Abstract
Many algorithms for sequence analysis rely on word matching or word statistics. Often, these approaches can be improved if binary patterns representing match and don’t-care positions are used as a filter, such that only those positions of words are considered that correspond to the match positions of the patterns. The performance of these approaches, however, depends on the underlying patterns. Herein, we show that the overlap complexity of a pattern set that was introduced by Ilie and Ilie is closely related to the variance of the number of matches between two evolutionarily related sequences with respect to this pattern set. We propose a modified hill-climbing algorithm to optimize pattern sets for database searching, read mapping and alignment-free sequence comparison of nucleic-acid sequences; our implementation of this algorithm is called rasbhari. Depending on the application at hand, rasbhari can either minimize the overlap complexity of pattern sets, maximize their sensitivity in database searching or minimize the variance of the number of pattern-based matches in alignment-free sequence comparison. We show that, for database searching, rasbhari generates pattern sets with slightly higher sensitivity than existing approaches. In our Spaced Words approach to alignment-free sequence comparison, pattern sets calculated with rasbhari led to more accurate estimates of phylogenetic distances than the randomly generated pattern sets that we previously used. Finally, we used rasbhari to generate patterns for short read classification with CLARK-S. Here too, the sensitivity of the results could be improved, compared to the default patterns of the program. We integrated rasbhari into Spaced Words; the source code of rasbhari is freely available at http://rasbhari.gobics.de/
Author Summary
We propose a fast algorithm to generate spaced seeds for database searching, read mapping and alignment-free sequence comparison. Spaced seeds—i.e. patterns of match and don’t-care positions—are used by many algorithms for sequence analysis; designing optimal seeds is therefore an active field of research. In sequence-database searching, one wants to optimize sensitivity, i.e. the probability of finding a region of homology; this can be done by minimizing the so-called overlap complexity of pattern sets. In alignment-free DNA sequence comparison, the number N of pattern-based matches is used to estimate phylogenetic distances. Here, one wants to minimize the variance of N in order to obtain stable phylogenies. We show that for spaced seeds, the overlap complexity—and therefore the sensitivity in database searching—is closely related to the variance of N. Our algorithm can optimize the sensitivity, overlap complexity or the variance of N, depending on the application at hand.
Introduction
k-mers, i.e. words of length k, are used in many basic algorithms for biological sequence comparison. Word matches are used, for example, as seeds in the hit-and-extend approach to database searching and read mapping [1–3]. Here, fast algorithms are applied to find pairs of identical or similar words between two sequences. A slower but more sensitive alignment method is then used to extend these word pairs to both directions, to distinguish biologically relevant homologies from spurious word matches. In alignment-free sequence comparison, sequences are often represented by word-frequency vectors to estimate distances or similarities between them, e.g. as a basis for phylogeny reconstruction [4–8], see [9] for a review. Similarly, word statistics are used to classify DNA or protein sequences [10–12], for datamining [13] and for remote homology detection [14].
It is well known that many word-based approaches produce better results if spaced words or seeds are used instead of the previously used contiguous words or word matches. That is, for a pre-defined binary pattern P representing match and don’t-care positions, one considers only those positions in a word of the same length that correspond to the match positions of P. Pattern-based word matching has been proposed for hit-and-extend database searching by Ma et al. [15], see also [16]. Spaced seeds are also routinely used in metagenome sequence clustering and classification [17, 18], protein classification [19], read mapping [20, 21], to find anchor points for multiple sequence alignment [22, 23] and for alignment-free phylogeny reconstruction [24]. Similarly, the average common substring approach to sequence comparison [25] could be improved by allowing for mismatches [26–30]. Brejova et al. extended the concept of spaced seeds to homologies among protein-coding regions [31] and introduced vector seeds [32]. In general, the advantage of pattern-based approaches is the fact that spaced-word occurrences at neighbouring sequence positions are statistically less dependent than occurrences of contiguous words [33, 34]. Often sets of patterns are used, instead of single patterns; such multiple spaced seeds are now a standard filtering step in homology searching [35, 36].
In pattern-based approaches, the underlying patterns of match and don’t-care positions are of crucial importance for the quality of the results. Generally, non-periodic patterns are preferred since they minimize redundancies between overlapping words or word matches and lead to a more even distribution of matches. This increases the probability of obtaining a hit between two homologous sequences in database searching and leads to more stable distance estimates in phylogeny reconstruction. Noé and Martin [37] defined a coverage criterion for multiple spaced seeds and showed that this criterion is related to the Hamming distance between two sequences. In the context of database searching, patterns or sets of patterns are often called seeds. (Originally, the word seed denoted a match of—contiguous or spaced—words between a query and a database sequence that would be extended to the left and to the right. But now seed often denotes the underlying pattern in pattern-based approaches).
In hit-and-extend database searching, the sensitivity of a pattern set is defined as the probability of finding at least one hit within a gap-free alignment of a given length L and probability p for a match between two residues. Since each hit is extended to a local alignment, the sensitivity is the proportion of homologies that will be found by a search program—under the above simple model of homology, and under the assumption that each extension of a hit in a homologous region will verify the homology. In database searching, the goal is thus to maximize the sensitivity of pattern sets.
Calculating the sensitivity of a pattern set is NP-hard [33]. The sensitivity can be approximated by dynamic programming [15, 38], but the run time of this algorithm is still exponential in the length of the pattern. In PatternHunter II, a greedy algorithm is used to find suitable patterns. In 2007, Ilie and Ilie introduced the overlap complexity of a pattern set and showed experimentally that—for a given number of patterns with a given length and number of match positions—minimizing the overlap complexity corresponds to maximizing the sensitivity in database searching [39]. In contrast to the sensitivity, however, the overlap complexity can be easily calculated. To find optimal pattern sets, Ilie and Ilie proposed a hill-climbing algorithm that minimizes the overlap complexity. They implemented their algorithm in a software tool called SpEED [40], which is several orders of magnitude faster than competing approaches and is now considered the state-of-the-art in seed optimization.
Recently, we proposed to use spaced-word frequencies instead of word frequencies for alignment-free sequence comparison [24, 41]. We showed that phylogenetic trees calculated from spaced-word frequencies are more accurate than trees calculated from contiguous-word frequencies. As in database searching, our results could be improved by using multiple patterns. In our original study, we used randomly generated multiple patterns of match and don’t-care positions. In a follow-up paper, we studied the number N of spaced-word matches between two DNA sequences for a set of binary patterns [34]. Our data suggest that minimizing the variance of N for pattern sets improves alignment-free phylogeny reconstruction.
In this paper, we first show that the variance of the number N of spaced-word matches is closely related to the overlap complexity of the underlying set of patterns. We propose a modified hill-climbing algorithm that can be used to generate pattern sets, either with minimal variance of N, or with minimal overlap complexity, or with maximal sensitivity in database searching, depending on the application at hand. While the algorithm proposed in [39] iterates over all patterns P in a set of patterns and all pairs of positions in P to improve , we calculate for each pattern how much P contributes to the variance or overlap complexity, respectively, of . We then modify those patterns first that contribute most to the variance or complexity.
The implementation of our approach is called rasbhari (Rapid Approach for Seed optimization Based on a Hill-climbing Algorithm that is Repeated Iteratively). Experimental results show that pattern sets calculated with rasbhari have a slightly higher sensitivity in database searching than pattern sets calculated with SpEED, while the run time of both programs is comparable. In alignment-free sequence comparison, we obtain more accurate phylogenetic distances if we use rasbhari to minimize the variance of N for the underlying pattern sets, than we obtained with the randomly generated pattern sets that we previously used. In a third application, we used pattern sets generated with rasbhari in the program CLARK-S [18] for short read classification. The sensitivity of the classification could be improved in this way, while rasbhari is substantially faster than the method that is used by default for pattern generation in CLARK-S.
A earlier version of this paper has been published at the preprint server arXiv [42].
Methods
Overlap complexity
We consider sets of binary patterns, where ℓr is the length of pattern Pr and ℓ = maxr ℓr. That is, each Pr is a word of length ℓr over the alphabet {1, 0}. A ‘1’ in a pattern Pr represents a match position, a ‘0’ a don’t-care position. For a single pattern Pr, the number of match positions is called its weight w. For simplicity, we assume that all patterns in a set have the same weight.
In [34], we considered for two patterns Pr, Pr′ and the number n(Pr, Pr′, s) of positions that are match positions of Pr or match positions of Pr′ (or both), if Pr′ is shifted by s positions to the right, relative to Pr. If s is negative, Pr′ is shifted to the left. For Pr = 101011, Pr′ = 111001, for example, if Pr′ is shifted by 2 positions to the right, relative to Pr, then there are 6 positions (marked by asterisks below) that are match positions of Pr or Pr′. Thus, for s = 2, we have n(P, Pr′, 2) = 6:
For the same situation, Ilie and Ilie [39] defined σ[s] = σr,r′[s] as the number of positions where Pr and Pr′ have a match positions, such as the positions marked by ‘$’ above. In the above example one would therefore have σ[2] = 2. The overlap complexity (OC) of a set of patterns is then defined in [39] as
| (1) |
Note that, since for any two patterns Pr, Pr′ and , the equality
holds, the overlap complexity of a set can be written as
| (2) |
Consequently, if we are looking at sets of m patterns with fixed weight w and lengths ℓr, then minimizing the overlap complexity of is equivalent to minimizing the sum
| (3) |
Ilie and Ilie showed experimentally that the OC is closely related to the sensitivity of a pattern set. More precisely, they showed that for pattern sets with a given number of patterns of given lengths and weight, minimizing the OC practically amounts to maximizing the sensitivity. Consequently, in order to find suitable pattern sets for hit-and-extend approaches in database searching, they proposed to search for pattern sets with minimal OC. The main advantage of this approach is the fact that the OC of a pattern set is much easier to calculate than its sensitivity.
Variance of the number of spaced-word matches
For a pattern P of length ℓ, we say that two sequences S1 and S2 have a spaced-word match with respect to P at (i, j), if the ℓ-mers starting at i and j have identical characters at all match positions of P, i.e. if one has S1(i + π − 1) = S2(j + π − 1) for all match positions π in P. The sequences below, for example, have a spaced-word match at (2, 4) with respect to the pattern P = 110101. Indeed, the 6-mers starting at positions 2 and 4 of the sequences are identical at all positions corresponding to a match position (‘1’) in P, while positions at don’t-care positions (‘0’) may be matches or mismatches.
In [34], we considered spaced-word matches between two sequences S1 and S2 with respect to a set of patterns, so-called -matches. Note that there can be up to m -matches at each pair (i, j) of positions of S1 and S2, one -match for each pattern Pr in . We studied the number of -matches between sequences S1 and S2 under a simplified model of evolution without insertions and deletions, with a match probability p for pairs of homologous positions and a background match probability of q. Thus, in our model we have
It is easy to see that, for a pattern set , the expected number of -matches depends only on the number m of patterns in and on their lengths ℓi and their weight w, i.e. number of match positions, but not on the particular sequence of match and don’t-care positions in . The variance of N, however, does depend on the sequence of match and don’t-care positions.
As discussed in [34], many alignment-free distance or similarity measures are—explicitly or implicitly—a function of the number N of (spaced) word matches. To obtain stable distance measures for phylogeny reconstruction, it is therefore desirable to use pattern sets with a low variance of N. For a given set of patterns of lengths ℓ1, …, ℓm and weight w, and with the above simple model of evolution, the variance of N can be approximated by
| (4) |
where L is the length of S1 and S2, respectively, and
is the range in which Pr′ is to be shifted against Pr [34]. Note that for different patterns Pr′ ≠ Pr we have to consider all shifts between 1 − ℓr′ and ℓr − 1 of Pr′ against Pr, for example:
By contrast, if a pattern Pr is shifted against itself, only shifts between 0 and ℓr − 1 need to be considered, to avoid double counting of shifts, for example:
In [34], we ignored this fact and gave a slightly different estimate for Var(N).
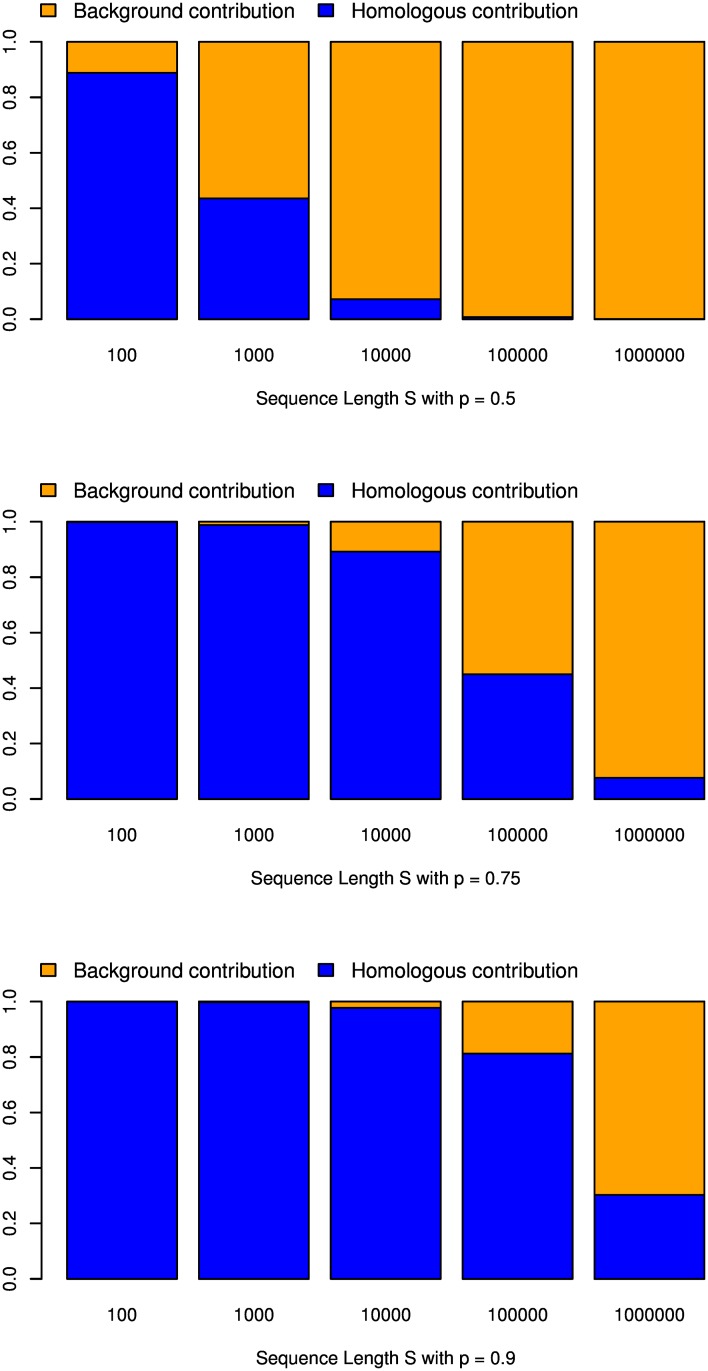
On the right-hand side of Eq (4), the first summand is the variance of the ‘homologous’ spaced-word matches (in a model without indels, these are spaced-word matches involving the same positions in both sequences), while the second summand comes from background matches. The relative weight of the background matches in Eq (4) depends on the match probability p and the sequence length L; for p >> q and small L, the Var(N) is dominated by the ‘homologous’ term, see Fig 1. Obviously, for large L, the background spaced-word matches dominate the ‘homologous’ ones, since the number of background matches grows quadratically with L, while the ‘homologous’ matches grow only linearly.
Fig 1. Homolgue and background contribution to the variance of the number N of spaced-word matches.
Contribution of the homologue and background variance to the total variance of the number N of spaced-word matches in eq (4) for different match probabilities p and sequence lengths L.
Note that, for L, ℓ and w fixed, minimizing the Var(N) amounts to minimizing
| (5) |
Comparison with Eq (2) shows that, in the special case of p = 1/2, the first summand of Eq (5) that corresponds to the homologous matches is almost identical with the overlap complexity defined by Ilie and Ilie (except for the range R(r, r) in which a pattern Pr is shifted against itself). For sequences of moderate length, the overlap complexity can therefore be seen as an approximation to the variance of the number of spaced-word matches.
In any case, the overlap complexity and the Var(N) for a set of pattern both have the form
| (6) |
with
| (7) |
Our optimization problem is therefore: for integers m, ℓ1, … ℓm, w, find a set of m patterns of lengths ℓ1, …, ℓm and weight w that minimizes the sum Eq (6).
Hill-climbing algorithms to find sets of patterns with minimal Var(N) or OC
Both SpEED and our new algorithm start with randomly generated pattern sets and use hill-climbing to gradually reduce the OC or Var(N). If one wants to find a pattern set with maximal sensitivity, the sensitivity is calculated for the pattern set that is produced by this procedure (this step is omitted, of course, if rasbhari is used to minimize Var(N) or OC). The whole procedure is repeated, and the pattern set with the overall highest sensitivity—or lowest Var(N) or OC, respectively—is returned.
Original hill-climbing algorithm
To improve the current pattern set , the hill-climbing algorithm implemented in SpEED looks at all triplets (r, i, j) where Pr is a pattern in , and i and j are a match position and a don’t-care position in Pr, respectively. For each such triplet (r, i, j), the algorithm considers the pattern set that would be obtained from by swapping i and j in Pr—i.e. by turning i into a don’t-care and j into a match position. The OC is calculated for all pattern sets that can be obtained in this way, and the one with the lowest OC is selected as the next pattern set . This is repeated iteratively.
There are O(m ⋅ ℓ2) triplets (r, i, j) to be considered to modify the current pattern set . For each of these triplets, the OC is to be calculated for the pattern set that would be obtained by swapping i and j in Pr. To this end, the modified pattern Pr has to be compared to the m − 1 remaining patterns in which, for each pattern comparison, involves O(ℓ) shifts of two patterns against each other. In each shift, the number of common match positions is to be counted, which takes again O(ℓ) time. Thus, calculating the OC of the pattern set obtained by swapping two positions i and j in a pattern Pr takes O(m ⋅ ℓ2) time, so finding an optimal triplet (r, i, j) to determine the next pattern set takes O(m2 ⋅ ℓ4) time. In SpEED, this step is repeated until the OC cannot be improved further, i.e. until a local minimum is reached. For the pattern set that is obtained by this hill-climbing routine, the sensitivity is calculated. This whole procedure is repeated 5,000 times, and finally the set with the best sensitivity is returned.
Modified hill-climbing algorithm
In our modified hill-climbing algorithm, we also swap a match position i with a don’t-care position j in some pattern Pr in each step of the algorithm, and we evaluate the OC or Var(N) of the pattern set that would be obtained by this operation. However, instead of looking at all possible triplets (r, i, j), we look at those patterns first that contribute most to the OC or Var(N), respectively, of the current pattern set . The contribution
| (8) |
of a pattern to the OC or Var(N) of can be calculated as a by-product, whenever OC or Var(N) is calculated, with αr,r′ as in Eq (7). We then sort the patterns in according to the values Cr, and we process them in descending order of Cr, i.e. we look at those patterns first that contribute most to the OC or Var(N) of .
For the current pattern in the list, we randomly select a match position i and a don’t-care position j. If swapping i and j does not improve the current pattern set, we move on to the next pattern in the list and proceed in the same way. This is repeated until we find a pattern where swapping the selected pair of random positions does improve . In this case, the modified pattern is accepted, all values Cr are updated, the patterns in are sorted accordingly, and we start again with the pattern Pr with maximum Cr. If we reach the last pattern in the list without obtaining any improvement, we start again with the first pattern, i.e. the pattern with the largest Cr, select new random positions i and j etc. Processing one pattern Pr in this way takes O(m ⋅ ℓ2) time, since we look only at one single pair (i, j) and calculate the OC or Var(N) of the pattern set that would be obtained by swapping i and j in Pr.
The hill climbing is continued until a user-defined number of pairs (i, j) have been swapped to improve the current pattern set; by default, 25,000 pairs are swapped. If we want to obtain a pattern set with maximal sensitivity, the described hill-climbing procedure is repeated 100 times, and for the pattern set with the lowest OC among the 100 obtained pattern sets, the sensitivity is calculated. To calculate the sensitivity, rasbhari uses program code from SpEED. Again, this whole process is repeated 5,000 times, so for a total of 5,000 pattern sets the sensitivity is calculated during one program run. This is similar to SpEED, but in SpEED the time-consuming sensitivity calculation is carried out after one round of hill climbing. By contrast, we run our faster hill-climbing routine 100 times before we calculate the sensitivity for the best pattern set from these 100 runs. The final output of our program is the pattern set with the highest sensitivity from the 5,000 iterations.
The number m of patterns and their weight w are to be specified by the user. If Var(N) is to be minimized for alignment-free sequence comparison, all patterns have the same length ℓ which is also to be specified by the user. If the sensitivity is to be maximized for database searching and read alignment, better results are achieved if the patterns in have different lengths. In this case, the maximum and minimum pattern lengths need to be specified. The program then selects lengths ℓ1, …, ℓm that are evenly distributed between these extreme values.
Results
Sensitivity in database searching
To evaluate rasbhari, we first applied it to generate pattern sets, maximizing the sensitivity for database searching and read mapping. For the number m and weight w of the patterns and for the length H and match probability p of the homology regions, we used the parameter settings from SHRiMP2 [43], PatternHunter II [38] and BFAST [44]. We and compared it to the sensitivity of pattern sets obtained with Iedera [45], SpEED [40], AcoSeeD [46], FastHC and MuteHC [47] as published by the authors of these programs; the results of this comparison are shown in Table 1. Here, the sensitivity values of AcoSeeD are average values over 10 program runs reported in [46].
Table 1. Sensitivity comparison of different programs.
| w | p | Iedera | SpEED | AcoSeeD | FastHC | MuteHC | rasbhari |
|---|---|---|---|---|---|---|---|
| SHRiMP2: 4 patterns (H = 50) | |||||||
| 10 | 0.75 | 90.6820 | 90.9098 | 90.9513 | 90.7312 | 92.6812 | 90.9614 |
| 0.80 | 97.7586 | 97.8337 | 97.8521 | 97.7625 | 98.3836 | 97.8554 | |
| 0.85 | 99.7437 | 99.7569 | 99.7614 | 99.7431 | 99.8356 | 99.7618 | |
| 11 | 0.75 | 83.2413 | 83.3793 | 83.4728 | 83.3068 | 83.4127 | 83.4679 |
| 0.80 | 94.9350 | 94.9861 | 95.037 | 94.9453 | 95.0194 | 95.0386 | |
| 0.85 | 99.2189 | 99.2431 | 99.2478 | 99.2250 | 99.2486 | 99.2506 | |
| 12 | 0.80 | 90.3934 | 90.5750 | 90.6328 | 90.4735 | 90.5820 | 90.6648 |
| 0.85 | 98.0781 | 98.1589 | 98.1766 | 98.1199 | 98.1670 | 98.1824 | |
| 0.90 | 99.8773 | 99.8821 | 99.8853 | 99.8771 | 99.8836 | 99.8864 | |
| 16 | 0.85 | 84.5795 | 84.8212 | 84.9829 | 84.6558 | 84.8764 | 84.969 |
| 0.90 | 97.2806 | 97.4321 | 97.4712 | 97.3556 | 97.4460 | 97.5035 | |
| 0.95 | 99.9331 | 99.9388 | 99.9419 | 99.9347 | 99.9424 | 99.9441 | |
| 18 | 0.85 | 72.1695 | 73.1664 | 73.27 | 72.9558 | 73.2209 | |
| 0.90 | 93.0442 | 93.7120 | 93.7778 | 93.6030 | 93.78 | ||
| 0.95 | 99.6690 | 99.7500 | 99.7599 | 99.7399 | 99.7557 | ||
| PatternHunterII: 16 patterns (H = 64) | |||||||
| 11 | 0.70 | 92.0708 | 93.2526 | 93.0585 | 93.4653 | ||
| 0.75 | 98.3391 | 98.6882 | 98.6352 | 98.7573 | |||
| 0.80 | 99.8366 | 99.8820 | 99.8750 | 99.8907 | |||
| BFAST: 10 patterns (H = 50) | |||||||
| 22 | 0.85 | 60.1535 | 60.8127 | 60.0943 | 60.9919 | ||
| 0.90 | 87.9894 | 88.5969 | 88.0426 | 88.8005 | |||
| 0.95 | 99.2196 | 99.3659 | 99.2923 | 99.4099 | |||
Sensitivity of pattern sets in hit-and-extend database searching, calculated with different programs. Parameter settings for the number m and weight w of patterns, the length H of the gap-free homology region between query and database sequences and the match probability p in the homology regions, are taken from three popular programs SHRiMP2, PatternHunter II and BFAST. Sensitivity values from rasbhari were calculated using program code from SpEED; results of all other programs are taken from their respective publications.
If pattern sets with maximal sensitivity are to be found, and if the lengths ℓr of the patterns are small, the run time of rasbhari is comparable to SpEED. In this case, the most time-consuming step in both programs is to calculate the sensitivity of pattern sets which, by default, is done 5,000 times per program run in each of the two programs. For longer patterns, however, SpEED can be much slower since it carries out hill-climbing until a local minimum is reached and, as explained above, each single step in the hill-climbing procedure of SpEED takes O(m2 ⋅ ℓ4) time. In contrast, rasbhari terminates this procedure after a given number of iteration steps, and it considers only a limited number of swaps of match and don’t-care positions in one iteration step.
Alignment-free phylogeny reconstruction
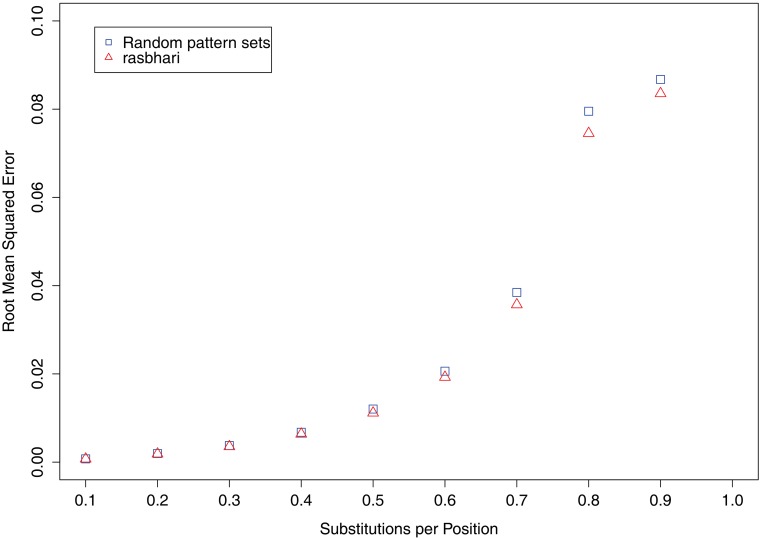
Next, we wanted to know how alignment-free phylogeny reconstruction can be improved with rasbhari. To this end, we simulated pairs of DNA sequences and estimated the distances between them using the Spaced Words approach described in [34]. We then measured the accuracy of the distance estimates for different underlying pattern sets. First, we used rasbhari to minimize the variance of the number N of spaced-word matches between two sequences. Since there is no other method to minimize Var(N), we compared the pattern sets from rasbhari with the randomly generated pattern sets that we previously used. The phylogenetic distances estimated with both types of pattern sets were compared to the ‘real’ distances between the sequences, i.e. the average number of substitutions per position. As test data, we generated nine data sets with 2,500 pairs of DNA sequences of length 100 kb each. The distances d of the sequence pairs ranged between 0.1 and 0.9 substitutions per position. For each program run, we used a set of m = 3 patterns of length 20 with 16 match and 4 don’t-care positions. Fig 2 shows the root mean square error of the estimated distances, compared to the ‘real’ distances d. The pattern sets generated with rasbhari were superior to the randomly generated pattern sets, especially for large distances.
Fig 2. Accuracy of phylogenetic distance estimates based on different pattern sets.
Nine sets of DNA sequence pairs were simulated with distances d between 0.1 and 0.9 substitutions per position. Distances were estimated based on the number N of spaced-word matches between them, using the alignment-free method published in [34]. We used two types of underlying pattern sets, (a) pattern sets generated with rasbhari, minimizing the variance of N, and (b) randomly generated pattern sets. The root mean square error of the estimated distances is plotted against the ‘real’ distances d.
Read classification with CLARK-S
As a third test case, we used different pattern sets for CLARK-S [18, 48], a recently developed tool for short read classification. We evaluated the accuracy of CLARK-S with three underlying pattern sets, namely (A) with the patterns used by default in the program, (B) with patterns from rasbhari minimizing overlap complexity and (C) with patterns from rasbhari maximizing sensitivity.CLARK-S uses sets of m = 3 patterns of length ℓ = 31 and with a weight of w = 22. Since SpEED is too slow to generate pattern sets with long patterns, the authors of the program generated pattern sets for CLARK-S by exhaustively searching over all single patterns with ℓ = 31 and w = 22. If the first and the last position in the patterns are required to be match positions, this approach has to evaluate possible patterns. The sensitivity of each of these patterns was calculated, and the three patterns with the highest sensitivity were selected. Note however, that maximizing the sensitivity of single patterns is only an approximation to finding a set of patterns with maximal total sensitivity.
Fig 3 shows the default pattern set from CLARK-S and the two pattern sets generated by rasbhari as described. The exhaustive procedure used by CLARK-S took 2 hours to generate the pattern set.rasbhari, by contrast, calculated pattern sets with the same parameters within 7.54 seconds with the slow version where the sensitivity is calculated, and within 0.068 seconds with the fast version where the overlap complexity is maximized without considering the sensitivity explicitly. The slow version of rasbhari is thus around 480 times faster than the exhaustive procedure in CLARK-S, while the fast version is around 52,000 times faster. The theoretical sensitivity of the three pattern sets is 0.999771 for the default patterns from CLARK-S, 0.999811 for the rasbhari patterns with minimized overlap complexity and 0.999822 for the rasbhari patterns with maximized sensitivity.
Fig 3. Pattern sets for short read classification.
Pattern sets used for short read classification: (A) as used by default in CLARK-S, (B) generated with rasbhari minimizing overlap complexity and (C) generated with rasbhari maximizing sensitivity.
To evaluate the classification accuracy of CLARK-S with these three pattern sets experimentally, we used five data sets from the literature, namely two sets, HC1 and HC2, from the MetaPhlAn project [49] and three sets, simHC, simMC and simLC, from the FAMeS databases [50]. For each of these data sets, we calculated precision and sensitivity of the classification at the species level as defined in [11]. That is, for a classification task where objects are to be assigned to classes, precision is defined as the fraction of correct assignments among the total number of assignments, while sensitivity is the ratio between the number of correct assignments and the number of objects to be classified. The two values are not the same since not every object is necessarily assigned to one of the classes; precision is always larger than or equal to sensitivity since the denominator in the definition of precision is smaller or equal to the denominator in the definition of sensitivity. Since this definition of sensitivity refers to the ability of a program to correctly classify objects, it is not to be confused with the sensitivity in database searching as discussed above. Table 2 summarizes precision and sensitivity of CLARK-S with its default pattern set and with a pattern set generated by rasbhari.
Table 2. Read classification with CLARK-S using different pattern sets.
| Default pattern set | rasbhari | ||||
|---|---|---|---|---|---|
| Dataset | #reads | Precision | Sensitivity | Precision | Sensitivity |
| HC1 | 999,998 | 97.69 | 90.36 | 97.69 | 90.44 |
| HC2 | 999,991 | 96.45 | 88.11 | 96.45 | 88.18 |
| simHC | 116,771 | 97.20 | 90.53 | 97.20 | 90.54 |
| simMC | 97,495 | 98.75 | 95.09 | 98.73 | 95.09 |
| simLC | 114,457 | 98.29 | 94.26 | 98.28 | 94.25 |
Read classification with CLARK-S [18] with the default pattern set of the program and with the pattern set from rasbhari for the same parameter values, namely n = 3 patterns of length ℓ = 31 and weight w = 21. Precision and sensitivity of the classification are reported at the species level for five data sets from the literature.
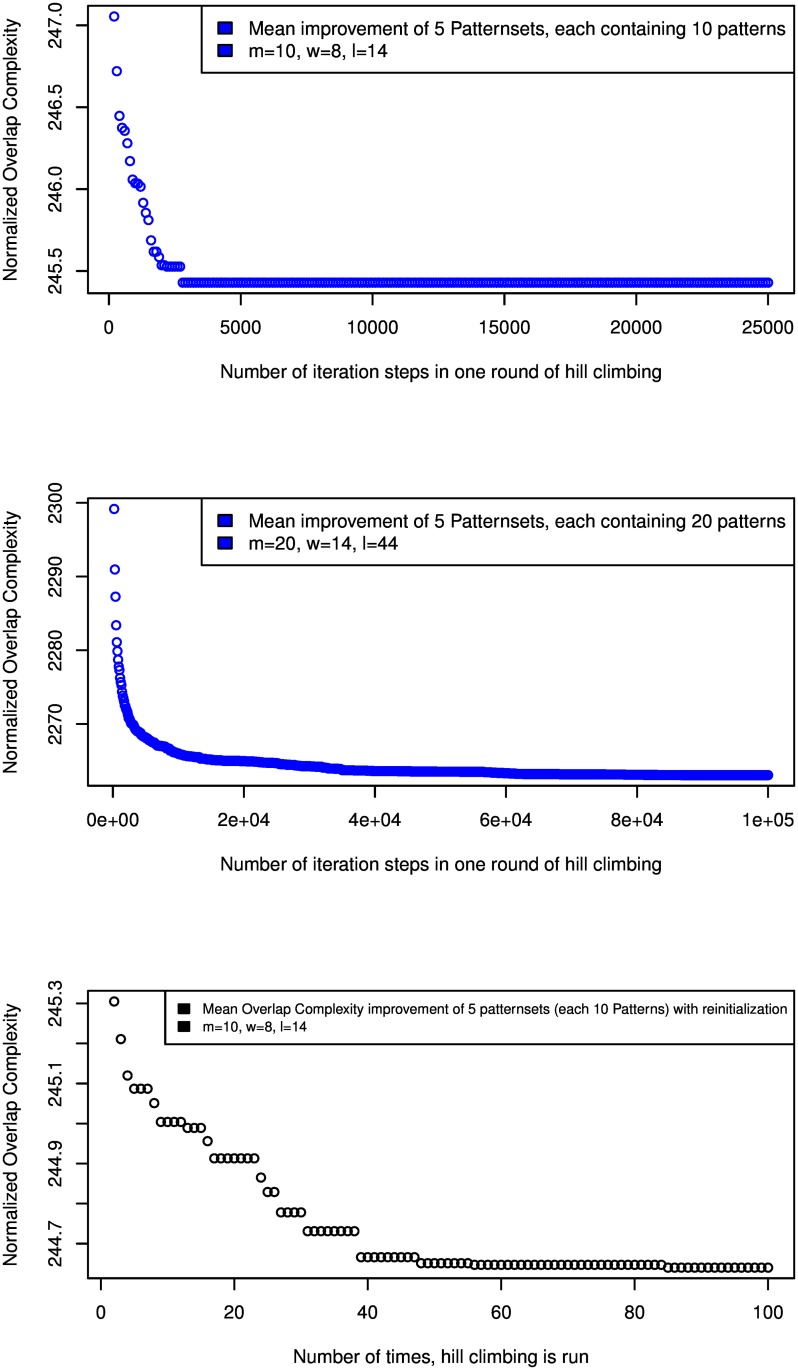
Fig 4 shows how the overlap complexity (OC) of pattern sets produced by rasbhari depends on the number of iteration steps carried out in the hill-climbing algorithm. For a set of m = 10 patterns of length ℓ = 14 and weight w = 8, a single run of the hill-climbing procedure converges after around 3,000 steps; for m = 20, ℓ = 44, w = 14, it converges after around 80,000 steps. The OC is further improved if the hill-climbing procedure is run multiple times and the best result of these runs is used.
Fig 4. overlap complexity of pattern sets in the hill-climbing algorithm.
Normalized overlap complexity (OC) of pattern sets depending on the number of iteration steps in our algorithm. The first two plots show how the OC is reduced in a single round of the hill-climbing algorithm for different parameters. For a set of m = 10 patterns of length ℓ = 14 and weight w = 8, the algorithm converges after around 3,000 iteration steps of hill-climbing (upper plot); for a set of m = 20 patterns of length ℓ = 44 and weight w = 14, it converges after around 80,000 steps (middle plot). The lower plot shows how the OC is improved if the hill-climbing algorithm is run multiple times and the best result of all runs is returned.
In the previous section, we mentioned that the OC is related to the variance of the number N of spaced word matches. Comparison of eqs (5) and (2) showed that, in the special case where p = 1/2 and the contribution of the ‘background’ spaced-word matches is small, minimizing the OC is equivalent to minimizing the variance of N. In general, however, this is not the case, as the following example shows. We applied rasbhari to generate two sets of m = 10 patterns each, with length ℓ = 20 and weight w = 8, one set by minimizing the OC and the other one by minimizing Var(N). When generating the second set, we used a match probability of p = 0.75 and a sequence length of L = 10,000. The pattern set that we obtained when we minimized the OC had an OC of 11,116, the set for which we minimized Var(N) had an OC of 11,195. Conversely, when we minimized Var(N), we obtained a pattern set with a variance of 156,061, while the variance was 156,152 when we minimized the OC. It thus makes a difference which one of these two parameters is minimized.
Discussion
We developed a program called rasbhari to calculate sets of binary patterns—or spaced seeds, as they are often called—for read mapping, database searching and alignment-free sequence comparison. For sequence-homology searching, rasbhari optimizes the sensitivity of pattern sets, i.e. the probability of obtaining at least one hit between a query and a database sequence sharing a gap-free homology of a given length and with a given match probability between nucleotides. Since the sensitivity of a pattern set is expensive to calculate, our algorithm optimizes the overlap complexity of the produced pattern sets which is closely related to its sensitivity. We use a hill-climbing algorithm, similar to the one used in SpEED, to minimize the overlap complexity. Unlike SpEED, however, our algorithm does not calculate the overlap complexity of all neighbours of a current pattern set, but modifies those patterns first that contribute most to the overlap complexity of the current pattern set. To maximize the sensitivity in database searching, we calculate the sensitivity of the current pattern set after a certain number of iterations. We repeat this procedure and, finally, we pick the pattern set with the highest sensitivity in all iterations.
Since calculating the sensitivity is time consuming, rasbhari can alternatively minimize the overlap complexity alone, without calculating the sensitivity of pattern sets. This option is useful in situations where large pattern sets are needed for which it would take too long to calculate the sensitivity. As a third option, rasbhari can minimize the variance of the number N of spaced-word matches in alignment-free sequence comparison which is used by various methods to estimate phylogenetic distances between sequences. We could show that, mathematically, the variance of N has a similar form as the overlap complexity of a pattern set, so the same optimization algorithm can be used to minimize both of them.
In both homology searching and read classification, pattern sets generated by rasbhari are more sensitive than alternative pattern sets, so more homologies can be detected and more reads can be correctly classified. At first glance, the increase in sensitivity that we obtained seems moderate; as shown in Table 1, the improvement is usually in the first or second digit after the decimal mark. In database searching and read mapping, however, even small improvements in sensitivity can lead to a large number of additional hits. Moreover, as these additional hits will be mostly in the ‘twilight zone’ of low sequence similarity, they may be of particular interest to the user.
In the context of read alignment, Ilie et al. pointed out that, with a 100-fold coverage of the human genome, a 1 percent improvement in pattern sensitivity would mean that 3 billion more nucleotides could be mapped [40], so the improvement that we achieved with rasbhari would still lead to tens or hundreds of millions of additionally mapped nucleotides. In database searching, the situation is similar. If we consider, for example, homology regions of length H = 64 with a match probability of p = 0.8 at the nucleotide level, then with w = 11, the sensitivity of rasbhari is improved by less than 0.01 percentage points compared to SpEED, see Table 1. Note, however, that these sensitivity values are already close to 100%, so the fraction of homologies that are not detected can be considerably reduced with the slight improvement in sensitivity obtained with rasbhari. In our example, the number of homologies that are missed is reduced by >7% if rasbhari is used instead of SpEED. With the same parameters, but with p = 0.7, the sensitivity of both programs is around 93%. Here, the number of missed homologies is still reduced by 3% with rasbhari, compared to SpEED.
For alignment-free sequence comparison, pattern sets produced by rasbhari lead to more accurate phylogenetic distances than the random pattern sets that we previously used. While this result may not be surprising, rasbhari is, to our knowledge, the first program that has been designed for this purpose and that can minimize the variance of the number of spaced-word matches. We therefore integrated rasbhari into our web server for alignment-free sequence comparison [41].
In read classification, the sensitivity of CLARK-S could be increased by 0.08 and 0, 07 percentage points, respectively, for the largest data sets that we used, HC1 and HC2. Each of these data sets contains around one million reads, so the improvement in sensitivity that we achieved with rasbhari means that 800 more reads from HC1 and 700 more from HC2 could be correctly classified by CLARK-S. This is remarkable, since the classification accuracy of CLARK-S is already very high, so it is hard to further improve the program. An interesting question in the context of CLARK-S is how the length and weight of the patterns influence its accuracy. So far, it was difficult to investigate this question systematically, since the exhaustive method that the program uses by default, is too time consuming. With the massive improvement in runtime that we achieved with rasbhari, it is now possible to systematically investigate how the accuracy of CLARK-S depends on the parameters of the underlying pattern sets.
In the hill-climbing procedure, our default of 25,000 iteration steps was sufficient to obtain stable results for the parameter settings that we used in our benchmark studies; we were unable to further improve these results by increasing the number of iterations. For different values of m, w, ℓ, p and H, however, it may be advisable to adapt the number of iteration steps. Fig 4 shows that, if the number of patterns or their length and weight are increased, a larger number of iteration steps can improve the results. The number of iterations within one round of hill climbing and the number of times the hill-climbing is carried out can be passed to rasbhari through the command line; the users can therefore adapt these parameter values for their particular applications if they do not want to use the default values of the program.
Acknowledgments
We would like to thank Laurent Noé for helpful discussions and for pointing out the similarity between the overlap complexity and the variance of the number of spaced-word matches. Lucian Ilie made useful comments on a previous version of this paper.
Data Availability
All relevant data are within the paper or it is mentioned how to download it with a given http-link.
Funding Statement
For BM, LH and CAL this work was funded by the budget of the Department of Bioinformatics, Institute of Microbiology and Genetics, University of Göttingen. Therefore there was no external funding. For SL this work was funded by the budget of the Department of Computer Science and Engineering, University of California, Riverside USA. For RO this work was funded by the graduate student support NSF IIS-1526742 and NSF IIS-1302134. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Altschul SF, Gish W, Miller W, Myers EM, Lipman DJ. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 2. Schbath S, Martin V, Zytnicki M, Fayolle J, Loux V, Gibrat JF. Mapping Reads on a Genomic Sequence: An Algorithmic Overview and a Practical Comparative Analysis. Journal of Computational Biology. 2012;19:796–813. 10.1089/cmb.2012.0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hauswedell H, Singer J, Reinert K. Lambda: the local aligner for massive biological data. Bioinformatics. 2014;30:i349–i355. 10.1093/bioinformatics/btu439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chor B, Horn D, Levy Y, Goldman N, Massingham T. Genomic DNA k-mer spectra: models and modalities. Genome Biology. 2009;10:R108 10.1186/gb-2009-10-10-r108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sims GE, Jun SR, Wu GA, Kim SH. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proceedings of the National Academy of Sciences. 2009;106:2677–2682. 10.1073/pnas.0813249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vinga S, Carvalho AM, Francisco AP, Russo LMS, Almeida JS. Pattern matching through Chaos Game Representation: bridging numerical and discrete data structures for biological sequence analysis. Algorithms for Molecular Biology. 2012;7:10 10.1186/1748-7188-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jun SR, Sims GE, Wu GA, Kim SH. Whole-proteome phylogeny of prokaryotes by feature frequency profiles: An alignment-free method with optimal feature resolution. Proceedings of the National Academy of Sciences. 2010;107:133–138. 10.1073/pnas.0913033107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allman ES, Rhodes JA, Sullivant S. Statistically-Consistent k-mer Methods for Phylogenetic Tree Reconstruction. arXiv:151101956 [q-bioPE];. [DOI] [PubMed]
- 9. Vinga S. Editorial: Alignment-free methods in computational biology. Briefings in Bioinformatics. 2014;15:341–342. 10.1093/bib/bbu005 [DOI] [PubMed] [Google Scholar]
- 10. Leslie CS, Eskin E, Cohen A, Weston J, Noble WS. Mismatch string kernels for discriminative protein classification. Bioinformatics. 2004;20:467–476. 10.1093/bioinformatics/btg431 [DOI] [PubMed] [Google Scholar]
- 11. Ounit R, Wanamaker S, Close TJ, Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 2015;16:236 10.1186/s12864-015-1419-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meinicke P. UProC: tools for ultra-fast protein domain classification. Bioinformatics. 2015;31:1382–1388. 10.1093/bioinformatics/btu843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meinicke P, Tech M, Morgenstern B, Merkl R. Oligo kernels for datamining on biological sequences: a case study on prokaryotic translation initiation sites. BMC Bioinformatics. 2004;5:169 10.1186/1471-2105-5-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lingner T, Meinicke P. Remote homology detection based on oligomer distances. Bioinformatics. 2006;22:2224–2231. 10.1093/bioinformatics/btl376 [DOI] [PubMed] [Google Scholar]
- 15. Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. 10.1093/bioinformatics/18.3.440 [DOI] [PubMed] [Google Scholar]
- 16.Califano A, Rigoutsos I. FLASH: a fast look-up algorithm for string homology. In: Computer Vision and Pattern Recognition, 1993. Proceedings CVPR’93., 1993 IEEE Computer Society Conference on; 1993. p. 353–359. [PubMed]
- 17. Břinda K, Sykulski M, Kucherov G. Spaced seeds improve k-mer-based metagenomic classification. Bioinformatics. 2015;31:3584–3592. 10.1093/bioinformatics/btv419 [DOI] [PubMed] [Google Scholar]
- 18.Ounit R, Lonardi S. Higher Classification Accuracy of Short Metagenomic Reads by Discriminative Spaced k-mers. In: Pop M, Touzet H, editors. Algorithms in Bioinformatics: 15th International Workshop, WABI 2015, Atlanta, GA, USA, September 10–12, 2015, Proceedings. Berlin, Heidelberg: Springer Berlin Heidelberg; 2015. p. 286–295.
- 19.Onodera T, Shibuya T. The gapped spectrum kernel for support vector machines. In: Perner P, editor. Machine Learning and Data Mining in Pattern Recognition. vol. 7988 of Lecture Notes in Computer Science. Berlin,Heidelberg: Springer Berlin Heidelberg; 2013. p. 1–15.
- 20. Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: Accurate Mapping of Short Color-space Reads. PLOS Computational Biology. 2009;5:e1000386+ 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noé L, Gîrdea M, Kucherov G. Designing efficient spaced seeds for SOLiD read mapping. Advances in Bioinformatics. 2010;2010:1–12. 10.1155/2010/708501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darling AE, Treangen TJ, Zhang L, Kuiken C, Messeguer X, Perna NT. Procrastination Leads to Efficient Filtration for Local Multiple Alignment. In: Bücher P, Moret BME, editors. Algorithms in Bioinformatics: 6th International Workshop, WABI 2006, Zurich, Switzerland, September 11–13, 2006. Proceedings. Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. p. 126–137.
- 23. Darling AE, Mau B, Perna NT. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLOS ONE. 2010;5:e11147+ 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leimeister CA, Boden M, Horwege S, Lindner S, Morgenstern B. Fast alignment-free sequence comparison using spaced-word frequencies. Bioinformatics. 2014;30:1991–1999. 10.1093/bioinformatics/btu177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ulitsky I, Burstein D, Tuller T, Chor B. The average common substring approach to phylogenomic reconstruction. Journal of Computational Biology. 2006;13:336–350. 10.1089/cmb.2006.13.336 [DOI] [PubMed] [Google Scholar]
- 26. Haubold B, Pierstorff N, Möller F, Wiehe T. Genome comparison without alignment using shortest unique substrings. BMC Bioinformatics. 2005;6:123 10.1186/1471-2105-6-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leimeister CA, Morgenstern B. kmacs: the k-mismatch average common substring approach to alignment-free sequence comparison. Bioinformatics. 2014;30:2000–2008. 10.1093/bioinformatics/btu331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thankachan SV, Chockalingam SP, Liu Y, Krishnan A, Aluru S. A greedy alignment-free distance estimator for phylogenetic inference (extended abstract). In: Computational Advances in Bio and Medical Sciences (ICCABS), 2015 IEEE 5th International Conference on; 2015. p. 1–1.
- 29. Thankachan SV, Chockalingam SP, Liu Y, Apostolico A, Aluru S. ALFRED: a practical method for alignment-free distance computation. Journal of Computational Biology, in press;. 10.1089/cmb.2015.0217 [DOI] [PubMed] [Google Scholar]
- 30. Thankachan SV, Apostolico A, Aluru S. A Provably Efficient Algorithm for the k-Mismatch Average Common Substring Problem. Journal of Computational Biology. 2016; 10.1089/cmb.2015.0235 [DOI] [PubMed] [Google Scholar]
- 31. Brejova B, Brown DG, Vinar T. Optimal spaced seeds for homologous coding regions. Journal of Bioinformatics and Computational Biology. 2004;1:595–610. 10.1142/S0219720004000326 [DOI] [PubMed] [Google Scholar]
- 32. Brejova B, Brown DG, Vinar T. Vector seeds: an extension to spaced seeds. Journal of Computer and System Sciences. 2005;70:364–380. 10.1016/j.jcss.2004.12.008 [DOI] [Google Scholar]
- 33.Li M, Ma B, Zhang L. Superiority and complexity of the spaced seeds. In: Proceedings of the Seventeenth Annual ACM-SIAM Symposium on Discrete Algorithm. SODA’06. Philadelphia, PA, USA: Society for Industrial and Applied Mathematics; 2006. p. 444–453.
- 34. Morgenstern B, Zhu B, Horwege S, Leimeister CA. Estimating evolutionary distances between genomic sequences from spaced-word matches. Algorithms for Molecular Biology. 2015;10:5 10.1186/s13015-015-0032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frith MC, Noé L. Improved search heuristics find 20 000 new alignments between human and mouse genomes. Nucleic Acids Research. 2014;42:e59 10.1093/nar/gku104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12:59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 37. Noé L, Martin DEK. A coverage criterion for spaced seeds and its applications to SVM string-kernels and k-mer distances. Journal of Computational Biology. 2014;12:947–963. 10.1089/cmb.2014.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li M, Ma B, Kisman D, Tromp J. PatternHunter II: highly sensitive and fast homology search. Journal of Bioinformatics and Computational Biology. 2004;02:417–439. 10.1142/S0219720004000661 [DOI] [PubMed] [Google Scholar]
- 39. Ilie L, Ilie S. Multiple spaced seeds for homology search. Bioinformatics. 2007;23:2969–2977. 10.1093/bioinformatics/btm422 [DOI] [PubMed] [Google Scholar]
- 40. Ilie L, Ilie S, Bigvand AM. SpEED: fast computation of sensitive spaced seeds. Bioinformatics. 2011;27:2433–2434. 10.1093/bioinformatics/btr368 [DOI] [PubMed] [Google Scholar]
- 41. Horwege S, Lindner S, Boden M, Hatje K, Kollmar M, Leimeister CA, et al. Spaced words and kmacs: fast alignment-free sequence comparison based on inexact word matches. Nucleic Acids Research. 2014;42:W7–W11. 10.1093/nar/gku398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn L, Leimeister CA, Morgenstern B.RasBhari: optimizing spaced seeds for database searching, read mapping and alignment-free sequence comparison. arXiv:151104001 [q-bioGN]. 2015;. [DOI] [PMC free article] [PubMed]
- 43. David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: Sensitive yet Practical Short Read Mapping. Bioinformatics. 2011;27:1011–1012. 10.1093/bioinformatics/btr046 [DOI] [PubMed] [Google Scholar]
- 44. Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLOS ONE. 2009;4:e7767+ 10.1371/journal.pone.0007767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kucherov G, Noé L, Roytberg M. A unifying framework for seed sensitivity and its application to subset seeds. Journal of Bioinformatics and Computational Biology. 2006;4:553–569. 10.1142/S0219720006001977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duc DD, Dinh HQ, Dang TH, Laukens K, Xuan HH. AcoSeeD: An ant colony optimization for finding optimal spaced seeds in biological sequence search. In: ANTS’12; 2012. p. 204–211.
- 47.Do PT, Tran-Thi CG. An improvement of the overlap complexity in the spaced seed searching problem between genomic DNAs. In: Proceedings of the 2nd National Foundation for Science and Technology Development Conference on Information and Computer Science (NICS); 2015. p. 271–276.
- 48. Ounit R, Lonardi S. Higher classification sensitivity of short metagenomic reads with CLARK-S. Bioinformatics. 2016; 10.1093/bioinformatics/btw542 [DOI] [PubMed] [Google Scholar]
- 49. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods. 2012;9:811–814. 10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nature Methods. 2007;4:495–500. 10.1038/nmeth1043 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper or it is mentioned how to download it with a given http-link.