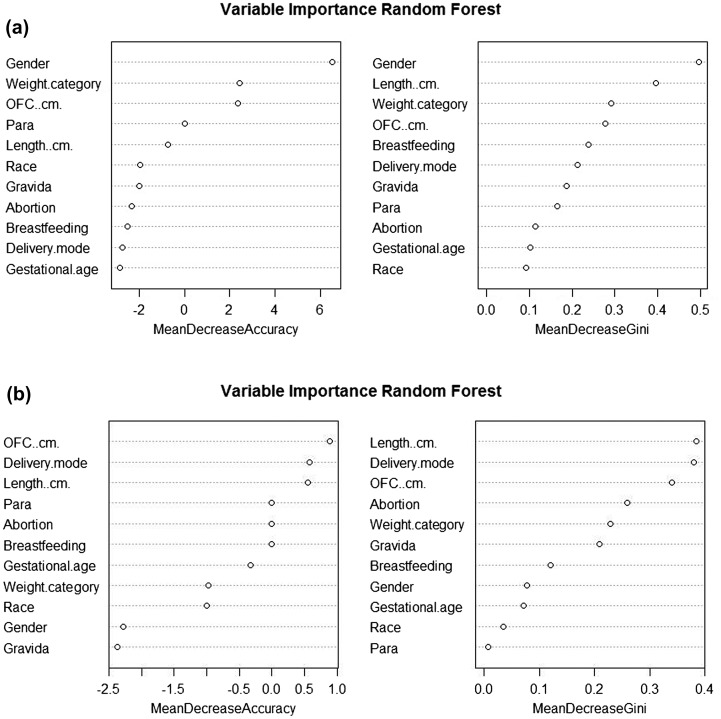
The risk factors for antibiotic resistance were assessed using random forest analysis. Using 11 demographic and clinical parameters, the predictive power of 62.64 and 81.82% was achieved in predicting the pattern of colistin and polymyxin resistance bacteria, respectively. Gender was found to be the most important predictor for the former while length, OFC, and delivery mode were showing greater predictive power in the latter (Fig. 3(A) and (B)).
Figure 3.
Variable importance based on random forest model. (A) Colistin resistance pattern, (B) Polymyxin B.
Discussion
Several multi-center studies have been carried out in Malaysia which included data from pediatrics and neonatal ICUs. Ariffin et al.7 reported on resistance patterns from three intensive care units of two hospitals in 2004. The isolates showed low susceptibility rates to the third-generation cephalosporins (47–62%) compared to imipenem (90%) and ciprofloxacin (99%).7 In our present study, reduced susceptibility of amoxicillin/clavulanic acid (18%) was also observed. ESBL production is a major mechanism of resistance to beta-lactam antibiotics. E. coli and K. pneumoniae are the most common ESBL producers and were identified as the causative agents of nosocomial infections. Third-generation cephalosporins resistant and ESBL selective media such as MacConkey agar plate containing 1 mg/L cefotaxime (CTX) could be used to detect ESBL/AmpC producing Enterobacteriaceae; however, this method was unable to detect Oxa 48-like carbapenemase producers and non-lactose fermenting E. coli could also be grown on the agar.26 The efficiencies of ESBL detection of five ESBL selective media were determined previously and the results showed that none of them were fully ESBL selective.27 Hence, in this study, CHROMagar was used as the primary screening medium as it provides good discrimination of common bacteria species in the mixed cultures isolated from clinical specimens.28 Then, disk diffusion test was employed after the primary screening. In addition, ESBL-producing colonies and non-ESBL-producing colonies could be found in the same culture passage as ESBL production is usually plasmid mediated. Therefore, it was suggested that several colonies must be picked and tested from the primary culture for optimal detection.29
All the results of antibiotic susceptibility tests in this study were reproducible. Based on the carbepenem susceptibility patterns, the isolates could be divided into two phenotypes namely the imipenem-resistant meropenem-susceptible phenotype and imipenem-susceptible meropenem-resistant phenotype. Weak hydrolysis of meropenem compared to imipenem has been previously reported mainly among the strains that harbored Oxa 54 and Oxa 48.30−32 On the other hand, the specific activity of Oxa 244 for imipenem, absence of ompK 35, and the defects in ompK36 was reported to play significant roles in the occurrence of imipenem-susceptible and meropenem-resistant phenotype.33,34 In this study, strain E1 with cephalosporins resistance and penicillin susceptible has been observed. Amoxicillin/clavulanate is one of the widely used antibiotics, which has good activity against non-beta-lactamase-producing strains as well as some beta-lactamase producing strains.35 Amoxicillin/clavulanate susceptible ESBL strains are rare, however, has been reported previously by Kumar et al.36 and Prakash et al.37
Nucleic acid-based molecular typing methods such as PFGE still remain as invaluable tools in establishing clonal patterns of isolates. PFGE enables the greatest discriminatory capacity compared to other techniques, making it a very robust tool in the study of epidemiology and public health.38 In this study, no direct association was established between the resistant phenotypes and XbaI-pulsotypes by cluster analysis of the K. pneumoniae and E. coli isolates. The E. coli isolates were highly clonal yet with different patterns of resistance to antibiotics. This might be due to the phenomenon where the clonal isolates harboring the same ESBL associated genes that were differentially expressed and the loss of porins or mutations which were not located at the PFGE restricted enzymes cutting sides.39−41 The finding is consistent with Benacer et al.42 which showed that PFGE could not distinguish similar pulsotypes with different resistance profiles. To increase the discriminatory power of PFGE, the use of more than one enzyme was previously recommended.43 Nevertheless, in this study, PFGE was able to discriminate isolates from the same individual that exhibited identical resistant profile but different pulsotypes.
Preterm infants are susceptible to Gram-negative bacilli and they are most likely to be infected during or shortly before birth from the colonized maternal genital tract.44 Based on the PFGE results, the strains that colonised the only pair of twins, B13 and B14 were clonal (KPXbal 8, KPXbal 10, KPXbal 15). Unfortunately, intrapartum antibiotic prophylaxis of the mother could not be traced and we also failed to obtain the high vaginal swab from the mother at the time of labor, hence pathway of colonization could not be determined. The results of this study will be more meaningful, especially if we were able to assess the risk of neonatal antibiotic resistant incidence in correlation to the antibiotic pressure to mothers, especially intrapartum antibiotic prophylaxis, which has been associated with neonatal sepsis or antimicrobial-resistant.44 In addition, similar strains (clonal) were also observed in different infants. For instance, the E. coli isolates of pulsotype ECXbal 1 that present in infants B8, B9, B13, and B14; K. pneumoniae of pulsotype KPXbal 10 in infant B8 and the twins (B13 and B14) and KPXbal 13 that observed for isolates from infants B9 and B8. These suggested the clonal spread of strains between the infants in the ward and physical contact might be most likely the mode of transmission.5
It has been considered that the intestinal tract and meconium were sterile at birth, and later being rapidly colonized with micro-organisms from the mother and the surrounding environment. However, several studies have shown that the meconium from healthy hosts is not sterile and gut colonization may have started before birth.45–47 A study on the bacterial diversity of meconium in preterm infants showed an association between low bacterial diversity in meconium and high risk to develop sepsis.48 In the present study, the isolation rate from meconium samples was low and only K. pneumoniae was isolated from meconium of infants B17 and B18. This is in agreement with Moles et al.49 who reported low isolation rates of Escherichia spp and Klebsiella spp in meconium of preterm infants. To overcome the limitation of culture-dependent techniques, a more robust culture-independent approach such as next generation metagenomics analysis should be applied in future to establish the bacterial communities in these infants. This may provide new clues on the initial gut colonizers in the Asian cohort which remains less studied in comparison to the western populations.
In this study, the risks of infection/colonization by the last treatment options for Gram-negative bacteria infections have been identified. For instance, gender was found to be an important factor for colistin resistance pattern. Apart from this study, male babies were reported to be more predominant to get early-onset neonatal sepsis previously.50 Moreover, our study also showed that delivery mode and neonate development (i.e. length and OFC) were important parameters for polymyxin B resistance pattern. A low worldwide colistin resistance at 0.9–3.3% was reported in the surveillance data recorded from 2001 to 2011.51 However, in contradiction with this previous finding, high resistance rate of colistin (6%) in this study is worrying. Furthermore, in the recent report, the identification of plasmid-mediated mcr-1 gene in polymyxin resistant in Enterobacteriaceae raises the possibility of widespread dissemination of resistance.52
In our present study, Gram-negative isolates detected from hospitalized preterm infants are still highly susceptible to gentamicin and meropenem despite their heavy usage in the particular unit studied. Many reports have associated the bacterial resistance with uncontrolled of antibiotic use and particularly in the treatment of neonatal sepsis.53 The highest rate of resistance is often observed in settings with the highest rates of antibiotic prescription.54,55 E. coli is one of the main sources of neonatal sepsis and highly resistant to commonly used antibiotics such as amoxicillin and ceftriaxone, and relatively more sensitive to less commonly used drugs like ceftazidime.56 However, our study provided evidence that reduced susceptibility of ceftazidime has been observed in E. coli isolates, which was in agreement with the findings of Yuan et al.57
Polymyxin B and colistin were the two polymyxins which have been used clinically against Gram-negative bacteria. Disk diffusion was reported to have an acceptable rate of errors and was useful for screening of polymyxin resistant strain, thus disk diffusion was used to detect polymyxin B resistant strains. For colistin resistant, E-test was further used after the initial screening using disk diffusion. Although low sensitivity of E-test on detecting colistin resistant strains has been reported recently,58 our results in this study was considered reliable as no zone of inhibition was observed for three colistin strains in two repeated experiments. However, the strains will be further subjected to the other MIC detection methods, such as broth macrodilution (TDS) and broth microdilution with polysorbate 80 (BMD-T) for further comparison and evaluation. In addition, a bigger sample size should be included in future studies to increase the robustness of the statistical analyses.
Notwithstanding the limitation, our results affirmed the prevalence of drug resistance of E. coli and K. pneumoniae in hospitalized preterm infants in Malaysia. A continuous surveillance of antimicrobial resistance in NICU should be carried out to identify the risk for infants and to assist in the empirical antibiotic therapy decision.
Funding
This study was supported by University of Malaya Research Grant (UMRG: RG535-13HTM) and Postgraduate Research Grant (PPP: PG179-2015A).
References
- 1.Couto RC, Carvalho EA, Pedrosa TM, Pedroso ER, Neto MC, Biscione FM. A 10-year prospective surveillance of nosocomial infections in neonatal intensive care units. Am J Infect Control. 2007. Apr;35(3):183–9. 10.1016/j.ajic.2006.06.013 [DOI] [PubMed] [Google Scholar]
- 2.Litzow JM, Gill CJ, Mantaring JB, Fox MP, MacLeod WB, Mendoza M, et al. High frequency of multidrug-resistant gram-negative rods in 2 neonatal intensive care units in the Philippines. Infect Control Hosp Epidemiol. 2009. Jun;30(6):543–9. 10.1086/600400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almuneef MA, Baltimore RS, Farrel PA, Reagan-Cirincione P, Dembry LM. Molecular typing demonstrating transmission of gram-negative rods in a neonatal intensive care unit in the absence of a recognized epidemic. Clin Infect Dis. 2001. Jan 15;32(2):220–7. 10.1086/318477 [DOI] [PubMed] [Google Scholar]
- 4.Waters V, Larson E, Wu F, San Gabriel P, Haas J, Cimiotti J, et al. Molecular epidemiology of gram-negative bacilli from infected neonates and health care workers’ hands in neonatal intensive care units. Clin Infect Dis. 2004. Jun 15;38(12):1682–7. 10.1086/386331 [DOI] [PubMed] [Google Scholar]
- 5.Mammina C, Di Carlo P, Cipolla D, Giuffre M, Casuccio A, Di Gaetano V, et al. Surveillance of multidrug-resistant gram-negative bacilli in a neonatal intensive care unit: prominent role of cross transmission. Am J Infect Control. 2007. May;35(4):222–30. 10.1016/j.ajic.2006.04.210 [DOI] [PubMed] [Google Scholar]
- 6.Millar M, Philpott A, Wilks M, Whiley A, Warwick S, Hennessy E, et al. Colonization and persistence of antibiotic-resistant Enterobacteriaceae strains in infants nursed in two neonatal intensive care units in East London, United Kingdom. J Clin Microbiol. 2008. Feb;46(2):560–7. 10.1128/JCM.00832-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariffin H, Navaratnam P, Kee TK, Balan G. Antibiotic resistance patterns in nosocomial gram-negative bacterial infections in units with heavy antibiotic usage. J Trop Pediatr. 2004. Feb;50(1):26–31. 10.1093/tropej/50.1.26 [DOI] [PubMed] [Google Scholar]
- 8.Zaidi AK, Huskins WC, Thaver D, Bhutta ZA, Abbas Z, Goldmann DA. Hospital-acquired neonatal infections in developing countries. Lancet. 2005. Mar 26–Apr 1;365(9465):1175–88. 10.1016/S0140-6736(05)71881-X [DOI] [PubMed] [Google Scholar]
- 9.Bizzarro MJ, Gallagher PG. Antibiotic-resistant organisms in the neonatal intensive care unit. Semin Perinatol. 2007. Feb;31(1):26–32. 10.1053/j.semperi.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 10.Beck S, Wojdyla D, Say L, Pilar Bertran AP, Meraldi M, Harris Requejo JH, et al. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ. 2010. Jan;88(1):31–8. 10.2471/BLT.00.00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Man P, Verhoeven BAN, Verbrugh HA, Vos MC, van den Anker JN. An antibiotic policy to prevent emergence of resistant bacilli. Lancet. 2000. Mar 18;355(9208):973–8. [DOI] [PubMed] [Google Scholar]
- 12.Pena C, Pujol M, Ardanuy C, Ricart A, Pallares R, Linares J, et al. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1998. Jan;42(1):53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boo NY, Ng SF, Lim VK. A case-control study of risk factors associated with rectal colonization of extended-spectrum beta-lactamase producing Klebsiella sp. in newborn infants. J Hosp Infect. 2005. Sep;61(1):68–74. 10.1016/j.jhin.2005.01.025 [DOI] [PubMed] [Google Scholar]
- 14.Ariffin N, Hasan H, Ramli N, Ibrahim NR, Taib F, Rahman AA, et al. Comparison of antimicrobial resistance in neonatal and adult intensive care units in a tertiary teaching hospital. Am J Infect Control. 2012. Aug;40(6):572–5. 10.1016/j.ajic.2012.02.032 [DOI] [PubMed] [Google Scholar]
- 15.Wan Hanifah W, Lee J, Quah B. Comparison of the pattern of nosocomial infection between the neonatal intensive care units of hospitals kuala terengganu and universiti sains malaysia, kelantan. Malays J Med Sci. 2000. Jan;7(1):33–40. [PMC free article] [PubMed] [Google Scholar]
- 16.Wong NA, Hunt LP, Marlow N. Risk factors for developing neonatal septicaemia at a Malaysian hospital. J Trop Pediatr. 1997. Feb;43(1):54–8. 10.1093/tropej/43.1.54 [DOI] [PubMed] [Google Scholar]
- 17.Halder D, Haque ME, Zabidi MH, Kamaruzzaman A. Nosocomial bacterial sepsis in babies weighing 1000-1499 g in Kelantan. Med J Malays. 1999. Mar;54(1):52–7. [PubMed] [Google Scholar]
- 18.Kong RYC, So CL, Law WF, Wu RSS. A sensitive and versatile multiplex PCR system for the rapid detection of enterotoxigenic (ETEC), enterohaemorrhagic (EHEC) and enteropathogenic (EPEC) strains of Escherichia coli. Mar Poll Bull. 1999. Dec;38(12):1207–15. 10.1016/S0025-326X(99)00164-2 [DOI] [Google Scholar]
- 19.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005. Aug;43(8):4178–82. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. M100-S25 Wayne, Pennsylvania: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 21.Tanwar J, Das S, Fatima Z, Hameed S. Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis. 2014;2014 Article ID 541340, 7 p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Centers for Disease Control and Prevention; 2013. Available from: http://www.pulsenetinternational.org/assets/PulseNet/uploads/pfge/PNL05_Ec-Sal-ShigPFGEprotocol.pdf [Google Scholar]
- 23.Barrett TJ, Gerner-Smidt P, Swaminathan B. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog Dis. 2006;3(1):20–31. 10.1089/fpd.2006.3.20 [DOI] [PubMed] [Google Scholar]
- 24.Graham W. Data mining with rattle and R. New York: Springer Verlag; 2011. [Google Scholar]
- 25.Breiman L. Random forests. Mach. Learn. 2001. Oct 1;45(1):5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 26.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012. Jul;67(7):1597–606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 27.Grohs P, Tillecovidin B, Caumont-Prim A, Carbonnelle E, Day N, Podglajen I, et al. Comparison of five media for detection of extended-spectrum beta-lactamase by use of the wasp instrument for automated specimen processing. J Clin Microbiol. 2013. Aug;51(8):2713–6. 10.1128/JCM.00077-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. Evaluation of CHROMagar orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J Clin Microbiol. 1996. Jul;34(7):1788–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coudron PE, Moland ES, Sanders CC. Occurrence and detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae at a veterans medical center: seek and you may find. J Clin Microbiol. 1997. Oct;35(10):2593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–8. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moquet O, Bouchiat C, Kinana A, Seck A, Arouna O, Bercion R, et al. Class D OXA-48 carbapenemase in multidrug-resistant enterobacteria, Senegal. Emerg Infect Dis. 2011;17(1):143–4. 10.3201/eid1701.100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. 10.1128/AAC.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayama S, Koba Y, Shigemoto N, Kuwahara R, Kakuhama T, Kimura K, et al. Imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae producing OXA-181 in Japan. Antimicrob Agents Chemother. 2015;59(2):1379–80. 10.1128/AAC.04330-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potron A, Poirel L, Dortet L, Nordmann P. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like beta-lactamase from Escherichia coli. Int J Antimicrob Agents. 2016;47(1):102–3. 10.1016/j.ijantimicag.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 35.White AR, Kaye C, Poupard J, Pypstra R, Woodnutt G, Wynne B. Augmentin (amoxicillin/clavulanate) in the treatment of community-acquired respiratory tract infection: a review of the continuing development of an innovative antimicrobial agent. J Antimicrob Chemother. 2004;53 Suppl 1:i3–20. 10.1093/jac/dkh050 [DOI] [PubMed] [Google Scholar]
- 36.Kumar D, Singh AK, Ali MR, Chander Y. Antimicrobial susceptibility profile of extended spectrum beta-lactamase (ESBL) producing Escherichia coli from various clinical samples. Infect Dis. 2014;7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash V, Lewis JS 2nd, Herrera ML, Wickes BL, Jorgensen JH. Oral and parenteral therapeutic options for outpatient urinary infections caused by Enterobacteriaceae producing CTX-M extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2009;53(3):1278–80. 10.1128/AAC.01519-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garland SM, Mackay S, Tabrizi S, Jacobs S. Pseudomonas aeruginosa outbreak associated with a contaminated blood-gas analyser in a neonatal intensive care unit. J Hosp Infect. 1996. Jun;33(2):145–51. 10.1016/S0195-6701(96)90099-7 [DOI] [PubMed] [Google Scholar]
- 39.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–86. 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice LB, Carias LL, Hujer AM, Bonafede M, Hutton R, Hoyen C, et al. High-level expression of chromosomally encoded SHV-1 beta-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000. Feb;44(2):362–7. 10.1128/AAC.44.2.362-367.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bidet P, Burghoffer B, Gautier V, Brahimi N, Mariani-Kurkdjian P, El-Ghoneimi A, et al. In vivo transfer of plasmid-encoded ACC-1 AmpC from Klebsiella pneumoniae to Escherichia coli in an infant and selection of impermeability to imipenem in K. pneumoniae. Antimicrob Agents Chemother. 2005;49(8):3562–5. 10.1128/AAC.49.8.3562-3565.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benacer D, Thong KL, Watanabe H, Puthucheary SD. Characterization of drug resistant Salmonella enterica serotype Typhimurium by antibiograms, plasmids, integrons, resistance genes and PFGE. J Microbiol Biotechnol. 2010. Jun;20(6):1042–52. [DOI] [PubMed] [Google Scholar]
- 43.Rudolph KM, Parkinson AJ, Roberts MC. Molecular analysis by pulsed-field gel electrophoresis and antibiogram of Streptococcus pneumoniae serotype 6B isolates from selected areas within the United States. J Clin Microbiol. 1998. Sep;36(9):2703–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore MR, Schrag SJ, Schuchat A. Effects of intrapartum antimicrobial prophylaxis for prevention of group-B-streptococcal disease on the incidence and ecology of early-onset neonatal sepsis. Lancet Infect Dis. 2003. Apr;3(4):201–13. 10.1016/S1473-3099(03)00577-2 [DOI] [PubMed] [Google Scholar]
- 45.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008. Apr;159(3):187–93. 10.1016/j.resmic.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 46.Hufnagel M, Liese C, Loescher C, Kunze M, Proempeler H, Berner R, et al. Enterococcal colonization of infants in a neonatal intensive care unit: associated predictors, risk factors and seasonal patterns. BMC Infect Dis. 2007;7:107. 10.1186/1471-2334-7-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3(8):e3056. 10.1371/journal.pone.0003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madan JC, Salari RC, Saxena D, Davidson L, O’Toole GA, Moore JH, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012. Nov;97(6):F456–62. 10.1136/fetalneonatal-2011-301373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One. 2013;8(6):e66986. 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhat YR, Baby LP. Early onset of neonatal sepsis: analysis of the risk factors and the bacterial isolates by using the bact alert system. J Clin Diagn Res. 2011;5(7):1385–8. [Google Scholar]
- 51.Poulakou G, Bassetti M, Righi E, Dimopoulos G. Current and future treatment options for infections caused by multidrug-resistant Gram-negative pathogens. Future Microbiol. 2014;9(9):1053–69. 10.2217/fmb.14.58 [DOI] [PubMed] [Google Scholar]
- 52.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016. Feb;16(2):161–8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 53.Teicher CL, Ronat JB, Fakhri RM, Basel M, Labar AS, Herard P, et al. Antimicrobial drug-resistant bacteria isolated from Syrian War-injured patients, August 2011–March 2013. Emerg Infect Dis. 2014. Nov;20(11):1949–51. 10.3201/eid2011.140835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahal JJ, Urban C, Segal-Maurer S. Nosocomial antibiotic resistance in multiple gram-negative species: experience at one hospital with squeezing the resistance balloon at multiple sites. Clin Infect Dis. 2002. Feb 15;34(4):499–503. 10.1086/338639 [DOI] [PubMed] [Google Scholar]
- 55.World Health Organization Antimicrobial resistance: global report on surveillance. World Health Organization; 2014. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en/ [Google Scholar]
- 56.Muhammad Z, Ahmed A, Hayat U, Wazir MS, Rafiyatullah WH. Neonatal sepsis: causative bacteria and their resistance to antibiotics. J Ayub Med Coll Abbottabad. 2010. Oct–Dec;22(4):33–6. [PubMed] [Google Scholar]
- 57.Yuan XY, Yu DY, Qu XH, Xiao XQ, Bi B, Sun SB, et al. Increased resistance rate to ceftazidime among blood culture isolates of ESBL-producing Escherichia coli in a university-affiliated hospital of China. J Antibiotics. 2016. March 16;69:169–172. [DOI] [PubMed] [Google Scholar]
- 58.Behera B, Mathur P, Das A, Kapil A, Gupta B, Bhoi S, et al. Evaluation of susceptibility testing methods for polymyxin. Int J Infect Dis. 2010;14(7):e596–601. 10.1016/j.ijid.2009.09.001 [DOI] [PubMed] [Google Scholar]