Abstract
The adhesion of osteoclasts (OCs) to bone and bone resorption require the assembly of specific F-actin adhesion structures, the podosomes, and their dense packing into a sealing zone. The OC-specific formation of the sealing zone requires the interaction of microtubule (MT) + ends with podosomes. Here we deleted cofilin, a cortactin (CTTN)- and actin-binding protein highly expressed in OCs, to determine if it acts downstream of the MT-CTTN axis to regulate actin polymerization in podosomes. Conditional deletion of cofilin in OCs in mice, driven by the cathepsin K promoter (Ctsk-Cre), impaired bone resorption in vivo, increasing bone density. In vitro, OCs were not able to organize podosomes into peripheral belts. The MT network was disorganized, MT stability was decreased and cell migration impaired. Active cofilin stabilizes MTs and allows podosome belt formation, whereas MT disruption de-activates cofilin via phosphorylation. Cofilin interacts with CTTN in podosomes and phosphorylation of either protein disrupts this interaction, which is critical for belt stabilization and for the maintenance of MT dynamic instability. Accordingly, active cofilin was required to rescue the OC cytoskeletal phenotype in vitro. These findings suggest that the patterning of podosomes into a sealing zone involves the dynamic interaction between cofilin, CTTN and the MTs plus-ends. This interaction is critical for the functional organization of OCs and for bone resorption. Key Words: Cofilin, Osteoclast, Bone, Podosome, Actin
(ii) INTRODUCTION
In bone, the balanced action of bone-resorbing osteoclasts (OCs) and bone-forming osteoblasts is essential for the maintenance of skeletal homeostasis. During bone resorption, OCs attach to the bone surface and seal off an extracellular compartment that they acidify via H+ transport through their ruffled-border apical membrane, dissolving the bone matrix mineral phase. OCs also secrete lysosomal enzymes, in particular cathepsin K, into the low pH bone resorbing compartment to degrade the organic bone matrix(1–7). A characteristic and critical feature of the OC is therefore the mechanism by which it attaches to the bone surface, allowing simultaneously sealing of the bone resorbing compartment and cell migration. These functions are both mediated by specialized adhesion structures, the actin-rich podosomes (also termed invadosomes) and their organization into a sealing zone(3, 8, 9). Migration and formation of the sealing zone require the constant and rapid assembly-disassembly of individual podosomes during their short life span (approx. 2-4 minutes)(10). Structurally, podosomes are organized in two distinct domains, an actin-rich core with actin polymerization regulators, including actin regulatory proteins like Wasp, Arp2/3 complex and cortactin (CTTN), surrounded by a ring of signaling molecules named “the cloud” that comprises proteins such as integrins, adaptors (Cbl, paxillin), kinases (Src, Pyk2), and Rho GTPases(11). Actin polymerization occurs at both the core and the cloud(10, 12, 13) but the molecular details of the regulation of actin polymerization in podosomes are not fully elucidated.
The formation of podosomes occurs early during OC differentiation. Individual podosomes are initially organized in clusters which evolve into dynamic rings that merge and form a peripheral belt, ultimately circumscribing the ruffled border and forming the sealing zone when OCs are attached to bone or dentin(10, 12, 14).
The transition between podosome clusters and the podosome belt/sealing zone constitutes a unique and reversible feature of the OC(14, 15). Furthermore, this transition is dependent on microtubules (MTs) and their dynamic properties(3, 16, 17). Proteins enriched at MT + ends interact with proteins in podosomes, allowing the physical interaction of MTs with podosomes to control their behavior, and in particular the transition to peripheral belt. We have previously shown that the interaction of MTs with individual podosomes involves the MT +end protein EB1 and CTTN in podosomes and that this interaction plays a key role in the transition of clusters and rings to belts. MT dynamics and MT+ ends proteins regulate CTTN phosphorylation by Src and CTTN phosphorylation and acetylation are dynamically and inversely related. Accordingly, CTTN-deleted OCs are defective in sealing-zone formation and bone resorption(3). Although these studies established the fact that MTs + ends interact with and dynamically regulate CTTN and podosomes, the precise mechanism by which information is transmitted from CTTN to actin polymerization, allowing the formation of the peripheral belt, is still elusive.
Cofilin, a small 19kDa protein that binds to both CTTN and F-actin, produces free barbed ends by severing actin filaments, used by Arp2/3 complexes to nucleate actin polymerization(18). Cofilin has therefore a central role in controlling actin dynamics by catalyzing actin polymerization and depolymerization through its severing activity. It is present in all eukaryotic cells with three forms in mammals: actin depolymerizing factor (ADF) also known as destrin, cofilin-1 (the major form in all cells, including OCs), and cofilin-2 (the major form in muscle)(19). In the present study we explored further the link between MTs, podosomes and bone resorption by focusing on cofilin 1, hereafter referred as cofilin, as it is highly expressed in OC. If it is known that cofilin is involved in invadosomes, affecting the migration, locomotion and metastatic capacity of cancer cells(20), its role in osteoclasts is only partially understood. Blangy et al (2012) have shown that activation of cofilin occurs as the result of activation of phosphatases by RANKL and plays a key role in podosome maturation into belts(21). We extended these studies and tested the hypothesis that cofilin may act downstream of the MT-CTTN axis to establish the link to actin polymerization and sealing zone formation in OCs.
Our findings show that in OCs, the patterning of podosomes into a peripheral belt involves the phosphorylation-dependent interaction between CTTN and cofilin downstream of the MTs + end proteins and that this interaction and the maintenance of MT dynamic instability are critical for the functional organization of podosomes into a sealing zone in OCs, and thereby for bone resorption.
(ii) MATERIALS AND METHODS
Animals
Generation of cofilinfl/fl (COFfl/fl) mice was described previously(22). OC targeted deletion of cofilin (COFOC−/−) was obtained by crossing COFfl/fl mice with CTSKcre/+ mice to generate CTSK-Cre:COFf/f mice. COFf/f and COFOC−/− were viable and fertile, with offspring being born at expected Mendelian ratio. All animal protocols were approved by the Harvard Medical School Institutional Animal Care and Use Committee policies.
Histomorphometry and micro-computed tomography
Offspring of the COFf/f × COFf/f; CTSKcre/+ crosses were used for analysis. Male littermates were injected 8- and 2- day before sacrifice with 20 mg/kg body weight of calcein and 40 mg/kg body weight of democlocycline (both from Sigma-Aldrich), respectively. At 12 weeks of age, right tibiae were collected and fixed in 70% ethanol for 3 days. Fixed tibiae were dehydrated and embedded in methylmethacrylate. Undecalcified 4-µm sagittal sections were obtained using microtome (RM2255, Leica Biosystems., IL, USA) and Von Kossa staining was performed following standard protocols. Dynamic and static bone histomorphometric analysis of the tibial metaphysis was performed using the OsteoMeasure analyzing system (Osteometrics Inc., Decatur, GA, USA). The structural, dynamic and cellular parameters were calculated and expressed according to standardized nomenclature(23). Right femurs were fixed in 70% ethanol for 3 days and scanned on high-resolution micro–computed tomography (µCT35; Scanco Medical, Brüttisellen, Switzerland) at 20 micron resolution. For analysis of femoral bone mass, a region of trabecular bone 2.1 mm wide was contoured, starting 280 microns from the proximal end of the distal femoral growth plate. Thresholds for trabecular bone and cortical bone were 373.8 mgHA/ccm and 589.4 mgHA/ccm, respectively. A Gaussian noise filter optimized for murine bone was applied to reduce noise in the 2D image. 3D reconstructions were created by stacking the thresholded, 2D images from the contoured regions.
Antibodies and reagents
The following primary and secondary antibodies were used in this study. Rabbit anti-cofilin, rabbit anti-acetylated lysine, rabbit anti-flag, rabbit anti-CTTN, rabbit anti-phospho CTTN (Tyr421), rabbit anti-mouse IgG, (Cell Signaling), rabbit anti-cofilin (Santa Cruz), rabbit anti-cofilin, rabbit anti-phosphoSer3cofilin, rabbit-anti EB1, mouse anti-alpha tubulin (Abcam), mouse anti actin (Millipore), goat anti-mouse and anti-rabbit IgG HRP conjugate (Promega) and mouse TrueBlot Ultra Ig HRP (eBioscience). Rhodamine phalloidin was from Invitrogen. Reagents used include nocodazole, taxol (Sigma), trichostatin A (TSA; Calbiochem) and tubacin (provided by Ralph Mazitschek and Stuart Schreiber). Minimum essential medium-alpha modification (α-MEM), Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA).
Plasmids
The following mammalian expression plasmids have been described: actin-eGFP (Clontech), EB1-GFP (Addgene plasmid 17234), CTTN-FLAG (gift from Dr Yingtao Zhang, University of Florida), pmRFP-N1 human cofilin WT (Addgene plasmid 50856), pmRFP-N1 human cofilin SA (Addgene plasmid 50857), pmRFP-N1 human cofilin WT (Addgene plasmid 50855), pEGFP-N1 human cofilin WT (Addgene plasmid 50859), pEGFP-N1 human cofilin SA (Addgene plasmid 50854), pEGFP-N1 human cofilin SE (Addgene plasmid 50861), HDAC6 Flag (Addgene plasmid 13823), pLenti PGK Puro DEST (Addgene, plasmid w529-2).
Flag-Cofilin was generated by cloning cofilin coding sequence from pLenti PGK Puro DEST (Addgene, plasmid w529-2) into pENTR4-Flag gateway entry vector (Clontech plasmid 17423) and subsequently to pcDNA™3.2-DEST destination vector (Clontech plasmid). Actin-mCherry was generated by cloning actin coding sequence from actin-eGFP into pmCherry-C1 (Clontech). Cortactin-tdTomato (CTNN-tdTom) was generated by cloning CTTN coding sequence from CTTN-eGFP into ptdTomato-N1 (Clontech). Cofilin K33, 44, 96, 144R (cofilin KR) (one construct with 4 point mutations) and cofilin K33, 44, 96, 144Q (cofilin KQ) (one construct with 4 point mutations) were generated from Flag-Cofilin using site directed or multi-site directed mutagenesis kits (QuickChange II).
Osteoclasts primary cultures
Mouse osteoclasts (OCs) were isolated from murine bone marrow macrophages (BMMs). Briefly, BMMs were cultured on untreated tissue culture plastic (Corning) in α-MEM for 3 days with M-CSF (30 ng/ml), lifted with EDTA (0.5 mM), replated and cultured on FBS-coated glass coverslips, glass bottom dishes (MatTek Corporation) or plastic dishes for an additional 3-5 days in a medium with M-CSF (R&D, 30 ng/ml) and RANKL (R&D, 10 ng/ml).
TRAP staining
TRAP staining was done according to the manufacturer’s protocol (Acid Phosphatase, Leukocyte (TRAP) Kit (Sigma, 387A). TRAP-positive multinucleated cells containing four or more nuclei were considered as osteoclasts, and counted for statistical study.
In Vitro Resorption Assay
Corning® Osteo Assay Surface (Corning) was used for resorption assay according to the manufacturer’s instructions. Briefly, BMMs were cultured on osteo assay plates for 5 days in the presence of M-CSF and RANKL. Bone resorption activity was evaluated by measuring the fluorescence intensity of the conditioned medium. Cells were then removed using 5% sodium hypochlorite and the area of pit formed was measured using Image J software.
Dentin Resorption Assay
Following culture with OCs, dentin slices were washed with PBS and sonicated in 1N NAOH for 1 minute to remove cells. Resorption pit images were captured using a 510 Meta laser scanning confocal microscope (Carl Zeiss) and the number and mean depth of selected pits with maximum depth larger than 2 µm was analyzed with Fiji software.
Transient transfections
293-VnR cells (HEK293 cells stably expressing the αvβ3 integrin(24)) were transiently transfected with Flag, Flag-cofilin, Flag-cofilin 4KQ, Flag-cofilin 4KR, HDAC6-Flag, RFP-alone, RFP-cofilin-WT, RFP-cofilin-S3A, RFP-cofilin-S3E using XtremeGENE HP (Roche) according the manufacturer's specifications. Transfected cells were maintained in α-MEM containing 10% fetal calf serum (FBS) for 24 or 36 hrs after transfection, lysed and used for co-immunoprecipitation assay.
Immunoprecipitation and immunoblot analysis
293-VnR cells and primary OCs generated from BMMs were lysed in modified radioimmune precipitation assay (mRIPA) buffer (50 mM Tris-Cl, 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate) containing protease and phosphatase inhibitors (Complete Mini and Phosphostop, Roche). For immunoprecipitation, cell lysates were incubated with primary antibody bound to Dynabeads Protein G (Life Technologies) or anti-Flag M2 beads for Flag-tagged constructs (Sigma Aldrich). After 24 hrs at 4 °C the beads were washed in washing buffer (PBS, 0.01% Tween-20) and immunoprecipitated proteins eluted by adding sample buffer (NuPAGE LDS Sample buffer and Sample Reducing Agent) (Life technologies) and heating at 95 °C for 10 min. Protein samples were resolved by SDS-PAGE and transferred electrophoretically onto nitrocellulose membranes by a semi-dry system (Bio-Rad). Membranes were incubated with primary antibodies overnight. Immunoreactive proteins were visualized using enhanced chemiluminescence reagents (GE Healthcare). Image J was used to compare the density of bands in western blots.
HDAC6 activity measurements
HDAC6 activity was assessed with the HDAC6 Fluorogenic assay kit (Bioscience). HDAC6 activity was measured using a unique fluorogenic substrate and developer combination. Measurements were performed according to standardized protocol.
Statistical Analysis
Statistical differences were assessed with the unpaired Student's t test. Calculations were performed using the Graphpad Prism statistical analysis software. Data are presented as mean ± SD, and p values smaller than 0.05 were considered statistically significant.
(iii) RESULTS
Targeted deletion of cofilin impairs bone resorption in vivo and in vitro
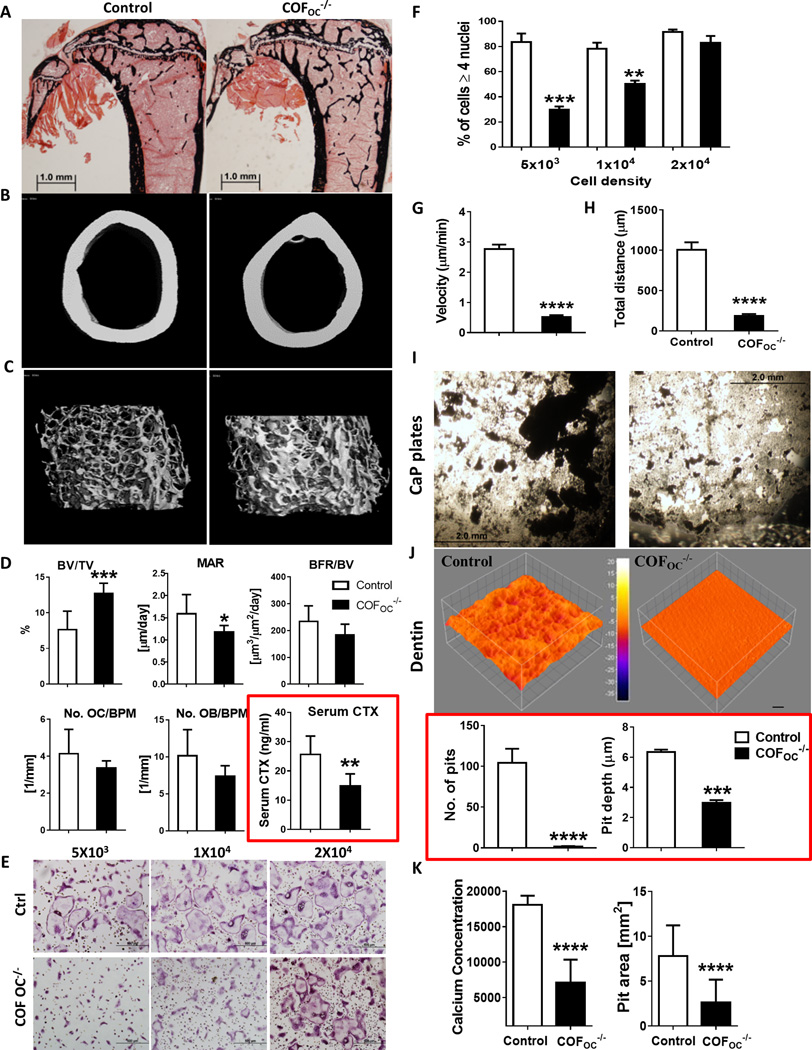
To investigate the role of cofilin in bone resorption, mice with targeted deletion of cofilin in OC (COFOC−/−) were used. COFfl/fl mice were used as control. As shown in Fig. S1A–B, cofilin was reduced in OCs by approximately 85% at days 3–5 of OCs differentiation in vitro. At 12-weeks of age, COFOC−/− male mice display a marked and significant increase in trabecular bone volume, as measured by microCT and histomorphometry (Fig. 1A–D and Table 1A and B). This increase was due to an increased trabecular number and thickness, whereas trabecular spacing was reduced (Table 1A and B). Cortical bone thickness was also increased significantly (Table 1B). There was no change in OC numbers, suggesting that OC differentiation is not significantly altered by deletion of cofilin (Figure 1D and Table 1A). Interestingly, osteoblast number and surface showed a trend towards a decrease and the mineral apposition rate (MAR) and bone formation rate (BFR) were decreased (Fig. 1D and Table 1A), suggesting that the deletion of cofilin in OCs also affects the osteoblasts. Despite the maintained number of osteoclasts, there was a marked decrease in bone resorption in vivo: serum levels of cross-linked C-telopeptide of type I collagen (CTX), a marker of osteoclast activity, measured in the same animals, demonstrated a 44.7 % (P ≤ 0.0011) reduction in COFOC−/− mice compared to control mice (Fig. 1D). These data therefore show that depletion of cofilin in OCs increases bone volume as a result of impaired OC function, and this despite a decrease in bone formation.
Figure 1.
Table. 1.
Histomorphometry and MicroCT data showing bone parameters for control and COFOC−/− mice (SD).
| A | Histomorphometry | ||
|---|---|---|---|
| Parameters | Cof f/f | COFOC−/− | p value |
| (n=6) | (n=8) | (t test) | |
| BV/TV (%) | 7.80±3.80 | 11.3±2.25 | 0.0341* |
| Tb.Th (um) | 31.5±1.92 | 34.4±2.67 | 0.0450* |
| Tb.N (/mm) | 2.44±0.88 | 3.28±0.42 | 0.0357* |
| Tb.Sp (um) | 424±159 | 275±34.4 | 0.0212* |
| Oc.S/B.Pm (%) | 10.4±3.01 | 8.78±1.04 | 0.1741 |
| N.Oc/B.Pm (/mm) |
4.14±1.31 | 3.38±0.37 | 0.1413 |
| ES/BS (%) | 3.07±1.41 | 2.14±0.56 | 0.1131 |
| Ob.S/B.Pm (%) | 13.2±4.40 | 9.64±1.69 | 0.0529 |
| N.Ob/B.Pm (/mm) |
10.1±3.52 | 7.37±1.41 | 0.0643 |
| OS/BS (%) | 8.47±4.40 | 5.23±2.61 | 0.1098 |
| O.Th (um) | 3.18±0.31 | 2.96±0.33 | 0.2226 |
| MAR (um/day) | 1.59±0.43 | 1.18±0.15 | 0.0277* |
| MS/BS (%) | 40.6±3.56 | 42.5±5.85 | 0.5213 |
| BFR/BV (%/year) |
1576±452 | 1074±211 | 0.0191* |
| BFR/BS (um3/um2/year) |
234±58.4 | 184±39.8 | 0.0907 |
| B | microCT data | ||
|---|---|---|---|
| Parameters | Cof f/f | COFOC−/− | p value |
| (n=8) | (n=8) | (t test) | |
| BV/TV (%) | 7.64±2.58 | 12.6±1.36 | 0.0002* |
| Tb.Th (um) | 33.9±1.51 | 42.9±4.43 | 0.108 |
| Tb.N (/mm) | 3.9±0.665 | 4.77±0.22 | 0.0031* |
| Tb.Sp (um) | 261±46.7 | 205±10.1 | 0.0031* |
| M.BV/TV (M) | 4.24±2 | 4.36±0.17 | 0.22 |
| C.Th (um) | 190±1.31 | 210±2.1 | 0.018* |
To gain insight into the cellular mechanisms by which cofilin deficiency leads to impaired OC function, we cultured bone marrow macrophages (BMMs) derived from COFOC−/− mice or control littermates in the presence of M-CSF and RANKL to generate OC precursors and OCs. OC precursors from COFOC−/− mice were able to form TRAP+ multinucleated cells on plastic but these cells appeared to be smaller than in controls: 83.4% of control OCs had 4 or more nuclei compared with only 29.6 % (P ≤ 0.0019) in COFOC−/− OCs (Fig. 1E and F). In contrast to cells depleted in dynamin(25), increasing the plating density of OC precursors rescued the phenotype seen at low cell density indicating that COFOC−/− OCs do not have a pronounced intrinsic fusion defect (Fig. 1E and F). Instead, COFOC−/− OCs showed a striking defect in migration as indicated by a significant reduction in both the distance traveled and the velocity measured by single-cell track analysis over a 6 hrs period (Fig. 1G–H), leading to slower fusion rates.
Our in vivo results strongly suggested that COFOC−/− OCs are impaired in their ability to resorb bone. This was confirmed in vitro: the average resorbed area per individual OC plated on calcium phosphate (CaP) disks (Fig. 1I) or dentin slices (Fig. 1J) was significantly reduced in the absence of cofilin, as indicated by quantification of the number and depth of resorption pits (Fig. 1J). The average OC activity measured as calcium released in the culture medium after culture on CaP disks, and pit area measured after culture on osteo assay plates, showed a similar pattern (Fig. 1K) demonstrating that deletion of cofilin impairs OC function, i.e. bone resorption.
Cofilin deletion alters podosome organization and dynamics
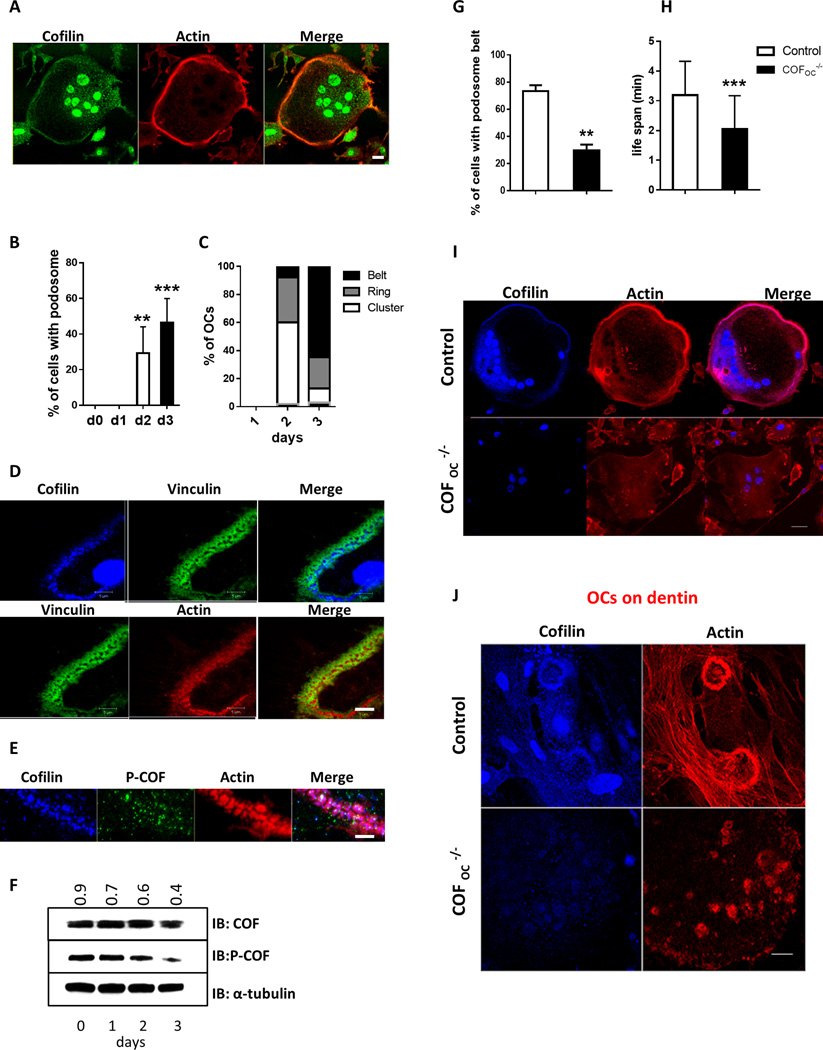
As previously shown(21), cofilin is primarily associated with the podosome belt in mature OCs (Fig. 2A–C and Supplemental Figure S1C). Cofilin exists in two pools: an active pool that is not phosphorylated and can bind to F- and G-actin, severing it and allowing actin polymerization, and an inactive pool that is phosphorylated(26). Total cofilin antibody recognizes both forms. In contrast, phosphorylated cofilin antibody (p-cofilin) is specific to serine 3 phosphorylation and recognizes only the phosphorylated/inactive form of cofilin. We confirmed that total cofilin is localized to the podosome belt and primarily in the core of individual podosomes (Fig. 2D and S1C). In contrast to total cofilin, the dephosphorylated form of cofilin was not abundant in the podosome belt (Fig. 2E). Consistent with Blangy et al.(21), this suggests that the dephosphorylated/active form of cofilin is primarily localized to the core of podosomes in the belt. To determine at which stages of podosome formation and assembly into rings and belt cofilin is activated, we analyzed total (active + inactive) and phosphorylated (inactive) cofilin protein levels at day 0, 1, 2 and 3 of OCs differentiation. While the total levels of cofilin remained constant during OC differentiation, inactivated cofilin levels decreased by 55.5% by day 3 of osteoclastogenesis (Fig. 2F) when more than 50% of OCs had formed podosome belts (Fig. 2B–C), suggesting that cofilin is predominantly activated at the stage of belt formation.
Figure 2.
To explore the mechanism by which deletion of cofilin impairs OC function, we then examined the organization of podosomes in COFOC−/− and control OCs. Deletion of cofilin led to a significant disruption of the podosome belt with the number of cells with a belt decreasing more than 59.5% ((P ≤ 0.0021) Fig. 2G and 2I). This was associated with a pronounced destabilization of the podosomes, their life span decreasing by 36% (P ≤ 0.0005) in the absence of cofilin (Fig. 2H). The podosome belt also failed to form in COFOC−/− OCs plated on dentin (Fig. 2J). Taken together, these data show that cofilin is required for the stabilization of podosomes, critical for the formation of the podosome belt, but not for the assembly of individual podosomes.
Cofilin interacts with MT+ end proteins to target MTs to podosomes
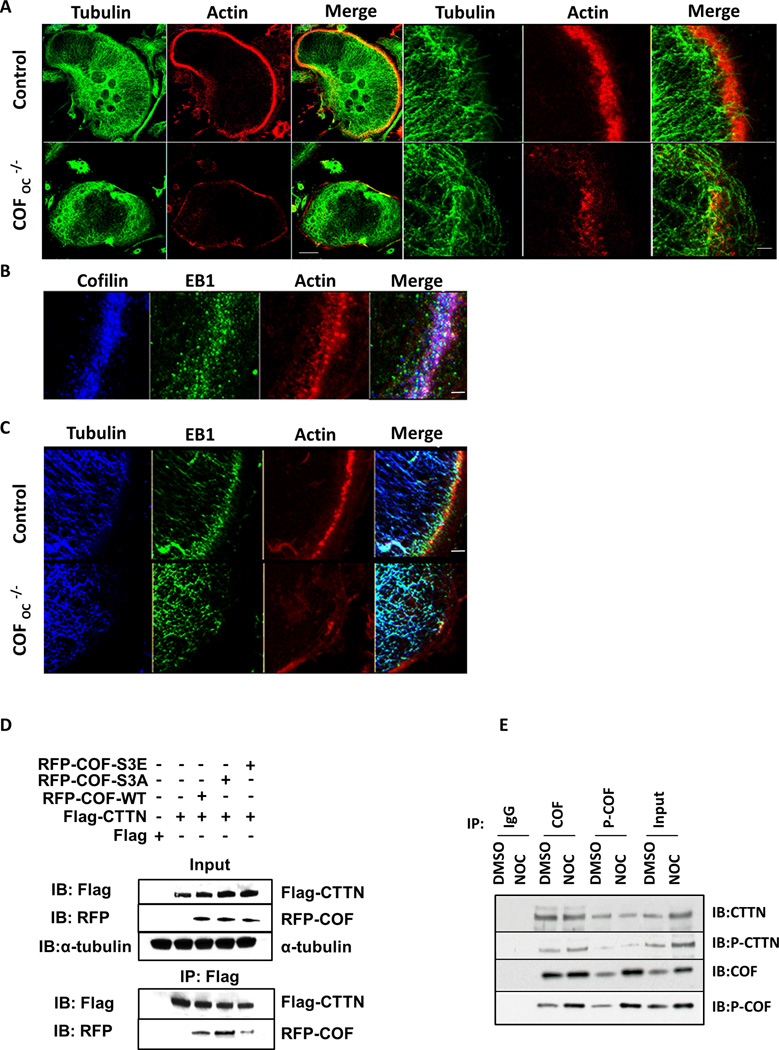
The transition from clusters to rings and to belt requires the interaction of individual podosomes with MT + ends(3, 16, 17). To determine whether deletion of cofilin led to alterations in the MT network and/or their interaction with podosomes, we examined the MT network in control and COFOC−/− OCs. As shown in Figure 3A, the MT network was markedly disrupted after deletion of cofilin. In control OCs the MTs + ends point towards the podosome belt and stop growing when they meet podosomes in the belt. In contrast, in COFOC−/− OCs, MTs lack directionality. Thus, the disruption of MTs in COFOC−/− may result from defective podosomes that fail to capture and stabilize the MT + end. CTTN interacts with EB1, a + end binding protein that functions to maintain MT dynamic instability and is essential for the formation and maintenance of the podosome belt(3). We found that cofilin also co-localizes with EB1 in OCs (Fig. 3B) and, similar to depletion of CTTN, depletion of cofilin results in the delocalization of EB1 from microtubules (Fig. 3C). This suggests that, like CTTN, cofilin plays a role in the interaction of MTs with podosomes, possibly affecting MT and podosome dynamics and stabilizing both the podosomes in the belt and their interaction with MTs.
Figure 3.
MT stability affects the activity of Cofilin and its interaction with CTTN
To examine the link between cofilin activity, MT dynamics and +TIP localization, we used nocodazole (a depolymerizing MT agent). This induced not only a striking disorganization of the podosome belt but also a significant increase in cofilin phosphorylation, i.e. an increase in inactivated cofilin, as indicated by immunofluorescence confocal microscopy and western blot analysis (Fig. S2A–B). Hence, MTs not only promote dynamically the stabilization of the podosome belt at the cell periphery but also regulate cofilin activity. We found that cofilin and CTTN co-localize with F-actin in the podosome belt (Fig. S3A) and interact in mature OCs, as shown by co-IP (Fig. S3B). Furthermore, cofilin is mislocalized in CTTNKO OCs where the podosome belt is disrupted (Fig. S3A) and, reciprocally, CTTN is mislocalized in COFOC−/− OCs where the podosome belt is also disrupted (Fig. S3C). Thus, both cofilin and CTTN are required for podosome belt formation and stabilization.
To assess the role of cofilin-CTTN interaction in MT dynamics and stability, we mutated RFP-tag cofilin constructs to mimic the cofilin-dephosphorylated/active form (where serine 3 is replaced with alanine (S3A)) and cofilin-phosphorylated/inactive form (where serine 3 is replaced by glutamic acid (S3E)) and tested their interaction with CTTN. In 293-VnR cells, CTTN interacts efficiently with RFP-COF-WT and dephosphorylated/active form of cofilin (RFP-COF-S3A), whilst it interacts much less with the phosphorylated/inactive form of cofilin(RFP-COF-S3E). These results suggest that it is the dephosphorylated/active form of cofilin that predominantly interacts with CTTN (Fig. 3D). We then determined whether the cofilin–CTTN interaction is affected when MT dynamics is altered in OCs by nocodazole. We found that cofilin is inactivated by phosphorylation when MTs are disrupted (Fig. 3E and Fig. S2A–B). This phosphorylated/inactive cofilin interacts less with CTTN, but loses its interaction with phosphorylated CTTN, suggesting that this pool of cofilin is independent of CTTN(18, 27).
The interaction between cofilin and CTTN is therefore very dynamic. It is regulated by phosphorylation and dephosphorylation events of both proteins and it is sensitive to, as well as determinant for, MTs dynamic instability. This regulated interaction appears to play an essential role in podosome stabilization into belts.
The interaction of cofilin with CTTN is regulated by acetylation and HDAC6-dependent deacetylation
The balance between CTTN phosphorylation and acetylation is essential in the dynamic interaction between MTs and podosomes(3). Having found that cofilin phosphorylation decreases its interaction with CTTN, we asked whether cofilin can be acetylated. Mass spectrometry data suggested that cofilin is acetylated at all 4 K residues (K33, K44, K96 and K144)) (Fig. S4A). To confirm this, we generated two mutants by replacing these 4 K residues with glutamine (Q) (cofilin 4KQ) to mimic the acetylated state or to arginine (R) (cofilin 4KR) to mimic the non-acetylated state. Flag-Cof-WT and Flag-Cof-KR mutant were transiently transfected in 293-VnR cells in the presence of DMSO (that acted as control) or TSA (a class I and II HDAC inhibitor) to prevent de-acetylation. Western analyses with anti-acetylated lysine and anti-cofilin antibodies, respectively, showed that immunoprecipitated Flag-cofilin is acetylated in vitro at one or more of the K residues (Fig. S4B). TSA markedly increased the level of acetylated Flag-COF in Flag-COF-WT but not Flag-COF-KR construct confirming that cofilin is indeed acetylated in vivo at K33, K44, K96 and/or K144 residues. Moreover, when the same samples were blotted with phospho-cofilin specific antibodies, we saw an increase in phospho-cofilin levels in cells transfected with non-acetylated cofilin mutants (Flag-COF-KR) compared to Flag-COF-WT. Importantly, the levels of phospho-cofilin in cells expressing Flag-COF-WT was significantly reduced, suggesting that, similar to CTTN, acetylation and phosphorylation of cofilin are mutually exclusive (Fig. S4B).
To assess whether the acetylation of cofilin affected its interaction with CTTN, we next co-transfected 293-VnR cells with CTNN-tdTom and either Flag-COF-WT or the cofilin mutants 4KQ or 4KR. The acetylated mimic of cofilin (4KQ) interacted efficiently with CTTN whereas nonacetylated cofilin (4KR) did not (Fig. S4C). Since the phosphorylated-inactive form of cofilin (S3E) showed a decreased interaction with CTTN (Fig. 3D–E), this suggests that acetylation of cofilin favors its interaction with CTTN whereas phosphorylation prevents it. Thus, there is an inverse relationship between the phosphorylation and acetylation status of cofilin and its ability to interact with CTTN.
Microtubule acetylation is altered in COFOC−/− OCs
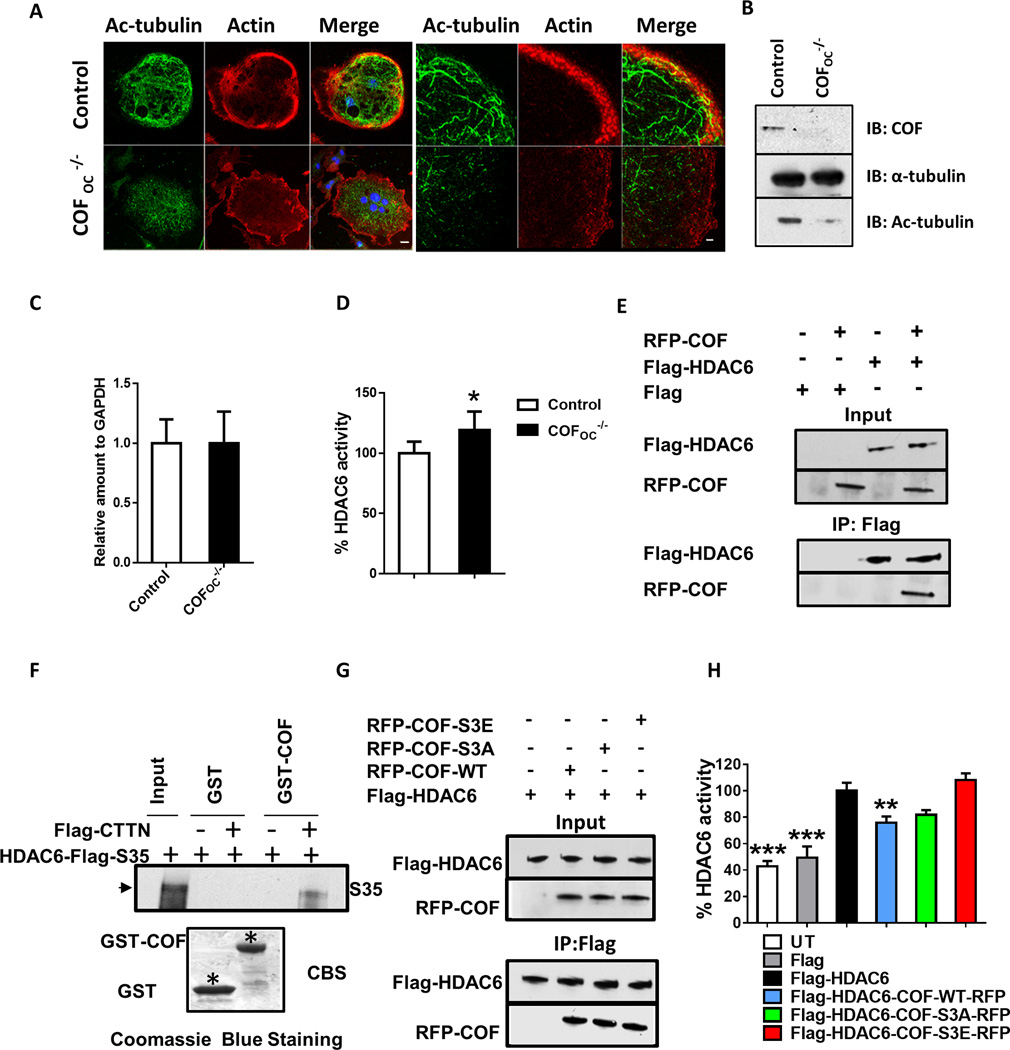
Acetylation also affects MT stability. Because the pool of MTs that accumulates during osteoclastogenesis is highly acetylated(16) we next assessed MT acetylation in control and COFOC−/− OCs. Consistent with their reduced stability, revealed by nocodazole, MT acetylation was reduced in COFOC−/− OCs (Fig. 4A). Similar results were obtained by immunoblotting demonstrating that acetylation of MTs is compromised in COFOC−/− OCs (Fig. 4B).
Figure 4.
HDAC6 is the main histone deacetylase in OCs(3, 16). We therefore asked whether the mechanism by which cofilin deletion destabilizes MTs involves an increased expression and/or activity of HDAC6, favoring HDAC6-catalyzed deacetylation of tubulin. The expression levels of HDAC6 were not changed in the absence of cofilin in OCs (Fig. 4C). In contrast, HDAC6 activity was higher in the absence of cofilin (Fig. 4D), and overexpression of cofilin significantly inhibited HDAC6 activity (Fig. 4H). To determine whether cofilin interacts with HDAC6, 293-VnR cells were transfected with Flag-HDAC6 alone, Flag-HDAC6-RFP-alone or Flag-HDAC6-RFP-COF. Flag immunoprecipitation revealed that cofilin and HDAC6 are found in the same immunoprecipitate in 293-VnR cells (Fig. 4E). GST-pulldown analysis indicated that cofilin interacts only indirectly with HDAC6, with CTTN acting as a scaffold between the two proteins (Fig. 4F).
To determine whether cofilin activity is important for HDAC6 inhibition we assessed the interaction of the RFP-COF-WT, dephosphorylated/active RFP-COF-S3A, and phosphorylated/inactive RFP-COF-S3E with HDAC6 (Fig. 4G) and then measured HDAC6 activity (Fig. 4H). HDAC6 interacted as well with phosphorylated/inactive or dephosphorylated/active forms of cofilin (Fig. 4G–H), and neither affected its activity.
Dephosphorylation and acetylation of cofilin are required to rescue the podosome phenotype in COFOC−/− OCs
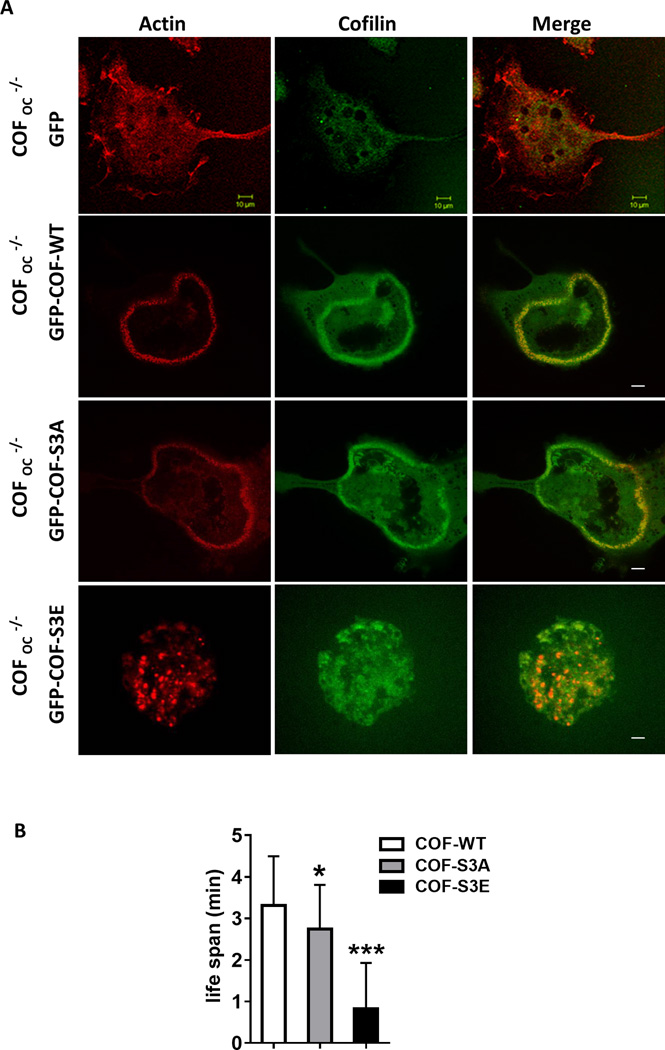
Finally, to determine whether active cofilin is required for podosome belt formation we overexpressed various mutants of GFP-COF along with the CTTN-tdTom plasmid and tested whether the podosome belt phenotype was rescued in COFOC−/− OCs. Podosome belt formation was rescued in COFOC−/− OCs by microinjecting GFP-COF-WT and CTTN-tdTom together (Fig. 5A–B) confirming the crucial role of cofilin in actin dynamics and podosome belt formation in OCs. Similar to wild-type cofilin, the dephosphorylated/active cofilin mutant (S3A), rescued podosome belt formation. In contrast, the phosphorylated-inactive cofilin mutant (S3E), did not rescue the phenotype (Fig. 5A–B), further confirming that cofilin’s activity is essential for podosome belt formation in OCs. The acetylated cofilin mutant (K33, 44, 96, 144Q), which is not phosphorylated and interacts efficiently with CTTN, also rescued podosome belt formation but the deacetylated cofilin mutant (K33, 44, 96, 144R) did not (Fig. S5). These data suggest that acetylated and dephosphorylated cofilin and its interaction with CTTN are critically important for podosome belt formation in OCs.
Figure 5.
(iv) DISCUSSION
The organization and dynamics of the actin cytoskeleton play a key role in several processes essential for OC activity and function, including podosome assembly and organization into a peripheral belt, ultimately sealing the bone resorbing compartment(3, 14), cell polarization(28), motility(29), and vesicular trafficking(30, 31). Our understanding of the molecular mechanism responsible for the regulation of actin dynamics in OCs, and in particular the mechanisms that govern the formation of the peripheral podosome belt/sealing zone, are only partially elucidated. We have previously shown that CTTN and its interaction with +TIP proteins at the +end of MTs are essential for the proper organization of podosomes in a peripheral belt and for bone resorption(3). Because cofilin is a central component of actin dynamics that catalyzes actin polymerization and/or depolymerization through its severing activity, we investigated its involvement in the interaction between MTs and podosomes, the formation of the peripheral belt and OC function.
We show that conditional deletion of cofilin in osteoclasts results in a significant impairment of bone resorption in vivo, leading to a marked increase in trabecular bone volume. Interestingly, but out of the scope of this manuscript, this targeted impairment of osteoclast function also leads to a decrease in bone formation, suggestive of a defective coupling in the bone remodeling process. Although remote, the possibility also exists that the Ctsk-driven deletion of cofilin affected osteocytes, shown to express cathepsin K under certain circumstances(32), affecting more directly bone formation.
Consistent with the observed marked decrease in the bone resorption marker sCTX, in vitro studies showed that COFOC−/− OCs are impaired in their ability to resorb mineralized substrates. In addition, COFOC−/− OCs fail to assemble podosomes into a peripheral belt. Podosome formation occurs in several steps: initially organized in clusters, podosomes evolve from clusters to small rings that ultimately merge to form podosome belts(10). These dynamic steps involve intense actin remodeling and reorganization. In agreement with previously published in vitro data(21) individual podosomes appeared to assemble normally and form clusters and some rings in the absence of cofilin but the formation of podosome belts was disrupted. Deletion of cofilin also impaired the migration of OC precursors and mature OCs, contributing to the decrease in their resorbing activity. A role of cofilin in migration has been previously demonstrated in other cell types.(33, 34).
At the individual level, podosomes are organized in two distinct domains: an actin-rich core that contains actin polymerization regulators, surrounded by a ring of signaling molecules, also referred to as the cloud(10, 15). In contrast to Kurita et al (2008)(35) in invadopodia, and Blangy et al (2012)(21) in podosomes in OCs, we found that dephosphorylated/active cofilin, and phosphorylated/inactive cofilin, are both present in the podosome core, with the dephosphorylated/active pool extending further out of the center than the phosphorylated/inactive cofilin. According to the 3 dimensional organization of podosomes described by Luxenburg et al (2007)(14) this would localize cofilin activity at the transition between the core and the cloud, where actin polymerization / depolymerization actually occurs.
It is intriguing that cofilin is found at highest levels in podosomes within the belt, and that its deletion does not prevent clusters but only their transition to peripheral belts (Fig. 2J). This could be due to the need to polymerize F-actin in order to stabilize podosomes into a three-dimensional dense network as found in the belt(14) or to the fact that the half-life of podosomes is shortest in the belt(10). It could also be specifically due to the fact that in order to establish a belt from clusters and rings, podosomes need to be interacting with microtubules(16) via their + ends(3). Indeed, as shown here, cofilin is essential to maintain this interaction and the dynamic instability of MT + ends. Consequently, the defect in podosome assembly into belts was associated with a disorganization of the MT network. Reciprocally, pharmacological disruption of the MT network also prevents podosome belt formation. Importantly, we found that this also leads to the phosphorylation/inactivation of cofilin. Thus, MTs regulate both cofilin and CTTN activity and cofilin and CTTN activity help maintain the interaction of MTs and podosomes as well as the MT dynamic instability required for plasticity of these short-lived structures(3).
The binding of cofilin to CTTN also negatively regulates cofilin activity, and this mechanism seems to be specific to invadopodia(18, 20). Initially, the release of cofilin from CTTN in invadopodia has been correlated with CTTN phosphorylation(18). We found that the interaction of cofilin with CTTN is highly dynamic, strongly affected by the phosphorylation of each protein and central to the maintenance of MT dynamic instability. The localization of cofilin and +TIP proteins was altered in CTTN-null cells and, conversely, CTTN and +TIP proteins localization was altered in COFOC−/−. Since disruption of MT stability leads to the mislocalization of CTTN, cofilin and +TIP proteins, we hypothesize that the interaction of MTs + ends with podosomes and their ability to allow the localization and stabilization of podosomes at the periphery of the cell require cofilin and CTTN. Similar to our findings with CTTN(3), the phosphorylation and acetylation of cofilin are mutually exclusive.
We show here that MT dynamics affects cofilin’s phosphorylation and therefore its activity. MTs also control the phosphorylation and acetylation of CTTN(3). We found that disruption of MTs by nocodazole leads to cofilin phosphorylation whereas stabilization of MTs by TSA leads to cofilin acetylation at one or more of the 4 K residues (K33, K44, K96 and/or K144). Importantly, active cofilin, i.e. dephosphorylated and acetylated cofilin, interacts more efficiently with CTTN, although according to Oser et al. (2009)(18) this should lead to cofilin inactivation through sequestration.
Meiler et al (2012)(36) showed that CTTN is regulated by acetylation and phosphorylation and these events are mutually exclusive: acetylated CTTN is not phosphorylated and vice versa. Furthermore, CTTN has also been shown to interact with HDAC6 through its repeated domain and this interaction prevents its interaction with F-actin(37). We show here that cofilin actually also gets acetylated, like CTTN, and that it regulates HDAC6 binding to CTTN and activity. Our rescue experiments have shown that cofilin’s activity is essential for podosome belt formation in OCs. The acetylated cofilin mutant (K33, 44, 96, 144Q), maintained active because it is not phosphorylated, interacts with CTTN and can also rescue podosome belt formation. In contrast, the deacetylated cofilin mutant (K33, 44, 96, 144R), that favors phosphorylation and decrease interaction with CTTN, failed to rescue the podosome belt (Fig. S5). These data suggest that active cofilin, which is dephosphorylated and acetylated, and its interaction with CTTN are critically important for podosome belt formation in OCs.
In summary, this study demonstrates that cofilin is present in the podosome core where its activity is required for the stabilization of podosomes within the peripheral belt and sealing zone. Accordingly, deletion of cofilin prevents formation of the sealing zone and impairs bone resorption. Cofilin’s activity is regulated by its interaction with CTTN and +TIP proteins at the +ends of microtubules, maintaining their dynamic instability. Cofilin is therefore an essential component of the podosome in osteoclasts, ensuring their dynamic interaction with microtubules, required for effective bone resorption.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH, NIAMS (AR062054 to R.B.), and a fellowship from the Harvard School of Dental Medicine Dean’s scholarship for medical research (to D.Z). We are very grateful to Kenichi Takeyama for his continuous help and support throughout the project and Sébastien Stephens for helping with microinjection
REFERENCES
- 1.Baron R, Neff L, Brown W, Courtoy PJ, Louvard D, Farquhar MG. Polarized secretion of lysosomal enzymes: co-distribution of cation-independent mannose-6-phosphate receptors and lysosomal enzymes along the osteoclast exocytic pathway. J Cell Biol. 1988;106(6):1863–1872. doi: 10.1083/jcb.106.6.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron R, Neff L, Louvard D, Courtoy PJ. Cell-mediated extracellular acidification and bone resorption: evidence for a low pH in resorbing lacunae and localization of a 100-kD lysosomal membrane protein at the osteoclast ruffled border. J Cell Biol. 1985;101(6):2210–2222. doi: 10.1083/jcb.101.6.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biosse Duplan M, Zalli D, Stephens S, Zenger S, Neff L, Oelkers JM, et al. Microtubule dynamic instability controls podosome patterning in osteoclasts through EB1, cortactin, and Src. Mol Cell Biol. 2014;34(1):16–29. doi: 10.1128/MCB.00578-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromme D, Okamoto K. Human cathepsin O2, a novel cysteine protease highly expressed in osteoclastomas and ovary molecular cloning, sequencing and tissue distribution. Biol Chem Hoppe Seyler. 1995;376(6):379–384. doi: 10.1515/bchm3.1995.376.6.379. [DOI] [PubMed] [Google Scholar]
- 5.Drake FH, Dodds RA, James IE, Connor JR, Debouck C, Richardson S, et al. Cathepsin K, but not cathepsins B, L, or S, is abundantly expressed in human osteoclasts. J Biol Chem. 1996;271(21):12511–12516. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 6.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4(8):638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 7.Troen BR. The role of cathepsin K in normal bone resorption. Drug News Perspect. 2004;17(1):19–28. doi: 10.1358/dnp.2004.17.1.829022. [DOI] [PubMed] [Google Scholar]
- 8.Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13(7):376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 9.Linder S, Kopp P. Podosomes at a glance. J Cell Sci. 2005;118(Pt 10):2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- 10.Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14(2):407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci. 2009;122(Pt 17):3037–3049. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luxenburg C, Parsons JT, Addadi L, Geiger B. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J Cell Sci. 2006;119(Pt 23):4878–4888. doi: 10.1242/jcs.03271. [DOI] [PubMed] [Google Scholar]
- 13.Ochoa GC, Slepnev VI, Neff L, Ringstad N, Takei K, Daniell L, et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150(2):377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, et al. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2007;2(1):e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurdic P, Saltel F, Chabadel A, Destaing O. Podosome and sealing zone: specificity of the osteoclast model. Eur J Cell Biol. 2006;85(3–4):195–202. doi: 10.1016/j.ejcb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118(Pt 13):2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 17.Gil-Henn H, Destaing O, Sims NA, Aoki K, Alles N, Neff L, et al. Defective microtubule-dependent podosome organization in osteoclasts leads to increased bone density in Pyk2(−/−) mice. J Cell Biol. 2007;178(6):1053–1064. doi: 10.1083/jcb.200701148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186(4):571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136(6):1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oser M, Condeelis J. The cofilin activity cycle in lamellipodia and invadopodia. J Cell Biochem. 2009;108(6):1252–1262. doi: 10.1002/jcb.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blangy A, Touaitahuata H, Cres G, Pawlak G. Cofilin activation during podosome belt formation in osteoclasts. PLoS One. 2012;7(9):e45909. doi: 10.1371/journal.pone.0045909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurniak CB, Perlas E, Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev Biol. 2005;278(1):231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, et al. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J Cell Biol. 2001;152(1):181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin NY, Choi H, Neff L, Wu Y, Saito H, Ferguson SM, et al. Dynamin and endocytosis are required for the fusion of osteoclasts and myoblasts. J Cell Biol. 2014;207(1):73–89. doi: 10.1083/jcb.201401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393(6687):805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 27.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14(7):405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi N, Ejiri S, Yanagisawa S, Ozawa H. Regulation of osteoclast polarization. Odontology. 2007;95(1):1–9. doi: 10.1007/s10266-007-0071-y. [DOI] [PubMed] [Google Scholar]
- 29.Novack DV, Faccio R. Osteoclast motility: putting the brakes on bone resorption. Ageing Res Rev. 2011;10(1):54–61. doi: 10.1016/j.arr.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng PY, Cheng TS, Zhao H, Ye S, Sm Ang E, Khor EC, et al. Disruption of the dynein-dynactin complex unveils motor-specific functions in osteoclast formation and bone resorption. J Bone Miner Res. 2013;28(1):119–134. doi: 10.1002/jbmr.1725. [DOI] [PubMed] [Google Scholar]
- 31.Pavlos NJ, Cheng TS, Qin A, Ng PY, Feng HT, Ang ES, et al. Tctex-1, a novel interaction partner of Rab3D, is required for osteoclastic bone resorption. Mol Cell Biol. 2011;31(7):1551–1564. doi: 10.1128/MCB.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qing H, Ardeshirpour L, Pajevic PD, Dusevich V, Jahn K, Kato S, et al. Demonstration of osteocytic perilacunar/canalicular remodeling in mice during lactation. J Bone Miner Res. 2012;27(5):1018–1029. doi: 10.1002/jbmr.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J Cell Biol. 2007;179(4):777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellenchi GC, Gurniak CB, Perlas E, Middei S, Ammassari-Teule M, Witke W. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev. 2007;21(18):2347–2357. doi: 10.1101/gad.434307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurita S, Watanabe Y, Gunji E, Ohashi K, Mizuno K. Molecular dissection of the mechanisms of substrate recognition and F-actin-mediated activation of cofilin-phosphatase Slingshot-1. J Biol Chem. 2008;283(47):32542–32552. doi: 10.1074/jbc.M804627200. [DOI] [PubMed] [Google Scholar]
- 36.Meiler E, Nieto-Pelegrin E, Martinez-Quiles N. Cortactin tyrosine phosphorylation promotes its deacetylation and inhibits cell spreading. PLoS One. 2012;7(3):e33662. doi: 10.1371/journal.pone.0033662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27(2):197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.