Abstract
Host bacterial associations have a profound impact on health and disease. The human gastrointestinal (GI) tract is inhabited by trillions of commensal bacteria that aid in the digestion of food and vitamin production and play crucial roles in human physiology. Disruption of these relationships and the structure of the bacterial communities that inhabit the gut can contribute to dysbiosis, leading to disease. This fundamental relationship between the host and microbiota relies on chemical signaling and nutrient availability and exchange. GI pathogens compete with the endogenous microbiota for a colonization niche (1, 2). The ability to monitor nutrients and combine this information with the host physiological state is important for the pathogen to precisely program the expression of its virulence repertoire. A major nutrient source is carbon, and although the impact of carbon nutrition on the colonization of the gut by the microbiota has been extensively studied, the extent to which carbon sources affect the regulation of virulence factors by invading pathogens has not been fully defined. The GI pathogen enterohemorrhagic E. coli (EHEC) gages sugar sources as an important cue to regulate expression of its virulence genes. EHEC senses whether it is in a gluconeogenic versus a glycolytic environment, as well as fluctuations of fucose levels to fine tune regulation of its virulence repertoire.
DYNAMICS OF INTESTINAL COLONIZATION BY PATHOGENIC BACTERIA
The mammalian gastrointestinal (GI) tract harbors a diverse collection of indigenous bacteria known as the microbiota. The number of bacterial cells within our bodies exceeds the number of our cells by one order of magnitude (3). Homeostasis of the microbiota is maintained by differential nutrient utilization and physical separation from the gut mucosa (4). However, environmental perturbations such as antibiotic treatment, changes in diet, and infection lead to substantial alterations in composition and structure of the microbiota, referred to as dysbiosis (5–8).
Efficient use of nutrient sources in the gut has a major impact on colonization by pathogenic bacterial species given that nutrient sources are scarce, and they compete with the exquisitely adapted commensal bacteria for these nutrients. According to Freter’s hypothesis, the ability of a pathogen to thrive during intestinal colonization depends on its ability to efficiently utilize nutrient sources and find a suitable niche for colonization (9). Competition for nutrient acquisition between enteric pathogens and the microbiota constitutes a protective mechanism against infection and is an important aspect of colonization resistance. Therefore, evolution of new nutrient acquisition mechanisms and metabolic diversification contributes to a pathogen’s survival and persistence and is an important determinant of the course of bacterial infections. Two important strategies employed by enteric pathogens are the alternative use of carbon sources (10) and utilization of byproducts of the microbiota’s metabolism.
Linking metabolism to the precise coordination of virulence gene expression is a key step in the adaptation of pathogens towards their colonization niches. In this chapter, we will discuss nutrient detection, acquisition, and utilization by enteric pathogens and the barriers against intestinal infection, highlighting the vital role played by the gut microbiota in these processes.
INTESTINAL BARRIERS AGAINST INFECTION
Enteric pathogenic bacteria face a series of barriers to colonizing the GI tract. The human gut is a very complex ecosystem that harbors a high number of commensal bacteria that compete with pathogens for nutrients and space. In addition, the intestinal epithelium is covered by a protective viscous mucus layer that impairs easy bacterial access to the epithelium (10, 11). Suffice it to say, tropism for the mammalian intestine co-evolved with several virulence traits that helped bacterial pathogens cross the aforementioned barriers. Some crucial virulence traits comprise the ability to attach to mucus and cell surface receptors; production of proteases and toxins; expression of flagella to swim across the mucus layer; invasion of epithelial cells; and quorum sensing (12–17). In addition to strict pro quo virulence factors, pathogens also require suitable nutrients. Therefore, nutrient-sensing systems play a major role in both early and late phases of infection.
The Intestinal Microbiota
The mammalian GI tract microbiota plays a fundamental role in human health. Ten trillion to 100 trillion microbes inhabit the distal segment of the human gut, with most belonging to the Bacteroidetes (Gram-negative) and Firmicutes (Gram-positive) phyla (3, 18, 19). Metagenomic studies have shown that the species composition of the microbiota is very diverse; nonetheless, there is conservation in the microbial phyla shared by all individuals (20, 21). In addition, the composition of the intestinal microbial community can vary according to the host genetic background (22, 23). The gut microbiota plays many roles in the host homeostasis and has been referred to as the “forgotten organ” (24). The genetic repertoire of this community is referred to as the microbiome and gives the human host metabolic capabilities not encoded in our genome (25, 26).
The Mucus Layer: A Source of Protection and Nutrients
The single layer of epithelial cells that separates the luminal contents from the GI mucosa is the target of many pathogenic bacteria (27, 28). Nonetheless, not all bacteria can directly interact with enterocytes. A gel-like mucus layer overlays the intestinal epithelial cells, shielding the colonic epithelium from bacteria (29). The mucus layer is in a dynamic state, being constantly synthesized and secreted by specialized goblet cells and degraded to a large extent by indigenous intestinal microbes (30, 31). In fact, mucus utilization has recently been proposed as a co-evolved adaptation of gut resident bacteria and their host (32).
The mucus is composed of mucin, antimicrobial peptides, glycoproteins, glycolipids and epithelial cell debris but 50% of it is made of polysaccharides (33). The major structural component of the mucus is mucin, a glyco-protein that has a protein backbone connected to hydrophilic and hygroscopic oligosaccharide side-chains, which form a gel-like tridimensional structure (34). Also, as part of the mucus composition, there are other goblet cell products such as trefoil peptides (TFF), resistin-like molecule β (RELMβ), and Fc-γ binding protein (Fcgbp), antimicrobial peptides (beta-defensin) and lysozymes secreted by the Paneth cells, and IgA secreted by enterocytes (34, 35). Moreover, the microbiota modulates mucin synthesis and secretion (36). O-linked glycans comprise 80% of the total weight of mucins and are a major nutrient source for bacteria (29). In addition, they provide attachment sites for commensal and pathogenic bacteria (29, 37). A diverse collection of 13 monosaccharides is part of the mucus composition: arabinose, fucose, galactose, gluconate, glucuronate, galacturonate, mannose, glucosamine, N-acetyl-glucosamine, galactosamine, N-acetyl-galactosamine, N-acetylneuramic acid, and ribose. All of these sugars are made available to pathogenic bacteria due to host epithelial cell turnover and the polysaccharide-degrading activity of commensal anaerobes. Hence, the mucus layer is an important habitat and source of nutrients for bacterial communities that colonize mucosal surfaces.
The highly glycosylated MUC2 mucin is synthesized by goblet cells in the small and large intestines and is a major component of the mucus layer (33). The intestinal epithelium also expresses membrane-bound mucins: MUC1, MUC3, MUC4, MUC12, MUC13, and MUC17. However, MUC2 is the predominant mucin in human and murine colons (33), and these mucins are constantly being degraded by the action of glycosidases produced by the anaerobic bacteria that dominate the colonic microbiota (38).
The important role of the mucus as a defensive barrier against infection and injury can be illustrated by the differential length and thickness of its structure along the GI tract. The mucus layer is progressively thicker towards the large intestine, the site where the highest numbers of commensal bacteria reside. The mucus is the front line of the innate host defense against pathogenic microbes (34). Alterations of the mucus layer, such as the lack of MUC2, renders mice susceptible to bacterial adhesion to the intestinal epithelium, resulting in disturbances of the intestinal physiology (33). It is noteworthy that deficiency in mucus production causes changes in the normal localization of commensal bacteria in the colon (39). In addition, recent evidence highlights the importance of mucin in the defense against bacterial infection, with MUC2-deficient mice developing severe life-threatening colitis when infected with the murine enteric pathogen Citrobacter rodentium (33). MUC2 has highly fucosylated glycans, and MUC17, a membrane-bound mucin expressed in the large intestine, protects against invasion by enteroinvasive E. coli (EIEC) (41).
The stratified mucus layer that protects the intestinal epithelium displays a highly complex structure. The mucus layer is spatially divided in two layers: an outer loose mucus layer facing the intestinal lumen and a thick inner mucus layer firmly attached to the epithelial cells (29). Using fluorescent in situ hybridization (FISH) staining for bacteria, Johansson et al elegantly demonstrated that commensal bacteria communities reside in the outer mucus layer, while the inner layer is virtually devoid of bacteria (39).
Gut commensals are not found in close contact with the epithelium lining, in contrast to pathogenic bacteria, which employ virulence factors to reach close proximity to enterocytes. EHEC produces the plasmid-encoded metalloprotease StcE, which targets intestinal mucins, therefore contributing to bacterial penetration towards the colonic epithelium (42). In fact, one of the theories of how commensal bacteria do not cause disease, even when sharing several common features with pathogenic bacteria, is their localization in the GI tract. Commensal bacteria are associated with the outer mucus layer but not at the interface of the intestinal epithelium, and this physical separation is thought to prevent dysbiosis (43). Both pathogenic and commensal bacteria can consume carbohydrates from the mucus as a carbon and energy source (44–47). In addition to being a nutrient source for indigenous and foreign bacteria, the mucus components can also be exploited as cues to trigger production of virulence factors by pathogens. MUC2 triggers virulence expression in Campylobacter jejuni, with MUC2 exposure leading C. jejuni to express cytolethal distending toxin JlpA, and flagellin (28).
The stratification of the mucus layer is important to sustain the symbiotic relationship between the microbiota and the host. It has been demonstrated that the lectin RegIII gamma, produced by Paneth cells of the intestinal epithelium, is important to the physical separation of the microbiota and the intestinal epithelium (4, 48). The physical separation of the intestinal microbiota and the colonic epithelium represents evidence that the mucus layers act as an effective barrier (11).
MICROBIOTA AND NUTRIENT GENERATION IN THE GUT
The consortium of the resident microbes that inhabit the human gut possess an incredible arsenal of glycolytic enzymes that allow the utilization of complex polysaccharides originated from the host itself or its diet (48–50). These polysaccharides are hydrolyzed into monosaccharides, which are subsequently utilized as carbon and energy sources after being released as free sugars in the intestinal lumen (51). Therefore, the metabolic activity of the microbiota is intrinsically related to the generation of the nutritional environment of the gut, which also impacts the survival and persistence of pathogenic bacteria. In this section, we will discuss the role of nutrient acquisition in the gut by pathogenic bacteria and how the microbiota modulates the availability of these nutrient sources.
Commensal Bacteria and Polysaccharide Degradation in the Gut
One of the major roles played by the microbiota is the manipulation of carbohydrate sources in the gut. The members of the microbiota community are fermenters with a broad range of metabolic capacity, being able to digest complex glycan structures originated from the host diet, host structures such as mucus, and cell-associated glycans otherwise not digested by invading microbes (50, 52–54). This plural metabolic capacity allows the microbiota to explore unique niches and survive in the human gut in homeostasis. Hence, the gut microbiota functions as a metabolic organ, helping the human host obtain energy from otherwise indigestible dietary sources (55).
The ability to degrade complex polysaccharides highlights the crucial role displayed by the gut commensals in metabolic pathways in the gut. Most of the knowledge of polysaccharide utilization by the microbiota derives from investigations of the glycophagic symbiont Bacteroides thetataiotaomicron. B. theta, a strict anaerobe, is the most abundant resident of the distal small intestine and colon of mice and humans (56, 57). The B. theta genome encodes an impressive arsenal of 246 glycolitic enzymes (58). This prominent commensal bacterium can degrade plant- and animal-derived complex glycans into monosaccharides, providing nutrient sources for other commensals, consequently playing a fundamental role in the supply of energy sources in the gut. Pathogenic bacteria exploit this B. theta-dependent nutrient availability. Therefore, the polysaccharide-degrading activity of commensal bacteria might affect colonization by pathogens by representing a source of competition but also a nutrient-generating machine capable of supporting pathogens to successfully grow and find a niche in the host. The ability of Bacteroides sp to use a diverse range of glycans is dependent on genes in its polysaccharide-utilization loci (PULs) (59).
Mucus utilization by primary fermenters such as B. theta involves the production of glycosyl hydrolases secreted into the environment. Consequently, monosaccharides are released from the complex glycan structures into the lumen, where they are accessible to other microbial species. B. theta dedicates about 18% of its genome to polysaccharide utilization, the PULs (58). Conversely, gut pathogens are not equally equipped to consume complex host-derived glycans. Although some pathogens, such as EHEC, are able to produce a mucus-degrading protease, StcE, it does not encode glycosidases. This means that gut pathogens rely on the glycosidase activity of commensal bacteria to access and import free monosaccharides for catabolism.
Certain members of the microbiota, such as Bifido-bacterium sp, may differ in their effects on nutrient generation to pathogenic bacteria. In contrast with B. theta, B. longum subspecies infantis (B.infantis) typically produces intracellular glycosidases to import complex glycans into the cell for digestion into monosaccharides (60, 61). B. bifidum, however, secretes glycosidases similarly to B. theta (62). Consequently, the access to free monosaccharides by microbial pathogens during polysaccharide degradation by certain species of Bifidobacteria is different, which may have interesting biological implications because Bifidobacteria are used as probiotics and may be able to control or prevent enteric infections (63).
A special relationship takes place between B. theta and its host. B. theta consumption of fucose may have a major impact on enteropathogens able to utilize this sugar as a carbon and energy source (64). B. theta induces the expression of fucosylated glycoconjugates by the host intestinal epithelium (65). Then B. theta produces fucosidases that harvest fucose from mucosal glycans (66). Fucose is abundant is intestinal glyco-conjugates, and it is usually a terminal α-linked sugar (67, 68). The triggering of intestinal fucosylation by B. theta depends on the bacterial density and the production of a B. theta-derived signal that remains elusive (69). In addition to using host-derived fucose as a carbon and energy source, B. theta also incorporates fucose into a capsular polysaccharide via an O-glycosylation system, which is believed to be important for competitive colonization of the gut (70). Nutrient utilization by B. theta can be modulated by diet: a diet rich in plant glycans triggers expression of genes involved in metabolism of dietary substrates by B. theta; conversely, on exposure to a diet devoid of complex glycans, B. theta switches its metabolism towards host glycans (50). The metabolic switch that B. theta undergoes during a change in diet might affect pathogen access to fucose, which could directly impact the outcome of bacterial infections.
Primary fermenters differ in their capacity to utilize carbohydrates, which is relevant in vivo because dietary changes cause alterations in the community structure of the microbiota, and likely, in the outcome of end-fermented products (71). Polysaccharide degradation has been recognized as a core function encoded within the microbiome (72). The prominent adult gut symbiont, B. theta encodes an arsenal of 261 glycosyl hydrolases (73). Other distal gut residents such as Akkermansia muciniphila have a mucin-degrading ability that may also lead to release of free monosaccharides that can be utilized by pathogenic enterobacteria (74).
A generation of free monosaccharides in the gut, an end product of extensive polysaccharide degradation by members of the microbiota, is a major modulator of the nutrient environment accessible to invading pathogens. Most pathogenic bacteria do not encode glycosyl hydrolases in their genomes and rely on simple monosaccharides or disaccharides as substrates for growth in vivo (32). Therefore, the enzymatic activity of the gut microbiota, to a certain degree, may render the host particularly susceptible to different infections. This concept could be further explored to design customized diets or probiotic interventions aimed at improving pathogen exclusion based on nutrient competition. This concept is exemplified by Deriu et al, who demonstrated that administration of the probiotic E.coli Nissle 1917 strain reduced murine colonization by Salmonella enterica Typhimiurium (75).
INTERPLAY BETWEEN COMMENSAL AND PATHOGENIC BACTERIA
Given the high content of commensal bacteria residing in the gut, the colonization site of several bacterial pathogens, it is not surprising that a complex relationship might arise from these interactions. Nonetheless, little is known of the mechanisms that govern the crosstalk between pathogens and the microbiota or the impact of the commensal bacteria on pathogenesis and infection outcomes. Elucidation of the processes involved in interactions among host, microbiota, and pathogens is of major importance in the design of novel therapeutic interventions (76–78).
Gut pathogens harbor several traits to maximize proliferation in the lumen, including motility, chemotaxis, and iron-scavenging and nutrient-sensing systems. In addition, pathogenic species can hijack carbohydrate utilization pathways of resident microbes for their own advantage by exploring the end-product of glycosidases produced by anaerobes from the microbiota to obtain monossacharides as carbon sources (38).
Competition for Nutrients and Colonization Resistance
While primary fermenters have a major impact on nutrient generation for bacterial pathogens, commensal bacteria displaying similar nutrient requirements pose a threat against pathogen survival during intestinal infection. By consuming similar carbohydrates, commensal E. coli competes with EHEC O157:H7 for nutrients, leading this pathogen to explore different niches to proliferate in the gut (79–81).
Commensal and pathogenic E. coli differs in the types of carbohydrates it preferably utilizes in vivo as carbon sources. EHEC can grow on mucus (82). Studies indicate that EHEC can grow in vitro on cecal mucus prepared from mice but cannot grow in the luminal content, suggesting that these bacteria colonize the mouse intestine by growing in the mucus layer that overlays the cecal epithelium (43). In addition, other studies provide evidence that carbohydrates derived from the mucus can support the growth of E. coli during murine colonization (44–47).
Commensal and pathogenic E. coli share their preferences for particular carbon sources they utilize during intestinal colonization, but they also present differences that reflect their spatial segregation inside the human gut. Commensal E. coli strains are found in the lumen and attached to the mucus layer, while pathogenic E. coli strains are able to cross the mucus layer and reach proximity to the IECs, which also exposes them to nutrients exclusively available at the epithelium interface (44, 83). Among the nutrient sources consumed by both commensal and pathogenic E. coli are monossaccharides and disaccharides. The fact that they consume similar carbon sources in vivo is the basis of colonization resistance that commensal E. coli imposes on pathogenic E. coli species.
Colonization of the mammalian intestine by EHEC requires precise coordination of metabolic and virulence factors. The infectious dose of EHEC is remarkably low compared with other enteric pathogens, highlighting important adaptations of EHEC to the human intestine. EHEC must expand its population to high numbers and find a niche in the colon, which is a major challenge considering the immense number of residing commensal bacteria adapted to live in the colon during millions of years of co-evolution with the human host (84).
An investigation of the carbohydrate utilization profile of EHEC in the bovine gut has revealed that this strain can catabolize mucus-derived carbohydrates inside the cattle gut and do so more rapidly than resident microbes, including commensal E. coli (85). Cattle are the major reservoir of EHEC O157:H7, which has tropism to the recto-anal junction (RAJ) (86). In vitro growth competition assays using WT and the EHEC sugar utilization mutant strains ΔmanA, ΔnagE, ΔnanAT, and ΔgalK, which are deleted for genes involved in catabolism of six major mucus-derived monosaccharides (galactose, N-acetyl-glucosamine [GlcNAc], N-acetylgalactosamine [GalNAc], fucose, mannose and N-acetyl neuraminic acid [Neu5Ac]), showed that the ability to consume mannose, GlcNAc, Neu5Ac, and galactose is important for EHEC growth, suggesting that metabolism of the aforementioned carbohydrates confers a growth advantage to EHEC in the bovine intestine (85).
In vivo carbon consumption was investigated using the streptomycin-treated mouse model and elucidated many aspects of the competition and nutrient utilization that allows EHEC to successfully colonize the mammalian intestine. Commensal and pathogenic E. coli share the ability to consume arabinose, fucose, and N-acetylglucosamine in the mouse intestine. EHEC is able to catabolize galactose, hexuronates, mannose, and ribose, while commensal E. coli exclusively catabolize gluconate and N-acetyl-neuramic acid (79). These data indicate that differential carbon nutrition in the gut contributes to niche adaptation of EHEC and helps avoid competition with commensal E. coli. According to the nutrient-niche hypothesis elaborated by Freter et al, better consumption of a limiting nutrient source than an organism’s competitors is imperative for successful colonization of the intestine (87, 89).
Recent evidence indicates that the ability to consume similar carbohydrate sources is an important factor that may influence bacterial infection. Kamada et al reported that competition with members of the gut microbiota for the same nutrients is necessary for pathogen clearance (88). This study represents a great advance on the investigation of the relationship among commensal and pathogenic bacteria, which has also shown that classic virulence traits such as the type 3 secretion system (T3SS), which is known to be required for host cell contact by EHEC, is also important for competition with gut microbiota (88).
EHEC does not significantly compete with B. theta for nutrient utilization during growth in mucus but competes with commensal E. coli for the same carbon sources during growth in the mammalian intestine (44, 47, 79, 89). One such carbon source is fucose, which is released into the lumen by glycophagic bacteria such as B. theta and can be utilized by E. coli, which itself cannot hydrolyze complex mucus carbohydrates (44, 47, 79). Because both EHEC and commensal E. coli compete for fucose utilization in the lumen, it would be counterproductive for EHEC to invest a lot of resources in the utilization of this carbon source in this compartment, where commensal E. coli are present (79, 89). However, EHEC can efficiently use other carbon sources, such as galactose, hexorunates, mannose, and ribose, which are not used by commensal E. coli in the intestine (79).
NUTRIENT SENSING IN THE GUT
Differential utilization of limiting nutrients is the basis for the coexistence of members of the gut microbiota. It has also a major impact on bacterial infection, as pathogens explore alternative nutrient sources to avoid competition with commensals. In cases of noncronical infections such as EHEC or EPEC, nutrient competition among commensal and pathogenic bacteria impacts the outcome of infection, leading to resolution of this infection and elimination of the intruder.
Fucose Sensing Regulates Intestinal Colonization by EHEC
EHEC is the causative agent of outbreaks of bloody diarrhea worldwide, with about 5% to 7% of the cases in any given outbreak developing a life-threatening complication known as hemolytic uremic syndrome (HUS) (90, 91). EHEC colonizes the human large intestine through the formation of attaching and effacing (AE) lesions on intestinal epithelial cells (92). Most genes necessary for AE lesion formation are clustered in a pathogenicity island (PAI) named the locus of enterocyte effacement (LEE) (93). The LEE region contains five major operons: LEE1-5 (94–96), which encodes a type III secretion system (TTSS) (12), an adhesin (intimin) (97) and its receptor (Tir) (98), and effector proteins (99–103). The LEE genes and the non–LEE-encoded effector, EspFu, are both required for the formation of AE lesions (104).
Cell-to-cell communication among bacteria in the intestine is a major mechanism that shapes bacteria-host relationships. Pathogenic bacteria such as EHEC can also cross-communicate with the host by detecting mammalian hormones (105). By virtue of its remarkably low infectious dose (50 CFU) (106), successful colonization of the human colon by EHEC relies largely on sensing multiple signals to coordinate the expression of virulence genes. EHEC exploits the autoinducer-3 (AI-3)/ epinephrine (Epi)/norepinephrine (NE) interkingdom signaling cascade to trigger expression of motility and AE lesion genes, two pathogenic traits that are crucial for colonization but are required at different time points during infection (105). The host hormones Epi/NE are specifically sensed by two histidine sensor kinases: QseC and QseE (107, 108). QseE is downstream of QseC in this signaling cascade, given that transcription of QseE is activated through QseC (109). In addition to sensing these host hormones, QseC also senses the bacterial signal AI-3 (110). QseE, however, does not sense AI-3, thereby discriminating between host- and bacterial-derived signals (108). QseC and QseE activate virulence gene expression and pathogenesis in vitro and in vivo in EHEC (108, 110, 111). On sensing these signals, QseC and QseE autophosphorylation increases, initiating a signaling cascade that promotes virulence gene expression. QseE exclusively phosphorylates the QseF response regulator (112). QseC, however, phosphorylates three response regulators: QseB, QseF, and KdpE.
Signal sensing by EHEC is crucial for colonization of the mammalian colon due to the orchestration of multiple virulence pathways that aim to promote intimate attachment of the bacteria to the apical portion of enterocytes (113). Also of major importance is the activation of pathways that allow suitable nutrition during infection. Recently, Pacheco et al (64) demonstrated that EHEC encodes for a two-component system (TCS) named FusKR, in which FusK is the sensor kinase and FusR is the response regulator (64). The FusKR TCS is repressed by the adrenergic-sensing QseBC and QseEF TCSs.
Investigation of the signal triggering fusKR transcription indicated that it was a component of the mucus. Using a combination of biochemical and genetic approaches, L-fucose was identified as the signal that activates the FusKR signaling cascade in EHEC. In fact, FusK specifically increases its autophosphorylation in response to fucose (64). FusKR signaling leads to repression of LEE and fucose utilization gene expression, allowing the pathogen to save energy by preventing unnecessary virulence gene expression while crossing the mucus layer and avoiding competition with the commensal E. coli for carbon sources, given that commensal E. coli preferentially catabolize fucose in the mammalian intestine (64).
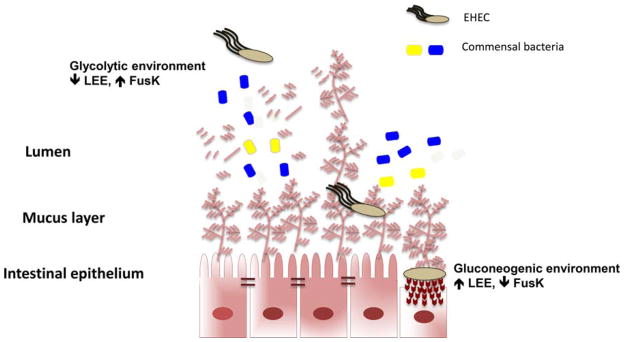
In vitro competition assays have demonstrated that the modulation of carbon availability by the prominent gut symbiont B. theta alters the effect of fucose utilization by EHEC on the expression of ler, the master regulator of the LEE genes. In the presence of free fucose in the media, B. theta has no effect on ler transcription, but this scenario changed on co-culture of B. theta and EHEC on mucin. During growth on mucin, EHEC relies on B. theta to access free fucose, and as the result of this relationship, expression of ler is reduced (64). Therefore, the interaction between EHEC and the commensal bacterium B. theta is able to change the pathogen’s virulence due to the nutrient modulatory activity of B. theta. In vivo competition assays using the infant rabbit model (114), which is able to reproduce several aspects of EHEC-mediated disease, show that the EHEC fusK mutant is attenuated for virulence, and regulation of ler by FusK plays a determinant role on EHEC fitness during intestinal colonization (Fig. 1) (64).
FIGURE 1.
Nutritional cues regulate the locus of enterocyte effacement (LEE) gene expression in enterohemorrhagic E. coli (EHEC). Glycophagic members of the microbiota such as B. theta make fucose from mucin accessible to EHEC, and EHEC interprets this information to recognize that it is in the lumen, where expression of its LEE-encoded type III secretion system (TTSS) is onerous and not advantageous. Using yet another nutrient-based environmental cue, EHEC also times LEE expression through recognition of glycolytic and gluconeogenic environments. The lumen is more glycolytic due to predominant glycophagic members of the microbiota degrading complex polysaccharides into monosaccharides that can be readily utilized by nonglycophagic bacterial species such as E. coli and C. rodentium. In contrast, the tight mucus layer between the lumen and the epithelial interface in the gastrointestinal (GI) tract is devoid of microbiota; it is known as a “zone of clearance.” At the epithelial interface, the environment is regarded as gluconeogenic. Hence, the coupling of LEE regulation to optimal expression under gluconeogenic and low-fucose conditions mirrors the interface with the epithelial layer environment in the GI tract, ensuring that EHEC will express only LEE at optimal levels to promote attaching and effacing lesion formation at the epithelial interface. doi:10.1128 /microbiolspec.MBP-0001-2014.f1
The genes encoding fusKR are clustered in a pathogenicity island (OI-20) only found in EPEC O55:H7 (the E. coli lineage that gave rise to EHEC O157:H7) (115, 116), EHEC O157:H7, and C. rodentium, AE GI pathogens that colonize the colon. EHEC’s ancestor, EPEC O55:H7 (116), is the only other serotype of E. coli to harbor fusKR, suggesting that acquisition of these genes is recent. The recent acquisition of OI-20 on EHEC evolution provided this pathogen with a novel signal transduction system, suggesting that expression of this TCS in mucus facilitates EHEC adaptation to the mammalian intestine.
The modulation of the nutrient supply in the gut by commensal microbes and its effects on bacterial virulence supports the use of probiotic interventions to control bacterial infections. It was demonstrated that the probiotic strain Lactobacillus casei consumes fucosyl-α-1,3- N-acetylglucosamine (Fuc-α-1,3-GlcNAc) as a carbon source and releases free L-fucose into the media (117). Given that fucose can repress EHEC virulence, it would be interesting to investigate the effects of a symbiotic approach (i.e., combination of a probiotic strain and a prebiotic diet) on preventing or treating EHEC infections.
Sensing of Glycolytic versus Gluconeogenic Environments
Glycolytic environments inhibit the expression of the LEE genes. Conversely, growth in a gluconeogenic environment activates expression of these genes. Part of this sugar-dependent regulation is achieved through two transcription factors: KdpE and Cra. KdpE and Cra interact to optimally and directly activate expression of the LEE genes in a metabolite-dependent fashion (118). EHEC competes with commensal E. coli (the predominant species in the γ-Proteobacteria) for the same carbon sources during growth in the mammalian intestine (1, 44, 47, 79, 89). EHEC uses glycolytic substrates for initial growth but is unable to effectively compete for these carbon sources beyond the first few days and begins to utilize gluconeogenic substrates to stay within the intestine (44). Hence, it is advantageous to coordinate expression of the LEE with these environmental conditions. Commensal E. coli can be found in the lumen, which is glycolytic due to the abundant sugar sources supplied by the glycophagic microbiota, while the interface with the epithelium is a more gluconeogenic environment. Hence, the KdpE/Cra-dependent activation of the LEE under gluconeogenic conditions ensures that these genes are optimally expressed only at the epithelium interface and not in the lumen (Fig. 1).
Ethanolamine Utilization
Ethanolamine is a breakdown product of phosphati-dylethanolamine, which is an abundant phospholipid of mammalian and bacterial cell membranes (119–121). Epithelial cell turnover and the gut microbiota are important sources of ethanolamine in the gut, which can be taken up and utilized as a carbon and/or nitrogen source by a number of bacterial species, including pathogenic bacteria such as EHEC and Salmonella. Exfoliation of intestinal cells also releases ethanolamine into the intestine (122–125). Ethanolamine is broken down into acetaldehyde and ammonia by the enzyme complex ethanolamine ammonia lyase, encoded by genes eutB and eutC. Ammonia is used as a nitrogen source, while acetaldehyde is converted into acetyl-CoA (126).
Ethanolamine consumption supports the growth of EHEC in vivo and also confers EHEC a competitive advantage over the indigenous microbiota in the bovine intestine (127). EHEC can utilize ethanolamine as a nitrogen source in the bovine small intestine, in contrast with commensal E. coli strains. Ethanolamine metabolism allows EHEC to flourish in the bovine gut (its main reservoir), which contributes to the spreading of EHEC infections (127).
In addition to its role as a nitrogen source, ethanol-amine functions as a signaling molecule that triggers EHEC virulence expression. Kendall et al demonstrated that EHEC senses ethanolamine partially via EutR, a previously known receptor for ethanolamine. On EHEC growth on M9 minimal medium with ethanolamine as the sole nitrogen source, expression of the LEE and Stx increases markedly, indicating that ethanolamine is a signal that triggers EHEC virulence gene expression (123). It was also shown that ethanolamine triggers transcription of the EHEC adrenergic sensors qseC and qseE, which are involved in cell-to-cell signaling and bacteria-host communication (123). These studies suggest that ethanolamine sensing may contribute to EHEC persistence in the mammalian gut, not only by supporting EHEC growth but also by controlling transcription of major virulence factors. The research conducted by Kendall et al also indicates that EHEC encodes an additional, yet unidentified, ethanolamine sensor (123).
While currently available data leave no doubt of the pivotal role played by ethanolamine during gut colonization by enteric pathogens, little is known of the sensory systems employed in ethanolamine detection. Future research is necessary to unravel the receptors involved in early ethanolamine sensing, which are critical steps in infection.
The Effects of Inflammation
Pathogen-promoted inflammation during enteric infection is now appreciated as a strategy to promote rather than a consequence of bacterial infection. Enteric pathogens such as C. rodentium and Salmonella can benefit from the inflammatory environment or the overall changes in the bacterial community that result from inflammation. By provoking intestinal inflammation, the murine pathogen C. rodentium reduces the overall number of commensal bacteria in the microbiota, which gives the pathogen a colonization advantage (76, 128). Although later stages of inflammation result in pathogen clearance from the gut, inflammation in the early stages of infection helps C. rodentium replicate and increase its population when competing with commensal microbes.
Destruction of intestinal integrity by inflammation promotes Salmonella Typhimurium persistence in the gut. Inflammation triggered by Salmonella releases a new electron acceptor, tetrathionate, which allows Salmonella to outcompete the gut microbiota and proliferate in the gut lumen (129). In addition, tetrathionate allows Salmonella Typhimurium to use ethanolamine as a carbon source in the inflamed intestine (124). The eutC mutant, which cannot grow anaerobically on ethanol-amine as a carbon source, was outcompeted by the wild-type (WT) strain only in the presence of tetrathionate, indicating that ethanolamine utilization and tetrathionate respiration likely occur concomitantly. Ethanol-amine levels present in colons of mice infected and uninfected were similar, indicating that ethanolamine consumption in the inflamed intestine was not due to release of ethanolamine due to epithelial cell destruction caused by inflammation. Interestingly, the growth advantage conferred by the ability to consume ethanol-amine in vivo relies on the ability to respire tetrathionate (124). In the absence of the electron acceptor tetra-thionate, respiration of ethanolamine does not support Salmonella growth in the mouse intestine. Therefore, the inflammatory response orchestrated by Salmonella during infection creates a nutritional environment that supports its replication in the gut lumen.
Nutrient Competition
Kamada et al (88) have demonstrated that the combined effects of virulence gene expression and competition with the microbiota are both crucial for pathogen clearance using the murine pathogen C. rodentium. C. rodentium is a natural mouse pathogen that causes colonic hyperplasia, and similarly to EHEC and EPEC, forms AE lesions on IECs. C. rodentium has been extensively used as a model for EHEC and EPEC infections (88). During infection of conventional mice, C. rodentium requires expression of the LEE to compete with indigenous microbes, while LEE expression is not necessary for C. rodentium colonization of germ-free mice. It was also shown that virulence gene expression (LEE) was triggered early but was reduced during late stages of infection, causing relocation of C. rodentium from the epithelium to the gut lumen, where the pathogen was exposed to commensal bacteria and had to compete for similar carbon sources for luminal growth. This shows that virulence and metabolism act in concert during bacterial infection, and both nutrient utilization and production of virulence traits are required to establish a successful colonization by pathogenic bacteria. A closer look at the nutrient competition between C. rodentium and commensal E. coli and B. tetha indicated that E. coli can outcompete C. rodentium due its ability to grow on monosaccharides, while B. theta does not outcompete C. rodentium because it can grow on polysaccharides. This work demonstrated that competition for similar nutrient sources is an important determinant of the outcome of bacterial infections of the mammalian intestine, reinforces Freter’s concept, and raises the possibility that shifting the commensal microbiota towards nutrient competition with pathogens may be an alternative to fight bacterial infections.
A recent study shows that EHEC colonization could be prevented by the probiotic strains E. coli Nissle 1917 and E. coli HS, based on the ability of these combined commensal strains to compete for the carbohydrate niches occupied by EHEC to colonize the mammalian gut. EHEC utilizes arabinose, galactose, and gluconate, carbohydrates also consumed by E. coli Nissle 1917 and E. coli HS. EHEC also competes with E. coli HS for ribose and N-acetylglucosamine, while it competes with E. coli Nissle 1917 for mannose (81).
CONCLUSIONS
Nutrient scavenging by pathogenic bacteria from microbiota-derived products is an emerging theme in bacterial pathogenesis. Dietary changes causing shifts in gut microbial populations are well established, although the consequences regarding infection by pathogenic agents are mostly unknown. The use of probiotic strains to reinforce colonization resistance may be a better alternative to treatment of infections for which antibiotic treatment is not advisable, such as EHEC and Salmonella. Future investigations on the relationships between indigenous microbiota members and pathogenic microorganisms are crucial for the development of new effective preventive and curative strategies for enteric infections.
References
- 1.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiennimitr P, Winter SE, Baumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2011;15:108–14. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon GL, Relman DA, Fraser-Liggett CM, Nelson KE. Meta-genomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 6.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamont JT. Mucus: the front line of intestinal mucosal defense. Ann N Y Acad Sci. 1992;664:190–201. doi: 10.1111/j.1749-6632.1992.tb39760.x. [DOI] [PubMed] [Google Scholar]
- 11.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis KG, Giron JA, Jerse AE, McDaniel EK, Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci U S A. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington SM, Sheikh J, Henderson LR, Ruiz-Perez F, Cohen PS, Nataro JP. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect Immun. 2009;77:2465–2473. doi: 10.1128/IAI.01494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Brien AD, LaVeck GD, Griffin DE, Thompson MR. Characterization of Shigella dysenteriae 1 (Shiga) toxin purified by anti-Shiga toxin affinity chromatography. Infect Immun. 1980;30:170–179. doi: 10.1128/iai.30.1.170-179.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinstein DL, Carsiotis M, Lissner CR, O’Brien AD. Flagella help Salmonella typhimurium survive within murine macrophages. Infect Immun. 1984;46:819–825. doi: 10.1128/iai.46.3.819-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finlay BB, Falkow S. Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J Infect Dis. 1990;162:1096–1106. doi: 10.1093/infdis/162.5.1096. [DOI] [PubMed] [Google Scholar]
- 18.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JL. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 20.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- 23.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 28.Tu QV, McGuckin MA, Mendz GL. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J Med Microbiol. 2008;57:795–802. doi: 10.1099/jmm.0.47752-0. [DOI] [PubMed] [Google Scholar]
- 29.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson G, Falk P, Hoskins LC. Degradation of human intestinal glycosphingolipids by extracellular glycosidases from mucin-degrading bacteria of the human fecal flora. J Biol Chem. 1988;263:10790–10798. [PubMed] [Google Scholar]
- 31.Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, Niedermeyer G. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest. 1985;75:944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: Bacterial consumption of host glycans in the gut. Glycobiology. 2013;23:1038–46. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, Vallance BA. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman N, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol. 1999;276:G941–950. doi: 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- 37.Robbe C, Capon C, Flahaut C, Michalski JC. Microscale analysis of mucin-type O-glycans by a coordinated fluorophore-assisted carbohydrate electrophoresis and mass spectrometry approach. Electrophoresis. 2003;24:611–621. doi: 10.1002/elps.200390071. [DOI] [PubMed] [Google Scholar]
- 38.Corfield AP, Wagner SA, Clamp JR, Kriaris MS, Hoskins LC. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun. 1992;60:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Resta-Lenert S, Das S, Batra SK, Ho SB. Muc17 protects intestinal epithelial cells from enteroinvasive E. coli infection by promoting epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2011;300:G1144–1155. doi: 10.1152/ajpgi.00138.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grys TE, Walters LL, Welch RA. Characterization of the StcE protease activity of Escherichia coli O157:H7. J Bacteriol. 2006;188:4646–4653. doi: 10.1128/JB.01806-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wadolkowski EA, Laux DC, Cohen PS. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun. 1988;56:1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004;72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peekhaus N, Conway T. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montagne L, Toullec R, Lalles JP. Calf intestinal mucin: isolation, partial characterization, and measurement in ileal digesta with an enzyme-linked immunosorbent assay. J Dairy Sci. 2000;83:507–517. doi: 10.3168/jds.S0022-0302(00)74910-1. [DOI] [PubMed] [Google Scholar]
- 47.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. Effects of Diet on Resource Utilization by a Model Human Gut Microbiota Containing Bacteroides cellulosilyticus WH2, a Symbiont with an Extensive Glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 51.Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Poly-saccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 56.Moore WE, Holdeman LV. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ushijima T, Takahashi M, Tatewaki K, Ozaki Y. A selective medium for isolation and presumptive identification of the Bacteriodes fragilis group. Microbiol Immunol. 1983;27:985–993. doi: 10.1111/j.1348-0421.1983.tb02929.x. [DOI] [PubMed] [Google Scholar]
- 58.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 60.Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifido-bacterium longum subsp. infantis. Anaerobe. 2012;18:430–435. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–664. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garrido D, Barile D, Mills DA. A molecular basis for bifido-bacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012;3:415S–421S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collins MD, Gibson GR. Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am J Clin Nutr. 1999;69:1052S–1057S. doi: 10.1093/ajcn/69.5.1052s. [DOI] [PubMed] [Google Scholar]
- 64.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 66.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 67.Bjork S, Breimer ME, Hansson GC, Karlsson KA, Leffler H. Structures of blood group glycosphingolipids of human small intestine. A relation between the expression of fucolipids of epithelial cells and the ABO, Le and Se phenotype of the donor. J Biol Chem. 1987;262:6758–6765. [PubMed] [Google Scholar]
- 68.Finne J, Breimer ME, Hansson GC, Karlsson KA, Leffler H, Vliegenthart JF, van Halbeek H. Novel polyfucosylated N-linked glycopeptides with blood group A, H, X, and Y determinants from human small intestinal epithelial cells. J Biol Chem. 1989;264:5720–5735. [PubMed] [Google Scholar]
- 69.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci U S A. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lozupone CA, Hamady M, Cantarel BL, Coutinho PM, Henrissat B, Gordon JI, Knight R. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci U S A. 2008;105:15076–15081. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 74.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 75.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 77.Curtis MM, Sperandio V. A complex relationship: the interaction among symbiotic microbes, invading pathogens, and their mammalian host. Mucosal Immunol. 2011;4:133–138. doi: 10.1038/mi.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keeney KM, Finlay BB. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr Opin Microbiol. 2011;14:92–98. doi: 10.1016/j.mib.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek. 2008;94:165–171. doi: 10.1007/s10482-008-9222-6. [DOI] [PubMed] [Google Scholar]
- 81.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snider TA, Fabich AJ, Conway T, Clinkenbeard KD. E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Vet Microbiol. 2009;136:150–154. doi: 10.1016/j.vetmic.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 83.Moller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, Krogfelt KA, Cohen PS. An Escherichia coli MG1655 lipopoly-saccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun. 2003;71:2142–2152. doi: 10.1128/IAI.71.4.2142-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, Cohen PS, Conway T, Forano E, Martin C. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ Microbiol. 2013;15:610–622. doi: 10.1111/1462-2920.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naylor SW, Roe AJ, Nart P, Spears K, Smith DG, Low JC, Gally DL. Escherichia coli O157 : H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology. 2005;151:2773–2781. doi: 10.1099/mic.0.28060-0. [DOI] [PubMed] [Google Scholar]
- 87.Freter R, Stauffer E, Cleven D, Holdeman LV, Moore WE. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect Immun. 1983;39:666–675. doi: 10.1128/iai.39.2.666-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS. L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 DeltafucAO and E. coli Nissle 1917 DeltafucAO mutants in the mouse intestine and in M9 minimal medium. Infect Immun. 2007;75:5465–5475. doi: 10.1128/IAI.00822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karmali MA. Prospects for preventing serious systemic toxemic complications of Shiga toxin-producing Escherichia coli infections using Shiga toxin receptor analogues. J Infect Dis. 2004;189:355–359. doi: 10.1086/381130. [DOI] [PubMed] [Google Scholar]
- 91.Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 92.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 93.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elliott SJ, Hutcheson SW, Dubois MS, Mellies JL, Wainwright LA, Batchelor M, Frankel G, Knutton S, Kaper JB. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol Microbiol. 1999;33:1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- 95.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 96.Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 97.Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of entero-pathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 99.McNamara BP, Donnenberg MS. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol Lett. 1998;166:71–78. doi: 10.1111/j.1574-6968.1998.tb13185.x. [DOI] [PubMed] [Google Scholar]
- 100.Kenny B, Jepson M. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol. 2000;2:579–590. doi: 10.1046/j.1462-5822.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 101.Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect Immun. 2001;69:4027–4033. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tu X, Nisan I, Yona C, Hanski E, Rosenshine I. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and entero-pathogenic Escherichia coli. Mol Microbiol. 2003;47:595–606. doi: 10.1046/j.1365-2958.2003.03329.x. [DOI] [PubMed] [Google Scholar]
- 103.Kanack KJ, Crawford JA, Tatsuno I, Karmali MA, Kaper JB. SepZ/EspZ is secreted and translocated into HeLa cells by the enteropathogenic Escherichia coli type III secretion system. Infect Immun. 2005;73:4327–4337. doi: 10.1128/IAI.73.7.4327-4337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Campellone KG, Robbins D, Leong JM. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Developmental cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 105.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tilden J, Jr, Young W, McNamara AM, Custer C, Boesel B, Lambert-Fair MA, Majkowski J, Vugia D, Werner SB, Hollingsworth J, Morris GJ., Jr A new route of transmission for Escherichia coli: infection from dry fermented salami. Am J Public Health. 1996;86:1142–1145. doi: 10.2105/ajph.86.8_pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reading NC, Rasko DA, Torres AG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci U S A. 2009;106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, Sperandio V. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol. 2007;189:2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 113.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect Immun. 2003;71:7129–7139. doi: 10.1128/IAI.71.12.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 116.Wick LM, Qi W, Lacher DW, Whittam TS. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol. 2005;187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rodriguez-Diaz J, Rubio-del-Campo A, Yebra MJ. Lactobacillus casei ferments the N-Acetylglucosamine moiety of fucosyl-alpha-1,3-N-acetylglucosamine and excretes L-fucose. Appl Environ Microbiol. 2012;78:4613–4619. doi: 10.1128/AEM.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. Virulence Meets Metabolism: Cra and KdpE Gene Regulation in Enterohemorrhagic Escherichia coli. MBio. 2012;3:e00280–00212. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bakovic M, Fullerton MD, Michel V. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP: phosphoethanolamine cytidylyltransferase (Pcyt2) Biochem Cell Biol. 2007;85:283–300. doi: 10.1139/o07-006. [DOI] [PubMed] [Google Scholar]
- 120.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 121.Dowhan W. Phosphatidylserine decarboxylases: pyruvoyl-dependent enzymes from bacteria to mammals. Methods Enzymol. 1997;280:81–88. doi: 10.1016/s0076-6879(97)80104-8. [DOI] [PubMed] [Google Scholar]
- 122.Cotton PB. Non-dietary lipid in the intestinal lumen. Gut. 1972;13:675–681. doi: 10.1136/gut.13.9.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kendall MM, Gruber CC, Parker CT, Sperandio V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. MBio. 2012;3:e00050–12. doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 128.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EEC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 129.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]