Abstract
The 2015 Nobel Prize in Physiology or Medicine has been awarded to William C. Campbell and Satoshi Omura, and Youyou Tu for the discovery of avermectins and artemisinin, respectively, therapies that revolutionized the treatment of devastating parasite diseases. With the recent technological advances, a New Golden Age of natural products drug discovery is dawning.
Natural products possess enormous structural and chemical diversity that cannot be matched by any synthetic libraries of small molecules and continue to inspire novel discoveries in chemistry, biology, and medicine. They are evolutionarily optimized as drug-like molecules and remain the best sources of drugs and drug leads (Newman and Cragg, 2012). The history of medicine is full of remarkable stories of how the discovery of a natural product profoundly impacted advances in biology and inspired drug discovery and therapy. The award of the Nobel Prizes in Physiology or Medicine to Sir Alexander Fleming, Ernst B. Chain, and Sir Howard Florey in 1945 for the discovery of penicillin and to Selman A. Waksman in 1952 for the discovery of streptomycin heralded the start of a period from 1950s to 1960s known as the Golden Age of natural product drug discovery.
Since the late 20th century, however, many pharmaceutical companies have significantly scaled back or abandoned their natural product programs, in part because of the phenomenal advances in both high throughput screening (HTS) and combinatorial synthesis, thereby the creation of enormous synthetic libraries of small molecules. In contrast, natural product libraries often consisted of extracts and partially purified fractions, in addition to pure compounds, and were perceived to be incompatible with contemporary HTS platforms. This de-emphasis in natural product drug discovery unfortunately correlated with the overall reduction in new leads in the drug development pipeline and the substantial decline in new drug approval (Li and Vederas, 2009).
The award of the 2015 Nobel Prize in Physiology or Medicine to William C. Campbell and Satoshi Omura, for the discovery of the microbial natural product avermectins, the derivatives of which, ivermectin (Fig. 1A), have radically lowered the incidence of onchocerciasis (also known as river blindness) and lymphatic filariasis (also known as elephantiasis), and to Youyou Tu for the discovery of the plant natural product artemisinin (Fig. 1B), a drug that has significantly reduced the mortality rates for patients suffering from malaria, therefore brought the greatest pride and optimism to the natural product community worldwide. Together with the recent advances in microbial genomics and metagenomics, synthetic biology, fundamental understanding of natural product biosynthesis, and bioinformatics and analytical technologies, we are surely entering a New Golden Age of natural products drug discovery.
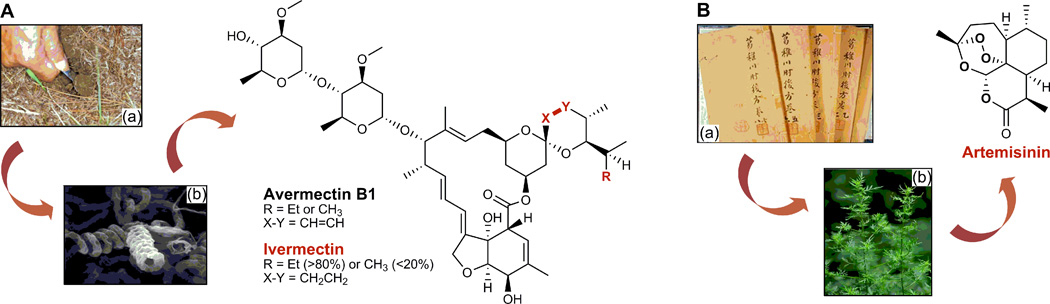
Figure 1. Discovery of avermectin from S. avermitilis and artemisinin from A. annua.
A. Soil samples Omura collected near a golf course, from which the avermectin producing S. avermitilis was isolated, and Campbell discovered avermectins and developed them into ivermectin. B. The ancient Chinese medicine “Handbook of Prescriptions for Emergencies” that inspired Tu to discovery artimisinin from the A. annua L. plants.
“Splendid gifts from microorganisms”
The discovery of the avermectins resulted from an intensive search for microbial natural products with anthelmintic activities and exemplified the best team effort from an academic and industry collaboration, led by Campbell, a parasitologist at the Merck Research Laboratories, and Omura, a microbial natural product chemist at the Kitasato Institute. The choice of natural products as the preferred source for initial screening was deeply rooted to Campbell’s belief that Nature constantly produces molecules that have not been thought of by humans and to Omura‘s personal research philosophy of exploiting the potential of microorganisms and microbial metabolites, “splendid gifts” as he calls them, to advance scientific progress and to accelerate and maximize improvement in human health and welfare worldwide (Omura, 2011).
In the early 1970s, Merck decided to search for new anthelmintics from microbial natural products. While microbial fermentations had been extensively screened for anticancer and antibacterial agents, parasitologists at the time had not tried this strategy, partly due to the lack of suitable assays. At the same time, the Kitasato Institute entered into a research agreement with Merck to screen for novel antibacterial agents from soil microbes isolated in Japan. This agreement was subsequently amended such that Kitasato Institute would send lyophilized cultures of the most chemically or morphologically “unusual” ones to Merck for any additional assays.
Initial examination of microbial cultures by several different in vitro assays was disappointing, primarily because of the large number of toxic compounds that had to be eliminated. This prompted Merck to develop an in vivo assay in mice with the hope that the mice would screen out the non-specific toxic compounds. Thus, mice, infected with the nematode, Nematospiroide dubius, were fed a diet mixed with the fermentation products to be tested and subsequently examined for the absence of eggs in fecal pellets and worms in small intestines. This assay turned out to be very successful, most importantly allowing the differentiation of nonspecific toxic activities from the true antiparasitic activity. Among the many thousands of cultures tested was one group of 54 cultures from the Kitasato Institute, including the now famous OS3153 strain isolated by Omura near a golf course. As fate would have it, OS3153 was almost missed in the initial assay because of substantial toxicity, but confirmatory tests firmly established an extremely potent anthelmintic acitvity at 0.0003% in diet.
OS3153 was identified as a novel species, named Streptomyces avermitilis, and the metabolites responsible for the activity were named avermectins. Despite decades of searching around the world, the S. avermitilis strain discovered by Omura remains the only avermectin-producing organism ever found, underscoring its rarity. The wild-type strain produced avermectins at a yield of ~9 mg/L. Subsequent strain improvement resulted in 50-fold increase in avermectin yield by the production strain.
The avermectins consisted of eight closely related congeners, comparison among which revealed insightful structure-activity relationship. 22,23-Dihydroavermectin B1, named ivermectin (Fig. 1A), was selected for development based on its exceptional potency against an unprecedented array of nematodes and anthropod parasites and a preferable safety profile. Its novel chemical structure, a 16-membered macrolide bearing two unusual deoxysugars, and mode of action, by hyperpolarizing neurons via the binding to receptors of glutamate-gated chloride ion channels, probably account for the lack of any cross-resistance to known antiparasitic agents. Since marketed in 1981, ivermectin has become the most revolutionary parasiticide in animal health.
Development of ivermectin as a human medicine for onchocerciasis originated with the early observations by Merck scientists that ivermectin was very effective against the skin-dwelling microfilariae of Onchocerca cervicalis in horse. Since O. cervicalis belongs to the same genus as Onchocerca vulvulus, the causative parasite for onchocerciasis in human, Campbell hypothesized that ivermectin would be effective against onchocerciasis. It was indeed found to be highly effective against the parasitic worms responsible for onchocerciasis and subsequently in combinatorial therapy for lymphatic filariasis (Campbell et al., 1983; Campbell 1993; Omura and Crump 2004, 2014). Today, under the brand name of Mectizan and via an unprecedented drug donation program, ivermectin provided us with a revolutionary therapy against onchocerciasis and lymphatic filariasis, mass distribution of which has become so successful that river blindness and elephantiasis are on the verge of eradication.
Deciphering the ancient Chinese medicine code
The discovery of artemisinin (qinghaosu ‘青蒿素’ in Chinese) came from an intensive search for plant natural products with antimalaria activity and represented a nationwide collaborative effort during the “Culture Revolution” period from the 1960s to 1970s in China. Tu, a phytochemist and the head of the malaria research group at the China Academy of Chinese Medical Sciences, was destined to make the Nobel Prize-winning discovery of artemisinin due to her unique trainings in both Western and Chinese medicines and her belief in the wisdom of Chinese medicine and determination to exploit “gifts from Chinese medicine” to conquer life-threatening diseases worldwide (Tu, 2011).
In the 1960s, malaria rebounded with a vengeance primarily due to resistance to chloroquine, the antimalaria drug of choice at the time. “Project 523”, initiated on May 23, 1967 by the Chinese government, enlisted more than 60 academic and military institutions and more than 500 researchers, with the estimation of an additional 2,000 to 3,000 people involved at one time or another, to discover a new antimalaria drug. Efforts in the first two years, included both screening known compounds (~40,000) for leads and combing the traditional Chinese medicine literatures for hints, however, were disappointing, yielding nothing of practical values.
Tu was asked to join the project in 1969. Knowing the setbacks in the previous two years, Tu and colleagues first surveyed traditional Chinese medicine records and literatures and visited many herbalists and rural healers all over China. As a result, they compiled a list of more than 200 traditional Chinese medicine receipts, consisting of more than 2,000 herbs, with indications of treating fever, thereby possible antimalaria activities. However, initial evaluations of ~380 extracts, made from ~200 selected herbs, were simply disappointing, with malaria inhibiton in a mouse model, infected with the malaria parasite Plasmodium berghei, straggling at 12–14%.
The breakthrough came from an extract, made from the plant Artemisia annua L. (also known as qinghao or sweet wormwood), that showed promising antimalaria activity in the mouse model, but this activity was not reproducible. Upon re-examining a book titled “Handbook of Prescriptions for Emergencies” by Ge Hong, a fourth-century healer, which called for “soaking one bunch of qinghao in two liters of water, squeezing and then drinking the juice”, Tu reasoned that the active ingredients may be not stable under the traditional Chinese medicine preparation by boiling. Inspired by this passage, Tu made, now known as the most important decision leading to the discovery of artemisinin, to experiment alternative methods of extraction at lower temperatures. Finally an extract made with diethyl ether afforded a reproducible antimalaria activity, from which artemisinin was finally isolated and showed 100% efficacy against malaria in the animal models. Within the genus Artemisia, we now know that A. annua is the only species that contains artemisinin, most of which is enriched in the leaves of plants harvested in the later summer and early fall. Artemisinin yields also vary, from 0.01 to 0.5% with the plants growing in Sichuan, China, in comparison to 0.06% with plants growing in the Washington, D.C., vicinity. The ancient code was finally deciphered.
The ensuing events leading to the clinical development of artemisinin into an antimalaria drug were by no means conventional either. At the time with most of the scientific establishments in China on standstill, Tu and two colleagues bravely tested the safety of the active compound, before knowing its chemical structure, on their own. Upon confirming that the compound was safe, they completed the very first clinical trials for 30 patients infected with both Plasmodium vivax and Plasmodium fulciparum, all of which were successful. It was only then that systematic characterizations of the chemistry and pharmacology for the active compound started.
The structure of the active compound, named artemisinin, was established. It belongs to the amorphane family of sesquiterpenes but features a rare peroxy bridge (Fig. 1B). Artemisinin was highly effective against the malaria parasites, including those that were resistant to chloroquine. It rapidly kills the malaria parasites at an early stage of their development, which explains its unprecedented potency in the treatment of severe malaria. It was estimated that during the three-year period between 1975 and 1978 alone, clinical trials on more than 6,550 patients were completed, all of which were successful. Thus was born, artemisinin, a completely novel class of antimalarias. Analogues of artemisinin, such as dihydroartemisinin, artemether, and artesunate, were sequently found to be more potent than artemisinin itself and are now completely changing the landscape of antimalaria chemotherapies (Klayman, 1985; Tu, 2011).
It is worth noting that the impact of artemisinin as a novel class of antimalaria drugs on world health was not realized until nearly 30 years after its effectiveness was firmly established in China. WHO started to adopt artemether as a front-line antimalaria drug in 2001 and switched to the artemisinin-based combination therapies in 2005, which now are estimated to reduce mortality from malaria by more than 20% overall, and by more than 30% in children.
A New Golden Age of natural products drug discovery
This year’s Nobel Prize-winning discoveries will surely inspire the world to continue the search for history-changing natural product drugs. There were many parallels between the discovery and development of avermectins and artemisinin into paradigm-shifting medicines that could enlighten the scientific community to reconsider the perceived incompatibility of natural products with contemporary HTS platforms for drug discovery. Both programs started with massive screening campaigns against libraries of known compounds and natural product extracts of microbial or plant origins, but the eventual breakthroughs both came from natural products. The in vivo models allowed differentiation between the true biological activity from non-specific toxicities, thereby allowing resources to be devoted preferentially to the most promising leads. While it is inconceivable today, in both cases the efficacy of the active compounds was firmly established in multiple animal models before pursuing chemical structure elucidation. For artemisinin, the initial trials on patients were carried out with the preparation without knowing its structure, though its safety was established and this was done more out of the historial circumstances rather than by choice. After more than 30 years of successful clinical application, the exact modes of action for both ivermectin and artemisinin remain poorly understood. While the structural complexity and rich functionalities have been credited for their great potency and exquisite selectivity, natural products often interact with multiple biological targets, challenging the prevailing paradigm of mechanism-based drug discovery. Investigations of the multitude modes of action for natural product drugs, therefore, promise to reveal the intricate interplay of biology, thereby providing new opportunities for drug discovery. The unprecedented scaffolds of the natural products have served as great inspiration for the next generation of therapeutics, as exemplified by the many analogues, with designer properties, derived from avermectins and artemisinin. It is precisely the novel chemical scaffolds of the avermectins and artemisinin that account for their lack of cross-resistance to many of the known drugs. This is particularly imperative for antibiotics since resistance is inevitable. The world would be better served when these lessons are taken into consideration in the quest for the next wave of wonder drugs.
It has been more than 30 years since the ivermectin and artemisinin families of drugs were first introduced as human therapies. Resistances to both drugs have been observed and continue to rise. The world has invested billions of dollars to search for novel anthelmintic and antimalaria drugs, but without a fundamental breakthrough. Is it a time to reconsider natural products?
The genome of the avermectin producing S. avermitilis strain has since been sequenced, revealing, among the many surprises, its potential to biosynthesize at least 36 other natural products, in addition to avermectins, and the 80-kb avermectin gene cluster, setting the stage to improve avermectin production and engineer avermectin analogues by manipulating its biosynthesis (Ikeda et al., 1999). Indeed, targeting genes encoding the specific steps in avermectin biosynthesis has already demonstrated the feasibility to produce selected avermectin congeners and improve overall yields. While decades of search around the world has yet to discover other avermectin producers, gene clusters homologous to avermectin biosynthesis have been discovered, providing outstanding opportunities to engineer novel avermectin analogues by exploring combinatorial biosynthesis strategies.
Reliable supplies of artemisinin based on massive cultivation of A. annua, given its low yield and regional and seasonal variations, have always been a concern. The artemisinin structure was unprecedented when it was first discovered nearly 40 years ago, but now, much of its biosynthesis is known. In a remarkable triumph of synthetic biology, artemisinic acid, the key precursor from which artemisinin and other artemisinin analogues can be readily prepared by chemical synthesis, can now be produced by fermentation of a recombinant yeast carrying genes hijacked from A. annua to convert farnesyl diphosphate, the universal precursor for sesquiterpenes, to artemisinic acid in a whopping yield of 25 g/L (Paddon et al., 2013).
We are surely entering a New Golden Age of natural products drug discovery. Recent advances in microbial genomics and metagenomics have definitively revealed that the vast majority of earth’s biodiversity is yet to be exploited for natural products discovery (Charlop-Powers et al., 2014). Innovations in microbial cultivation (Ling et al., 2015) and synthetic biology (Paddon et al., 2013) have radically expanded the chemical space of natural products and demonstrated the feasibility of engineered production of natural products from uncultivated species or species on the verge of extinction in model heterologous hosts. Fundamental understandings of natural product biosynthesis have allowed rational manipulation of Nature’s biosynthetic machinery for the production of both the natural products and their engineered variants. Genome mining for Nature’s myriad so-called “cryptic” natural product biosynthetic pathways has started to awaken these “sleeping beauties”. Finally, emerging strain prioritization, bioinformatics, and analytical technologies have revolutionized natural product dereplication, enabling structural prediction, rapid detection, and isolation of the most promising natural products in a rapid pace (Bouslimani et al., 2014; Rudolf et al., 2016). Nature has been extremely generous in offering mankind live-saving therapies, the next great drug may be just around the corner, and are we ready to seize the opportunity?
REFERENCES
- Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Nat. Prod. Rep. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell WC. Med. Res. Rev. 1993;13:61–79. doi: 10.1002/med.2610130103. [DOI] [PubMed] [Google Scholar]
- Campbell WC, Fisher MH, Stapley EO, Albers-Schonberg G, Jacob TA. Science. 1983;221:823–828. doi: 10.1126/science.6308762. [DOI] [PubMed] [Google Scholar]
- Charlop-Powers Z, Owen JG, Reddy BVB, Ternei MA, Brady SF. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klayman DL. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- Li JW-H, Vederas JC. Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- Ling L, Lewis K, et al. Nature. 2015;517:455–459. [Google Scholar]
- Newman DJ, Cragg GM. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S. Tetrahedron. 2011;67:6420–6459. [Google Scholar]
- Omura S, Crump A. Nat. Rev. Microbiol. 2004;2:984–989. doi: 10.1038/nrmicro1048. [DOI] [PubMed] [Google Scholar]
- Omura S, Crump A. Trands Parasitol. 2014;30:445–455. doi: 10.1016/j.pt.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Newman JD, et al. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Rudolf JD, Yan X, Shen B. J. Ind. Microbiol. Biotechnol. 2016 doi: 10.1007/s10295-015-1671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. Nat. Medicine. 2011;17:1217–1220. doi: 10.1038/nm.2471. [DOI] [PubMed] [Google Scholar]