Abstract
Necroptosis is a well-known form of caspase-independent cell death. Necroptosis can be triggered by various extrinsic stimuli, including death ligands in the presence of receptorinteracting protein kinase 3 (RIPK3), a key mediator of necroptosis induction. Our recent studies have revealed that C-terminus HSC-70 interacting protein (CHIP), an E3 ligase, can function as an inhibitor of necroptosis. CHIP−/− mouse embryonic fibroblast showed higher sensitivity to necrotic stimuli than wild-type mouse embryonic fibroblast cells. Deleterious effects of CHIP knockout MEFs were retrieved by RIPK3 depletion. We found that CHIP negatively regulated RIPK3 and RIPK1 by ubiquitylation- and lysosome- dependent degradation. In addition, CHIP−/− mice showed postnatal lethality with intestinal defects that could be rescued by crossing with RIPK3−/− mice. These results suggest that CHIP is a negative regulator of RIPK1 and RIPK3, thus inhibiting necroptosis. [BMB Reports 2016; 49(5): 247-248]
Keywords: CHIP, Lysosome, Necroptosis, RIPK3, Ubiquitylation
The cell death pathway can be divided into caspase-dependent and caspase-independent cell death pathways. Necroptosis, a form of programmed necrosis, is an extensively studied caspase-independent cell death pathway. It is considered as an alternative form of cell death pathway developed in the host to fight against viruses and bacteria. Necroptosis is triggered by the activation of death receptors, including tumor necrosis factor (TNF) receptor, Toll-like receptors (TLR), and interferon receptors (IFNRs). Once activated, death receptors recruit a variety of signaling molecules to form primary complexes such as TNF complex I/II or death-inducing signaling complex. When caspase-8 activity is low or inhibited, necrosome complexes are formed. Necrosomes are composed of phosphorylated receptor-interacting protein kinase 1 (RIPK1) and RIPK3. Activated RIPK3 induces the phosphorylation of mixed lineage kinase domain-like protein (MLKL) which leads to MLKL oligomerization. Oligomerized MLKL moves to the plasma membrane and disrupts the membrane integrity directly or indirectly. While MLKL might be the final effector molecule of necroptosis, other unknown factors affecting these final steps cannot be excluded. In contrast to apoptosis, necroptosis induces the release of damage-associated molecular patterns (DAMP) such as high-mobility group box 1 (HGMB1) that can lead to inflammation by activating innate immune cells. Recent studies have shown that several disease models related to apoptosis such as ischemia-reperfusion injury and neurodegeneration are also associated with necroptosis. While more detailed in vivo studies are necessary, these observations indicate that necroptosis involves functionally integrated pathways and causes a variety of human diseases.
Our study revealed that C-terminus HSC70-interacting protein (CHIP) could function as a negative regulator of necroptosis. CHIP is known as an UBOX-containing E3 ligase with tetratricopeptide as a chaperone-binding motif. CHIP regulates various tumor suppressor proteins such as p53, phosphatase and tensin homolog (PTEN), thus protecting cells from apoptosis. However, the function of CHIP in extrinsic or non-apoptotic cell death signaling remains unclear. Since hyperactivation of necroptosis is linked to the disruption of tissues or lethality, we evaluated whether non-apoptotic cell death was activated in CHIP−/− mice. To test this, we first employed CHIP knockout mouse embryonic fibroblasts (MEFs) to study necroptosis.
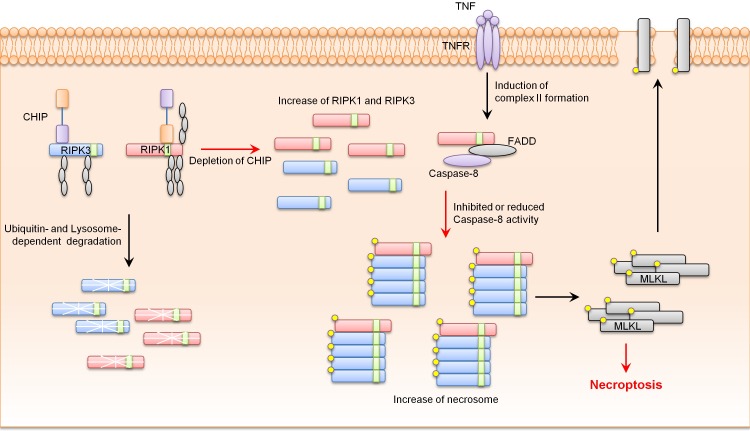
Based on our results, CHIP−/− MEFs showed higher sensitivity to necroptosis and promoted necrosome complex formation in response to necrotic stimuli. Western blotting data revealed that CHIP−/− MEFs had higher RIPK1 and RIPK3 protein levels and stabilities compared to wild-type MEFs. We also showed that CHIP could degrade RIPK1 and RIPK3. In addition, CHIP directly ubiquitylated RIPK1 and RIPK3 under exogenous and endogenous conditions. Finally, CHIP-mediated ubiquitylation of RIPK1 and RIPK3 occurred at lysine 571, 604, and 627 of RIPK1 and lysine 55 and 363 of RIPK3, leading to lysosomal localization of RIPK1 and RIPK3 (Fig. 1).
Fig. 1. CHIP inhibits necroptosis by ubiquitylation- and lysosome- dependent RIPK1 and RIPK3 degradation.
Recently, intestinal epithelial cell-specific knockout of Fas-associated death domain (FADD) or caspase-8 has been shown to be able to induce massive cell death and inflammatory phenotypes in mouse intestinal epithelial cells. These phenotypes were rescued by treatment with necrostatin-1, a necroptosis inhibitor, or crossing with RIPK3 knockout mice, indicating the critical role of necroptosis in intestinal defects. Our analysis showed that CHIP−/− mice died within 4-8 weeks with a high rate of cell death in the intestinal epithelium, a typical phenotype of necroptotic animal models. These phenotypes of CHIP−/− mice were rescued by crossing with RIPK3−/− mice. There was no change in cleaved-caspase-3 protein levels in the intestines of CHIP−/− mice, indicating that intestinal defects in CHIP−/− mice occurred in an apoptosis-independent manner. RIPK1 and RIPK3 protein levels were increased in the intestines of CHIP−/− mice, suggesting that CHIP deficiency could lead to the activation of necroptosis by up-regulating the levels of RIPK1 and RIPK3 proteins. Although further analyses of organs in CHIP−/− mice related to necroptosis are required, our data suggest that CHIP can negatively regulate necroptosis in the intestinal epithelium by ubiquitylation-mediated and lysosome-dependent degradation of RIPK1 and RIPK3.
Acknowledgments
This work was supported by a grant (NRF-2015R1A3A2066581 to J. Song) from the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning, Republic of Korea.