Abstract
RNA polymerase II C-terminal domain phosphatases are newly emerging family of phosphatases that contain FCPH domain with Mg+2-binding DXDX(T/V) signature motif. Its subfamily includes small CTD phosphatases (SCPs). Recently, we identified several interacting partners of human SCP1 with appearance of dephosphorylation and O-GlcNAcylation. In this study, using an established cell line with inducible CTDSPL2 protein (a member of the new phosphatase family), proteomic screening was conducted to identify binding partners of CTDSPL2 in nuclear extract through immunoprecipitation of CTDSPL2 with its associated. This approach led to the identification of several interacting partners of CTDSPL2. This will provide a better understanding on CTDSPL2. [BMB Reports 2016; 49(6): 319-324]
Keywords: CTD phosphatase, CTDSPL2 inducible HEK293T cell line, CTDSPL2, Immunoprecipitation, Nuclear interactome
INTRODUCTION
C-terminal domain (CTD) of DNA dependent RNA polymerase II (Pol II) is conserved from fungi to humans. Coordinately switched phosphorylation status of CTD is not only critical for the recruitment and assembly of transcription complexes, but also important for the regulation of transcription and mRNA processing (1). A methodical approach searching for genome-wide distribution of CTD phosphorylation has indicated delicate interplays between CTD kinase and phosphatases at global mode for all genes (2). Up to date, only a few mammalian CTD phosphatases have been defined. TFIIF-associating CTD phosphatase 1 (FCP1), the 1st identified CTD phosphatase, contains a Brca-1 C-terminal (BRCT) domain and catalytic N-terminal Fcp1 homology (FCPH) domain with DXDX(T/V) motif. It is a highly conserved and pivotal enzyme for modifying the CTD of RNA Pol II (3). While searching for FCP1 homologs, proteins containing a region sharing homology with the catalytic domain of the 1st identified and characterized FCP1, but not the BRCT domain, have been identified, including small CTD phosphatase 1 (SCP1), SCP2, SCP3, CTD small phosphatase like protein 2 (CTDSPL2), mitochondrial import inner membrane translocase subunit TIM50 (TIMM50), and UBLCP (4-7). SCPs are transcriptional regulators that can silence neuronal genes in non-neuronal tissue in a global manner. They are also cardiac hypertrophy inducer with help from its intronic miRNAs (7). Although the catalytic mechanisms of SCPs and basis for their enzymatic specificity are well characterized, the identities of physiological substrates that could explain the biological activities of SCPs are massively unknown. Although it is well known that CTDSPL2 contains a CTD phosphatase catalytic domain (8, 9) with function of dephosphorylating phosphoSer5 and phosphoSer2 within GST-CTD in vitro (10), the biological function of CTDSPL2 gene is totally unclear. Recently, it has been demonstrated that CTDSPL2 gene can modify the expression of ε- and γ-globin genes in certain cells derived from umbilical cord blood and that CTDSPL2 as a SMAD phosphatase plays an important role in BMP-induced signaling system (6, 9). However, this phosphatase failed to dephosphorylate phosphoSer5 in the C-terminal domain of RNA Pol II (6).
In this study, we established a HEK293T cell line with inducible expression of CTDSPL2 and analyzed the interacting partners of CTDSPL2 protein in the nucleus. Although a set of proteins can function as single monomeric unit, a considerable portion of proteins function by interacting with their partners or function as components of macromolecular complexes or as enzyme-substrate(s). Multi-protein complexes are essential for various cellular functions. Protein complexes are more than just the sum of individual components. They reflect a basic level of integration of information encoded by individual genes (11). To date, proteomics techniques have been used to investigate protein expression profile, protein quantification, protein-protein interactions, and post translational modification (PTM). Proteomics techniques have been increasing used in various biological studies (12, 13). Therefore, the aim of this study was to use proteomics approach (co-immunoprecipitation, 1D electrophoresis, mass spectrometry) to identify interacting partners of CTDSPL2 in the nucleus using a cell line that could specifically express CTDSPL2 protein in order to understand the physiological function and/or role of CTDSPL2 in the nucleus.
Here, we provided some new insights regarding the physiological role of CTDSPL2.
RESULTS AND DISCUSSION
Inducible cell line and subcellular localization of CTDSPL2
CTDSPL2 was fused with Flag sequence at its N-terminus and subcloned into pTRE-IRES-EGFP inducible expression vector (14). After transfection of Flag-CTDSPL2 inserted to pTRE-IRES-EGFP (placing together pEF1-Puro-Tet-On), several candidate clones were selected based on their antibiotics resistance and microscopic appearance for EGFP as well as immunoblot analysis with α-DYKDDDDK antibody for Flag-CTDSPL2 (Fig. 1). Increased expression of CTDSPL2 was only detected at 12 h post induction when a specific inducer doxycycline was used. After cells were treated with doxycycline for the indicated period of time, when cells were re-plated onto new culture devises with fresh medium without doxycycline, the expression of CTDSPL2 was totally reverted to no expression status (Fig. 1A). Therefore, the expression of CTDSPL2 in this cell line was under a tight control of doxycycline, thus suitable for characterizing CTDSPL2 and identifying CTDSPL2 interactome. The established CTDSPL2 inducible HEK293T cell was designated as HEK293T/Flag-CTDSPL2.
Fig. 1. Establishment of the CTDSPL2 inducible HEK293T cells. (A) The expression levels in 60 μg of total protein treated with or without doxycycline for the indicated periods of time were monitored by immunoblot. To check reversion, cells were treated with doxycycline for the indicated periods. After replacing old media with fresh medium, cells were allowed to grow for the indicated periods of time without the inducer doxycycline. (B) Immunoblot analysis of CTDSPL2 in the nuclear and cytoplasmic fraction of HEK293T/Flag-CTDSPL2 cells. α-Tubulin and lamin B were used as controls to confirm the presence of cytosol and nuclei, respectively. (C) Expression of wild type and inactive mutant type SCP1 from HEK293T/Flag-SCP1/Wt and HEK293T/Flag-SCP1/D96N cells was monitored. (D) Fluorescence microscopic assays to determine the expression of CTDSPL2 in CTDSPL2 inducible cells.
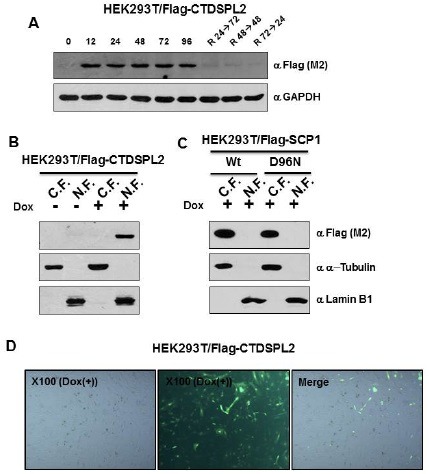
It is well known that CTDSPL2 is a nuclear protein containing FCPH domain and DXDX(T/V) motif. To examine the localization of CTDSPL2 in different cellular compartments, purified nuclei was used as a starting material for immunoblot analysis (Fig. 1B). Cytoplasmic and nuclear extracts of cells were prepared at 72 h post induction. When α-Flag immunoblots were performed for cytoplasmic and nuclear extracts, the inducible CTDSPL2 protein was only detected in the induced nuclear fraction (Fig. 1B). When the localization of SCP1 inducible HEK293T cells HEK293T/Flag-SCP1/Wt (N-terminal Flag-tagged wild-type) and HEK293T/Flag-SCP1/D96N (inactive mutant SCP1) was analyzed, SCP1s were only significantly detected in the cytoplasmic fraction (Fig. 1C). Fluorescence microscopic investigation of CTDSPL2 in HEK293T cells also demonstrated its subcellular distribution (Fig. 1D), consistent with results from nuclear staining.
CTDSPL2 does not dephosphorylate phosphoSer5 in the CTD of RNA Pol II, although it is structurally similar to CTD phosphatase FCP1 (6). Enzymatic activities of immunocomplexes from inducible expression of Flag-tagged CTDSPL2 showed phosphatase activity against p-NPP (15). Therefore, HEK293T/Flag-CTDSPL2 could be used as an effective tool to dissect the cellular and physiological functions of CTDSPL2 in vivo and for the identification of CTDSPL2 nuclear interactome.
Purification of immunocomplexes and identification of partner protein components in the nuclear interactome
Identification of protein-protein interactions is the key to understand the specificity and fidelity of many physiological reactions. Immune-affinity purification combined with mass spectrometry has been successfully used to identify interacting molecules that are directly or indirectly associated with a protein of interest (12, 13, 16, 17). Large-scale immunoprecipitation can allow us identify protein partners of CTDSPL2 with high confidence. The usefulness of Tet-On inducible system has been previously demonstrated (12, 14, 17-19). A schematic schema of the strategy used to identify CTDSPL2 interacting proteins is shown in Fig. 2A. Nuclei were lysed with conventional RIPA buffer. CTDSPL2 immunocompelexes were then affinity-purified through immunnoprecipitation using α-Flag antibody (Fig. 2B). Nuclear cell extracts (30 mg) were prepared from CTDSPL2 induced cells after 72 h of induction. Flag-CTDSPL2 proteins and any interacting protein molecules were recovered by immunoprecipitation (Fig. 2B). The immunocomplexes immobilized onto Protein A Agarose beads were then removed using 2X Laemmli buffer and separated with 6-15% gradient SDS-PAGE gels. These proteins were then subjected to tryptic digestion and LC-MS/MS analysis. As a negative control, the same procedure was done by using un-induced nuclear extract (Fig. 2B). A lot of proteins were co-immunoprecipitated with Flag-CTDSPL2 from the nuclear fraction. Protein lanes were evenly cut to 10 equivalent gel slices. Immunoprecipitation lanes from both positive induction and negative control were subjected to LC-MS/MS analysis after staining, destaining, in-gel digestion, and peptide extraction. Proteins identified in the CTDSPL2 immunocomplex lane with two or more peptides and a confidence level of more than 95% (more than 42 Probability based Mowse Score) but absent in the negative control lane of un-induced nuclear fraction were regarded as specific nuclear interactome of CTDSPL2 (Table 1). A total of 33 proteins were identified from the immunoprecipitation lane of induced CTDSPL2. The accession number, protein name, protein ID, description, number of tryptic peptides, and protein probability score from all three replicates are summarized in Table 1. Based on the Panther Database (http://www.pantherdb.org), these identified proteins were grouped systematically. Their main functional categories in the cells were: chromatin/chromatin-binding protein, RNA-binding protein, DNA-binding protein, transcription factor, transporter, non-receptor type protein kinase, DNA ligase, and unclassified proteins (Table 1). The identification of these proteins suggests a unique biological role of CTDSPL2 in vivo. Cellular functions and signaling networks are the consequences of coordinated action of individual proteins in macromolecular complexes. Despite these intriguing and critical findings, little progress has been made on the characterization of CTDSPL2. Its activity and function in cells remain largely unknown. For that reason, analyzing protein complex composition is a critical starting point to solve the cellular physiology and signaling networks of a biological system (17, 18). Our approach is efficient for the identification of macromolecular complexes formed under native conditions. Although further study is needed to understand the biochemical basis for the assembly of these complexes and their biological roles, protein composition of CTDSPL2 nuclear interactome could provide some clue for their specific cellular and physiological roles.
Fig. 2. Affinity purification of CTDSPL2-interacting proteins. (A) Schematic diagram of the strategy used to identify CTDSPL2 interacting proteins. (B) Proteins purified from the nuclear extracts of HEK293T/Flag-CTDSPL2 cells induced with or without doxycycline were resolved by SDS-PAGE. Proteins were visualized after colloidal Coomassie blue staining.
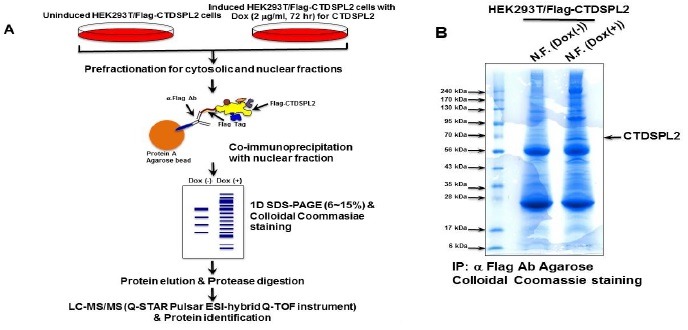
Table 1. Identification of proteins recovered from CTDSPL2 tandem affinity purification from HEK293T/Flag-CTDSPL2a.
| Accession No. | Protein ID | Peptide No. | Mass (Da) | Protein scorebb | Description | Biological functioncc |
|---|---|---|---|---|---|---|
| gi:1147813 | AAA85135 | 4 | 331571 | 150 | Desmoplakin I | Non-motor actin binding protein |
| gi:28243 | CAA37495 | 8 | 280586 | 134 | Unnamed protein product | |
| gi:1082356 | PH02683 | 3 | 73218 | 130 | Epidermal autoantigen 450K (Clone pE450-C/D) | |
| gi:4760577 | BAA77349 | 3 | 245394 | 46 | IDN3-B | Chromatin/chromatin-binding protein |
| gi:30268331 | CAD89954 | 33 | 154064 | 790 | Hypothetical protein | |
| gi:189054178 | BAG36698 | 7 | 65980 | 134 | Unnamed protein product | |
| gi:14249928 | AAH08345 | 3 | 107235 | 84 | Unknown (protein for IMAGE:3531356) | |
| gi: 5262568 | CAB45727 | 3 | 81727 | 63 | Hypothetical protein | |
| gi:10435899 | BAB14698 | 2 | 87829 | 42 | Unnamed protein product | |
| gi:119623341 | EAX02936 | 2 | 140679 | 42 | hCG1999854, isoform CRA_a | |
| gi:190267 | AAA60155 | 3 | 113063 | 107 | Poly(ADP-ribose) polymerase | DNA ligase |
| gi:167234419 | NP_005110 | 3 | 108601 | 134 | NcoA3, Nuclear receptor coactivatore 3 | Acetyltransferase/ transcription factor |
| gi:119588286 | EAW67880 | 2 | 141940 | 95 | Nucleoporin 160kDa, isoform CRA_c | Transporter |
| gi:28317 | CAA32649 | 2 | 59492 | 58 | Unnamed protein product | |
| gi:5031755 | NP_005817 | 3 | 70899 | 167 | Heterogeneous nuclear ribonucleoprotein R-2 | |
| gi:5762315 | AAD51099 | 3 | 95325 | 108 | Nuclear factor associated with dsRNA NFAR-2 | RNA-binding protein |
| gi:7021912 | BAA91433 | 2 | 37824 | 53 | Unnamed protein product | |
| gi:6841480 | AF161478_1 | 10 | 53096 | 183 | HSPC129 | Phosphatase |
| gi:7023323 | BAA91924 | 2 | 41713 | 162 | Unnamed protein product | |
| gi:187281 | AAA36192 | 4 | 77555 | 82 | hnRNA binding protein M4 | RNA-binding protein |
| gi:194384236 | BAG64891 | 2 | 59806 | 80 | Unnamed protein product | |
| gi:35222 | CAA36061 | 2 | 70810 | 80 | Unnamed protein product | |
| gi:1040970 | AAA79948 | 2 | 53345 | 65 | FUS/TLS | Transcrption factor, DNA-binding Protein, mRNA splicing factor |
| gi:532313 | AAA20993 | 4 | 44669 | 169 | NF45 protein | NF-AT DNA binding protein |
| gi:9802306 | AF261087_1 | 3 | 32871 | 127 | DNA-binding protein TAXREB107 | DNA-binding protein |
| gi:36102 | CAA29922 | 3 | 34131 | 114 | Unnamed protein product | |
| gi:6678271 | NP_031401 | 4 | 44711 | 172 | TAR DNA binding protein 43 | DNA-binding protein |
| gi:34039 | CAA68556 | 2 | 44079 | 62 | Unnamed protein product | |
| gi:4502709 | NP_001777 | 2 | 34074 | 84 | Cdc2 p34 | Non-receptor type protein kinase |
| gi:4885377 | NP_005311 | 6 | 22336 | 69 | Histone H1.3 | Chromatin/chromatin-binding protein |
| gi:4758302 | NP_004441 | 4 | 12251 | 87 | Enhancer of rudimentary homolog | Transcription factor |
| gi:31979 | CAA40416 | 2 | 13898 | 84 | Histone H2A.2 | Chromatin/chromatin-binding protein |
| gi:119571409 | EAW51024 | 2 | 18209 | 69 | hCG1643342, isoform CRA_a | |
aBolded proteins represent those validated by co-immunoprecipitation with CTDSPL2.
bIons score is -10 log(P), where P is the probability of a random event of the observed match. Individual ion scores > 42 indicate identity or extensive homology (P < 0.05).
cBased on Panther Database (http://www.pantherdb.org).
Validation of some novel nuclear CTDSPL2 interacting proteins
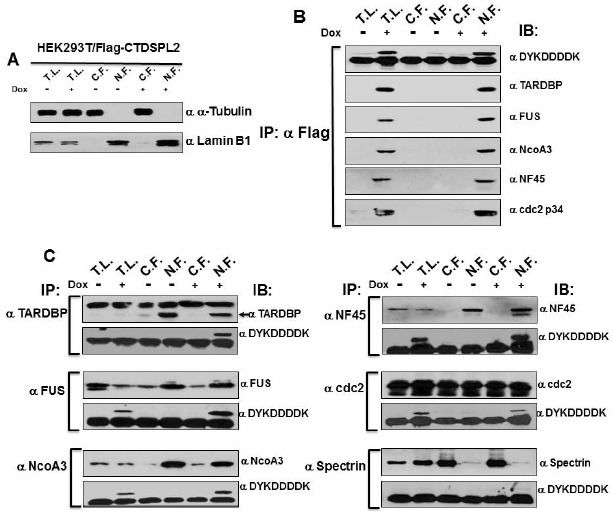
To validate the proteins identified by LC-MS/MS peptide matching as components of the CTDSPL2 nuclear interactome, immunoblot analysis was performed using total cell lysate and CTDSPL2 immunoprecipitation complexes from the cytoplasmic and nuclear fractions of induced and un-induced CTDSPL2 expressing cells. Five proteins (fused in sarcoma/translocated in liposarcoma (FUS/TLS), TAR DNA binding protein (TARDBP), nuclear factor of activated T cells 45 kDa (NF45), cdc2 p34, and nuclear receptor coactivator 3 (NcoA3) were chosen for immunoprecipitation using commercially available antibodies. As shown in Fig. 3B, CTDSPL2 protein expression was identified in the immunocomplexes obtained from total cell lysate, especially in the nuclear fraction of doxycycline-induced cells, but not in the total cell lysate or the nuclear extracts of un-induced cells or the cytoplasmic fraction of induced HEK293T/Flag-CTDSPL2. To further validation the interaction and the efficiency of immunoprecipitation, a reciprocal immunoprecipitation was carried out with CTDSPL2 expressing cell line. The associated complexes were subjected to immunoblot analysis to determine the presence of inducible CTDSPL2, FUS/TLS, NF45, TARDBP, cdc2 p34, and NcoA3 (Fig. 3C). Our results revealed that these five proteins were co-immunoprecipitated with the inducible CTDSPL2 protein.
Fig. 3. Co-immunoprecipitation using α-Flag antibody and specific primary antibodies directed against TARDBP, FUS/TLS, NcoA3, NF45, cdc2 p34, and spectrin using total cell lysates, pre-fractionated nuclear fraction, or pre-fractioned cytoplasmic fractions of cells with or without the inducer. (A) Nuclear-Extract Kit was used to prepare nuclear, cytoplasmic, and total-cell extracts from cultured HEK293T/Flag-CTDSPL2 cells with or without inducer. (B) Co-immunoprecipitation using α-Flag antibody with total cell lysates, nuclear, or cytoplasmic fractions. Each extract was incubated with α-Flag-Protein A Agarose, washed, and subjected to 6-15% gradient SDS-PAGE. Gels were then immunoblotted with specific antibody. (C) Co-immunoprecipitation using specific antibodies against TARDBP, FUS/TLS, NcoA3, NF45, cdc2 p34, and spectrin. Spectrin is used as a false-negative control for immunoprecipitation.
FUS/TLS is a nuclear RNA binding protein. It interacts with DNA binding domains of nuclear steroid receptors. It is also present in subpopulations of TFIID complexes, indicating its potential role in the processing of primary transcripts that are generated in response to hormone-induced transcription (20). NF45 is a transcription factor that can interact with NF90 (ILF3, DRBP76) to control gene expression. It affiliates to the double-stranded RNA-binding protein family. It is a substrate of dsRNA-activated protein kinase PKR (21). NcoA3 is a member of key family of co-activators involved in the regulation of steroid receptor-mediated transcription. TARDBP, also known as TDP-43, has been identified as a major constituent of proteinaceous inclusions characteristic of most forms of amyotrophic lateral sclerosis and frontotemporal lobar degeneration (22). The 43-kDa transactivation response DNA binding protein (TDP43), FUS/TLS, heat shock protein 70, and β-amyloid have been reported to be induced and involved in cerebral ischemia, amyotrophic lateral sclerosis, and Alzheimer's disease (23). However, their involvements in ischemic tolerance have never been examined, although they could be involved in endogenous neuroprotection under ischemic preconditioning (23).
Our proteomic analysis of CTDSPL2 interacting partners also identified various unknown proteins. These may represent novel proteins that are either regulators or substrates of CTDSPL2 phosphatase. Some of these proteins identified by mass spectrometry were validated by CTDSPL2 immunoprecipitation. Our finding demonstrated the robustness of this proteomic approach for identifying protein partners of CTDSPL2 phosphatase. This will open new doors for understanding the functions and mechanisms of CTDSPL2 in physiological settings.
MATERIALS AND METHODS
Cell culture and establishment of inducible CTDSPL2-expressing cell line
HEK293T human embryonic kidney cells were purchased from ATCC (Bethesda, MD, USA) and maintained in high glucose (25 mM) DMEM (Life Technologies, Burlington, ON, USA) supplemented with 10% fetal bovine serum (Life Technologies) and antibiotics (Sigma-Aldrich Co. LLC, St. Louis, MO, USA) in a humidified incubator at 37℃ with 5% CO2. A tetracycline-inducible cell line for specific CTDSPL2 protein expression was established as previously described (18). The expressed CTDSPL2 protein was tagged with Flag epitope in the N-terminal region of the CDTSPL2 cDNA obtained from Korea Human Gene Bank (Daejeon, Korea). After antibiotics selection with puromycin and microscopic screening assays based on IRES-driven expression of EGFP, individual puromycin-resistant clones were selected, amplified, and screened through immunoblot analysis to check inducible expression (with or without doxycycline) of CTDSPL2. The purified cell clone was named HEK293T/Flag-CTDSPL2.
Preparation of cell lysates and subcellular fractionation
Cells were washed with ice-cold PBS and lysed with a lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 containing protease inhibitor cocktails) or conventional RIPA buffer containing protease inhibitors. Preparation of cytoplasmic and nuclear fractions from cultured HEK293T/Flag-CTDSPL2 with or without doxycycline induction was performed with Nuclear Extraction Kit from Cayman Chemical Co. (Ann Arbor, MI, USA) according to the manufacturer’s instruction. The efficiency of cellular fractionation was analyzed through immunoblot analysis using primary antibodies of α tubulin for cytoplasmic fraction or lamin B for nuclear fraction.
Immunoblot analysis and immunoprecipitation
Proteomic screenings were performed using immunocomplexes immunoprecipitated from total cell lysates, cytoplasmic, or nuclear fractions of induced and un-induced HEK293T/Flag-CTDSPL2 cells. Procedures for immunoblot analysis and immunoprecipitation were performed as described previously (14, 17). For immunoprecipitation assay and protein identification with mass spectrometry, pre-fractionated nuclear fractions containing 30 mg of total protein from HEK293T/Flag-CTDSPL2 cells with or without inducer doxycycline were precleared using appropriate isotype IgG antibody followed by mixing α-Flag antibody conjugated to Protein A Agarose (A2220, Sigma-Aldrich) and incubation at 4℃ overnight with gentle shaking. Immunoprecipitates were subjected to 6-15% gradient SDS-PAGE and Coomassie blue G250 staining. Images of stained gels were acquired by scanning using a high resolution scanner (GS-800 Calibrated Imaging Densitometer: BioRad Laboratories, Hercules, CA, USA). Protein concentration was determined using PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. For co-immunoprecipitation to identify protein partners of CTDSPL2, total cell lysate or pre-fractionated cytoplasmic or nuclear extracts (2 mg of total protein each) from cells cultured with or without the inducer were incubated with α-Flag antibody conjugated with Protein A Agarose (30 μl) and appropriate primary antibodies (5 μg). Immunoprecipitated proteins were separated by SDS-PAGE and analyzed as described previously. The following primary antibodies were used: α-Flag (Sigma-Aldrich), α-DYKDDDK (BioLegend Inc., San Diego, CA, USA), α-GAPDH (Young In Frontier Co., Seoul, Korea), α-NcoA3 (Santa Cruz Biotech. Inc., Santa Cruz, CA, USA), α-cdc2 p34 (Santa Cruz), α-lamin B (Santa Cruz), α-spectrin (Santa Cruz), α-TARDBP (Santa Cruz), α-NF45 (Santa Cruz), α-FUS/TLS (Santa Cruz), and α-α tubulin (Santa Cruz). Secondary antibodies were purchased from KPL, Inc. (Gaithersburg, MD, USA) or AbCam plc. (Cambridge, UK). For reprobing, blots were washed in 1X TBS to remove chemiluminescent substrate and incubated with BioFresh Western Blot Stripping Reagent (SignaGen Laboratories, Gaithersburg, MD, USA).
In-gel trypsin digestion and mass spectrometry analysis
For in-gel digestion procedure, the evenly excised 10 Coomassie Blue stained protein lanes were destained with 50% acetonitrile in 25 mM NH4HCO3 and dried in a vacuum centrifuge. Tryptic digested samples were analyzed by LC-ESI-MS/MS as described previously (14, 17). Peak lists of MS/MS spectra were processed using Analyst QS software (v.1.1, Applied Biosystems, Foster City, CA, USA) and searched against International Protein Index protein database version 3.20 (European Bioinformatics Institute, Hinxton, UK), NCBI- nonredundant protein databases, EST database, and other database using Mascot operating on a local server (24). The criteria for positive identification of proteins were set as follows: (i) at least two matching peptide masses, (ii) 50 ppm or better mass accuracy, and (iii) Mr of identified proteins should match estimated values obtained.
Acknowledgments
This work was supported by the DGIST R&D Program of the Ministry of Science, ICT and Technology of Korea (15-HRLA-02 and 15-BD-06) (J.H.K.) and by grants from the National Research Foundation of Korea (No. NRF-2010-0024199) (Y.Y.B.).
References
- 1.Licciardo P, Amente S, Ruggiero L. The FCP1 phosphatase interacts with RNA polymerase II and with MEP50 a component of the methylosome complex involved in the assembly of snRNP. Nucleic Acids Res. (2003);31:999–1005. doi: 10.1093/nar/gkg197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bataille AR, Jeronimo C, Jacques PE. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. (2012);45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Chambers RS, Dahmus ME. Purification and characterization of a phosphatase from HeLa cells, which dephosphorylates the C terminal domain of RNA polymerase II. J Biol Chem. (1994);269:26243–26248. [PubMed] [Google Scholar]
- 4.Kemmer D, Podowki RM, Arenillas D. Novel-Fam3000-uncharacterized human protein domains. BMC Genomics. (2006);7:1471–2164. doi: 10.1186/1471-2164-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsin J-P, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. (2012);26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Xiao M, Sun B, et al. C-terminal domain (CTD) small phosphatase-like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via smad dephosphorylation. J Biol Chem. (2014);289:16441–16450. doi: 10.1074/jbc.M114.568964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harikrishna RR, Kim H, Noh K, Kim YJ. Diverse roles in RNA polymerase II C-terminal domain phosphatase SCP1. BMB Rep. (2014);47:192–196. doi: 10.5483/BMBRep.2014.47.4.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamenski T, Heilmeier S, Meinhart A, et al. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol Cell. (2004);15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 9.Ma YN, Zhang X, Yu HC, Zhang JW. CTD small phosphatase like 2 (CTDSPL2) can increase ε- and γ-globin gene expression in K562 cells and CD34+ cells derived from umbilical cord blood. BMC Cell Biol. (2010);11:75–86. doi: 10.1186/1471-2121-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian H, Ji C, Zhao S, et al. Expression and characterization of HSPC129, a RNA polymerase II C-terminal domain phosphatase. Mol Cell Biochem. (2007);303:183–188. doi: 10.1007/s11010-007-9472-z. [DOI] [PubMed] [Google Scholar]
- 11.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. (1998);92:291–294. doi: 10.1016/S0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 12.Koo JH, Bahk YY. In vivo putative O-GlcNAcylation of human SCP1 and evidence for possible role of its N-terminal disorder structure. BMB Rep. (2014);47:593–598. doi: 10.5483/BMBRep.2014.47.10.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blalock W, Piazzi M, Bavelloni A, et al. Identification of the PKR nuclear interactome reveals roles in ribosome biogenesis, mRNA processing and cell division. J Cell Physiol. (2014);229:1047–1060. doi: 10.1002/jcp.24529. [DOI] [PubMed] [Google Scholar]
- 14.Bahk YY, Cho IH, Kim TS. A cross-talk between oncogenic Ras and tumor suppressor PTEN through FAK Tyr861 phosphorylation in NIH/3T3 mouse embryonic fibroblasts. Biochem Biophys Res Comm. (2008);377:1199–1204. doi: 10.1016/j.bbrc.2008.10.157. [DOI] [PubMed] [Google Scholar]
- 15.Maehama T, Taylor GS, Slama JT, Dixon JE. A sensitive assay for phosphoinositide phosphatases. Anal Biochem. (2000);279:248–250. doi: 10.1006/abio.2000.4497. [DOI] [PubMed] [Google Scholar]
- 16.Kwon Y, Vinayagam A, Sun X, et al. The Hippo signaling pathway interactome. Science. (2013);342:737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahk YY, Lee J, Cho IH, Lee HW. An analysis of an interactome for apoptosis factor, Ei24/PIG8, using the inducible expression system and shotgun proteomics. J Proteome Res. (2010);9:5270–5283. doi: 10.1021/pr100552y. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Lee YZ, Bahk YY. A proteomic approach for protein-profiling the oncogenic ras induced transformation (H-, K-, and N-Ras) in NIH/3T3 mouse embryonic fibroblasts. Proteomics. (2008);8:3082–3093. doi: 10.1002/pmic.200800106. [DOI] [PubMed] [Google Scholar]
- 19.Kim YJ, Bahk YY. A study of substrate specificity for a CTD phosphatase, SCP1, by proteomics screening of binding partners. Biochem Biophys Res Comm. (2014);448:189–124. doi: 10.1016/j.bbrc.2014.04.089. [DOI] [PubMed] [Google Scholar]
- 20.Powers CA, Mathur M, Raaka BM, Ron D, Samuels HH. TLS (translocated-in liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol Endocrinol. (1998);12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 21.Zhao G, Shi L, Qiu D, Hu H, Kao PN. NF45/ILF2 tissue expression, promoter analysis, and interleukin-2 transactivating function. Exp Cell Res. (2005);305:312–323. doi: 10.1016/j.yexcr.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 22.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. (2006);314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 23.Sun M, Yamashita T, Shang J, et al. Acceleration of TDP43 and FUS/TLS protein expressions in the preconditioned hippocampus following repeated transient ischemia. J Neurosci Res. (2014);92:53–63. doi: 10.1002/jnr.23301. [DOI] [PubMed] [Google Scholar]
- 24.Perkins DN, Pappin DJ, Cressy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. (1999);20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]


