Abstract
Low von Willebrand factor (VWF) levels are associated with bleeding symptoms and are a diagnostic criterion for von Willebrand disease, the most common inherited bleeding disorder. To date, it is unclear which genetic loci are associated with reduced VWF levels. Therefore, we conducted a meta-analysis of genome-wide association studies to identify genetic loci associated with low VWF levels. For this meta-analysis, we included 31 149 participants of European ancestry from 11 community-based studies. From all participants, VWF antigen (VWF:Ag) measurements and genome-wide single-nucleotide polymorphism (SNP) scans were available. Each study conducted analyses using logistic regression of SNPs on dichotomized VWF:Ag measures (lowest 5% for blood group O and non-O) with an additive genetic model adjusted for age and sex. An inverse-variance weighted meta-analysis was performed for VWF:Ag levels. A total of 97 SNPs exceeded the genome-wide significance threshold of 5 × 10−8 and comprised five loci on four different chromosomes: 6q24 (smallest P-value 5.8 × 10−10), 9q34 (2.4 × 10−64), 12p13 (5.3 × 10−22), 12q23 (1.2 × 10−8) and 13q13 (2.6 × 10−8). All loci were within or close to genes, including STXBP5 (Syntaxin Binding Protein 5) (6q24), STAB5 (stabilin-5) (12q23), ABO (9q34), VWF (12p13) and UFM1 (ubiquitin-fold modifier 1) (13q13). Of these, UFM1 has not been previously associated with VWF:Ag levels. Four genes that were previously associated with VWF levels (VWF, ABO, STXBP5 and STAB2) were also associated with low VWF levels, and, in addition, we identified a new gene, UFM1, that is associated with low VWF levels. These findings point to novel mechanisms for the occurrence of low VWF levels.
Introduction
Von Willebrand factor (VWF) is a multifunctional glycoprotein, which is secreted by endothelial cells and released upon endothelial cell activation. VWF initiates the adherence of platelets to the injured vessel wall, and the subsequent platelet aggregation facilitates adequate hemostasis.1, 2
Plasma levels of VWF antigen (VWF:Ag) are characterized by a large interindividual variation and range from 0.60 to 1.40 IU/ml in healthy individuals.3 Various environmental and lifestyle factors affect VWF:Ag levels, but ~60% of the variability in VWF:Ag levels can be explained by genetic factors.4
The necessity of maintaining normal VWF levels in the circulation is illustrated by two clinical manifestations that may occur when VWF exceeds its normal range. High VWF:Ag levels are associated with an increased risk of venous thrombosis and arterial thrombosis.5, 6, 7, 8 Conversely, low VWF:Ag levels are associated with an increased bleeding tendency and are a characteristic of von Willebrand disease (VWD). VWD is the most common inherited bleeding disorder in humans and is caused by a quantitative deficiency of VWF (type 1 and 3 VWD) and/or a qualitative defect of VWF molecules (type 2 VWD).9
Most severe forms of type 1 VWD are caused by dominant-negative family-based variations in the VWF gene (VWF).10, 11 However, in individuals with moderately decreased VWF:Ag levels, VWF variations are often not found and linkage with the VWF locus is rarely seen.10, 11 Hence, it is difficult to differentiate between subjects with physiologically low VWF:Ag levels and subjects with low VWF:Ag levels because of VWD.12, 13 However, as VWF:Ag levels are strongly genetically determined, it is expected that more common genetic variations in genes other than VWF are likely to be involved in the occurrence of low VWF:Ag levels and therefore in the etiology of type 1 VWD. We have previously shown that several loci outside the VWF gene are indeed associated with VWF:Ag levels and that the VWF decreasing alleles are more frequently observed in individuals diagnosed with VWD.13 To identify common genetic loci that are associated with low VWF:Ag levels, related to an increased bleeding tendency, we performed a meta-analysis of genome-wide association studies in 11 large population-based cohort studies.
Methods
Study populations
This meta-analysis was conducted in the CHARGE Consortium,14 which includes data from several population-based cohort studies. VWF:Ag measurements were available in four of these: the Rotterdam Study (RS) I and II, the Framingham Heart Study (FHS) and the Atherosclerotic Risk in Communities (ARIC) study. In addition, we included data from seven other studies that had VWF:Ag measurements and genome-wide data available: the British 1958 Birth cohort (B58C) study, the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER), the Prevention of Renal and Vascular Endstage Disease (PREVEND) study, Lothian Birth Cohort 1921 and 1936 (LBC1921, LBC1936), Vis Croatia Study (CROATIA-Vis) and ORKNEY complex Disease Study (ORCADES) (see Supplementary Tables 1 and 2). The designs of the studies have been described previously.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25
Genome-wide scans and VWF:Ag measurements were available for analysis in 31 149 individuals. Eligible participants were not using a coumarin-based anticoagulant at the time of VWF:Ag measurement and were of European ancestry by self-report. All studies were approved by their respective institutional review committee. In addition, written informed consent was obtained from all participants, as well as permission to use their DNA for research purposes.
Baseline measurements and VWF measures
Baseline measures of clinical and demographic characteristics were obtained at the time of cohort entry for ARIC, CROATIA-Vis, ORCADES, PROSPER, PREVEND and RS, and at the time of phenotype measurements for B58C, LBC1921, LBC1936 and FHS. Measures were obtained using standardized methods as specified by each study and included measures of height and weight, as well as self-reported treatment of diabetes and hypertension, current alcohol consumption and prevalent cardiovascular disease (history of myocardial infarction, angina, coronary revascularization, stroke or transient ischemic attack). Blood group antigen phenotypes (O and non-O) were reconstructed using genotype data of rs687289:C>T, which is a marker for the O allele.26
VWF:Ag was measured in all cohorts using enzyme-linked immunosorbent assays (ELISA) (Supplementary Table 3).
Genotyping
For the genotyping, DNA was collected from phlebotomy from all studies except B58C, which used cell lines. Genome-wide assays of SNPs were conducted independently in each cohort using various Affymetrix and Illumina panels (Supplementary Table 3). Each study conducted genotype quality control and data cleaning, including assessment of Hardy–Weinberg equilibrium and variant call rates. Details on genotyping assays have been described in detail previously and are provided in Supplementary Table 3.14
For this analysis, we investigated genetic variation in the 22 autosomal chromosomes.27 Genotypes were coded as 0, 1 and 2 to represent the number of copies of the coded alleles for all chromosomes.27 Each study independently imputed its genotype data to the ≈2.6 million SNPs identified in the HapMap Caucasian (CEU) sample from the Centre d'Etude du Polymorphisme Humain.28, 29, 30 Imputation software, including MACH, BIMBAM or IMPUTE, were used to impute unmeasured genotypes with SNPs that passed quality control criteria based on phased haplotypes observed in HapMap. Imputation results were summarized as an ‘allele dosage', which was defined as the expected number of copies of the minor allele of that SNP (a continuous value between 0 and 2) for each genotype. Each cohort calculated the ratio of observed to expected variance of the dosage statistics for each SNP. This value, which generally ranges from 0 to 1 (ie, poor to excellent), reflects imputation quality.
Public Repository: Our data are available on the European Genome-phenome Archive (https://ega.crg.eu, accession number: EGAS00001001341).
Statistical analysis
Genotype–phenotype data were analyzed independently by each study. VWF:Ag measurements were used as dichotomous variable (low versus normal) with low VWF defined as the lowest 5% within blood groups, that is, blood group O and non-O. All studies used logistic regression with an additive genetic model adjusted for age and sex to conduct analyses of all directly genotyped and imputed SNPs and their association with dichotomous VWF:Ag measures. FHS used generalized estimation equations to account for familial correlation. ARIC and PROSPER adjusted for field site, additionally. B58C adjusted for sex, date and time of sample collection, postal delay and the nurse who performed the inclusion, which also adjusts for the region of residence. Age adjustment was not necessary in B58C, as all cohort members were born in 1 week.
An inverse-variance weighted meta-analysis was performed using METAL software (http://www.sph.umich.edu/csg/abecasis/Metal/index.html) with genomic control correction being applied at the cohort level.31
The a priori threshold of genome-wide significance was set at a P-value of 5.0x10−8. When more than one SNP clustered at a locus, the SNP with the smallest P-value was selected to represent the locus.
Results
For this meta-analysis 31 149 participants of European ancestry were included. The sample size and participant characteristics from each cohort are displayed in Supplementary Table 1. The mean age ranged from 45 years in B58C to 87 years in LBC1921 and on average 48% of the participants was female.
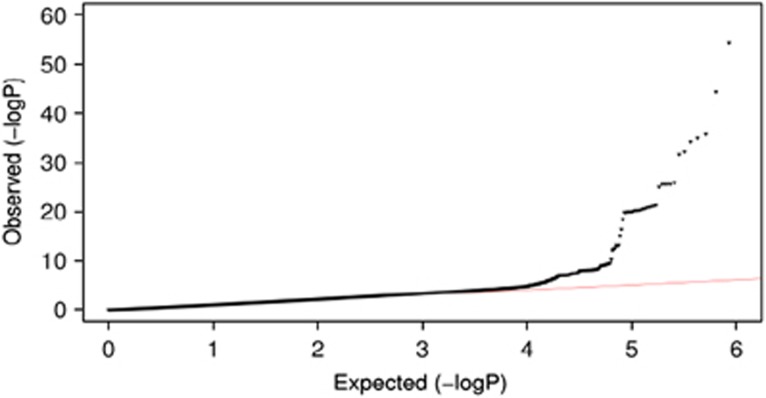
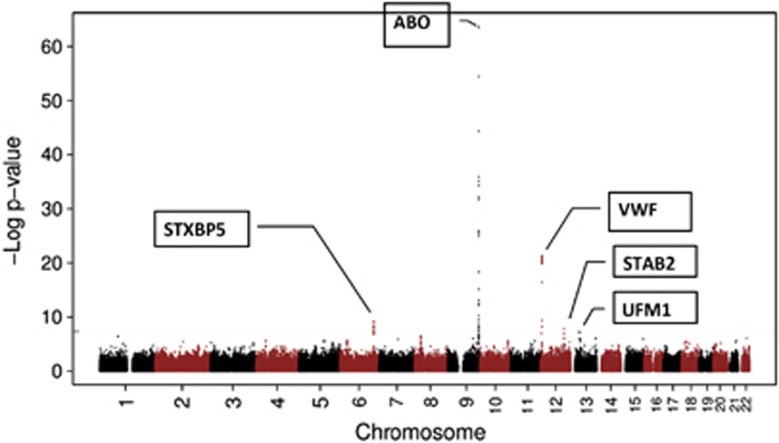
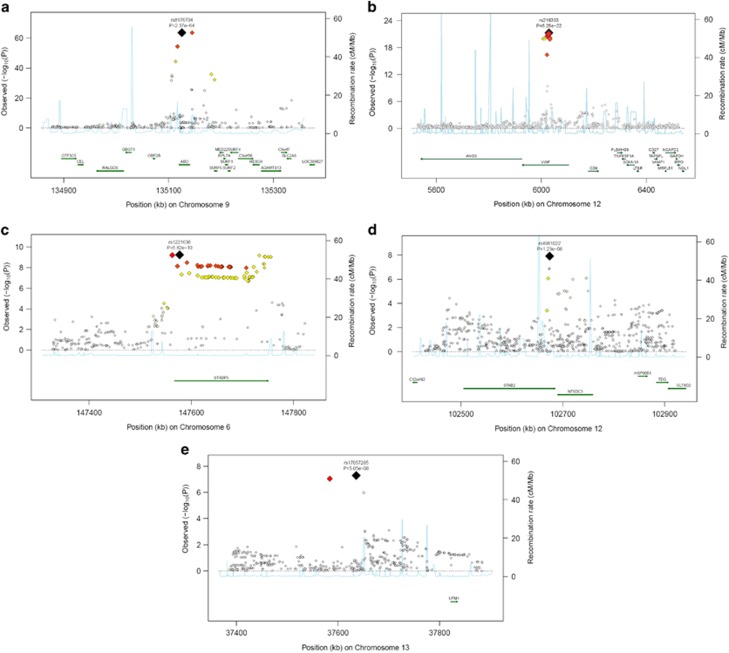
A quantile–quantile plot of the observed P-value from meta-analysis against expected P-value distribution is shown in Figure 1. Figure 2 illustrates the primary findings from the meta-analysis and presents P-values for each of the interrogated SNPs across the 22 autosomal chromosomes. A total of 97 SNPs exceeded the genome-wide significance threshold of 5 × 10−8 and clustered around five genetic loci on four different chromosomes (Table 1 and Figure 3). The SNP with the strongest signal was rs8176704:A>G, which is located at 9q34 (intron) in the ABO blood group gene (P=2.4 × 10−64). The odds ratio (OR) for having VWF levels in the lowest 5% was 2.83 (95% CI 2.52; 3.18). In addition, we performed a conditional analysis. Based on this analysis, we found three independent signals at 9q34. The analysis shows that rs579459 and rs8176747 are independently significant after taking into account the LD structure and their correlation with rs817704. The second most significant locus was marked by rs216303:T>C, which is located at 12p13 (intron) in the VWF gene (OR 0.57; 95% CI: 0.51; 0.64, P=5.3 × 10−22). The third genome-wide significant signal at chromosomal position 6q24 (intron) was within STXBP5 (Syntaxin Binding Protein 5). Rs1221638:A>G was associated with the smallest P-value (5.8 × 10−10) in this region (OR 1.28; 95% CI: 1.19; 1.39). The fourth statistical significant signal was marked by rs4981022A>G, which is located at 12q23 (intron) in STAB2 (stabilin-2) (OR 0.79; 95% CI: 0.73; 0.85, P=1.2 × 10−8). The final genome-wide significant locus was marked by rs17057285:A>C (OR 0.41; 95% CI: 0.30; 0.56, P=2.6 × 10−8), which is 200 kb upstream from UFM1 (ubiquitin-fold modifier 1). There are two SNPs close to rs17057285. The first one is rs17057209, which is 52 kb far from rs17057285 and is in complete LD with rs17057285 (R2=1). Both these SNPs are missing in 5 studies (VIS, ORKNEY, PREVEND, LBC1921, LBC1936) out of 11 studies that contributed to the study. The third SNP is rs7323793, which is 67 kb far and is partly in LD with rs17057285 (R2=0.496). Rs7323793 is missing only in the PREVEND study.
Figure 1.
Quantile–quantile plot of the observed and expected distribution of P-values for all ~2.6 million SNPs and their association with low VWF levels based on meta-analyzed data.
Figure 2.
−Log10 P-values for each of the ~2.6 million tests performed as part of the GWA analysis of low VWF levels. The gray dashed horizontal line marks the 5 × 10−8 P-value threshold of genome-wide significance.
Table 1. Genome-wide significant association of five loci with low VWF levels.
| SNP | |||||||
|---|---|---|---|---|---|---|---|
| Most significant | Region | MAF | P-value | OR (95% CI)a | Het I2 | Het P-value | Gene |
| Rs8176704 hg18:g.13512373A>G | 9q34 | 0.07 | 2.4 × 10−64 | 2.83 (2.52; 3.18) | 46.4 | 0.01 | ABO (intronic) |
| Rs216303 hg18:g.6029306T>C | 12p13 | 0.09 | 5.3 × 10−22 | 0.57 (0.51; 0.64) | −12.6 | 0.39 | VWF (intronic) |
| Rs1221638 hg18:g.147576998A>G | 6q24 | 0.43 | 5.8 × 10−10 | 1.28 (1.19; 1.39) | 0 | 0.85 | STXBP5 (intronic) |
| Rs4981022 hg18:g.102674004A>G | 12q23 | 0.32 | 1.2 × 10−08 | 0.79 (0.73; 0.85) | −0.5 | 0.29 | STAB2 (intronic) |
| Rs17057285 hg18:g.37635821A>C | 13q13 | 0.005 | 2.6 × 10−08 | 0.42 (0.30; 0.56) | 23.2 | 0.10 | 200 kb from UFM1 |
Abbreviations: SNP, single-nucleotide polymorphism; MAF, minor allele frequency; P, statistical significance level; OR, odds ratio; Het, heterogeneity.
Reference sequence Hg 18 (NCBI build 36).
OR represents the risk of having VWF:Ag levels in the lowest 5% relative to the upper 95%.
Figure 3.
Regional plots of top marker loci associated with low VWF levels. (A–E) The association P-values (−log10 transformed, indicated by the left y axis) for SNPs in a 60-kb region of each of the five loci (ABO, VWF, STXBP5, STX2, UFM1) are plotted against their chromosome positions (NCBI build 3) on x axis. The top SNPs are presented as a large diamond in red font and neighboring variants are presented in different colors based on linkage disequilibrium based on HapMap Caucasian data: red: 1≤r2>0.8; orange: 0.8≥r2>0.6; yellow: 0.6≥r2>0.3; green: 0.3≥r2>0.1; blue: 0.1≥r2>0.05; light blue: 0.05≥r2>0.0. The left y axis is the P-value on the −log10 P-value scale and the gray line marks the threshold of genome-wide significance (P=5 × 10−8). Shown in light blue are the estimated recombination rates in HapMap with values indicated by the right y axis. Regional genes and their direction of transcription are depicted with green arrows. The full colour version of this figure is available at EJHG Journal online.
In addition to our five genome-wide significant loci, five other loci demonstrated multiple-SNP hits with P-values below 1.0 × 10−6: rs10848820:A>G (P=1.2 × 10−7) within TSPAN9 (tetraspanin 9), rs4276643:T>C (P=3.4 × 10−7) within SCARA5 (scavenger receptor class A, member 5), rs17398299:A>C (P=4.1 × 10−7) close to 1 gene, LPHN2 (latrophilin 2), rs5995441:T>C (P=8.3 × 10−7) within CARD10 (caspase recruitment domain family, member 10) and rs3750450:T>G (P=9.6 × 10−7) within EPB41L4B (erythrocyte membrane protein band 4.1 like 4B).
Discussion
In this meta-analysis of GWA data from 11 population-based cohorts comprising 31 149 individuals of European ancestry, we identified five genetic loci that are associated with low VWF levels: ABO, VWF, STXBP5, STAB2 and UFM1.
The most significant signal in our study came from a well-known determinant of VWF:Ag levels, the ABO locus. The presence of blood group A and B antigens on VWF molecules leads to a decreased clearance of VWF molecules. Consequently, individuals with blood group O have 25% lower VWF plasma concentrations compared with individuals with blood group non-O.32 Although we used a different cutoff point for low VWF levels for blood group O and non-O separately to minimize the effect of blood group, the ABO locus still reached a very high level of statistical significance. This implies that blood group O versus non-O explains not the total ABO locus effect, and that A or B antigens also determine VWF levels. Indeed, carriers of the B antigen have higher VWF levels compared with carriers of the A antigen and carriers of both antigens have the highest VWF levels.33, 34
The second locus is within the VWF gene. It has been well established that common genetic polymorphisms in the VWF gene contribute to the variability in VWF:Ag levels.35, 36, 37 The most significant SNP that marked the VWF locus was rs216303:T>C, which is located within an intronic region. Until recently, intronic polymorphisms were often considered less relevant for disease development and regulating protein levels in plasma. However, there is now an increasing recognition that intronic variants can contribute by, for example, influencing the form and efficacy of gene splicing and mRNA stability.37 Another possibility is that SNPs in the intronic regions are in high LD with functional SNPs in adjacent regions.
The third locus is within the STXBP5 gene, which encodes the syntaxin binding protein 5. STXBP5 can bind to Soluble N-ethylmaleimide-sensitive factor (NSF) Attachment protein Receptor (SNARE) proteins, among which syntaxin-2 and syntaxin-4. Syntaxin-4 has been shown to be involved in Weibel Palade Body exocytosis,38 the well-known mechanism for the secretion of VWF molecules from endothelial cells. We have previously shown in a well-defined cohort of young patients with a first event of arterial thrombosis that genetic variation in STXBP5 is associated with VWF:Ag levels.13, 39 The LD between rs1221638:A>G and the SNP that had the highest significance in the previous meta-analysis is D′=0.90 and R2=0.67.
The fourth locus was marked by rs4981022:A>G, which is located in STAB2. STAB2 is a transmembrane receptor protein and is primarily expressed in liver and spleen sinusoidal endothelial cells. STAB2 can bind various ligands, such heparin, LDL, bacteria and advanced glycosylation products, and subjects them to endocytosis.40 STAB2 variation might be important in the regulation of VWF levels via the clearance of VWF molecules.
The final genome-wide significant locus was marked by rs17057285:A>C, which is upstream from UFM1. UFM1 encodes the ubiquitin-fold modifier 1, which has been recently identified as a novel protein-conjugating system.41 Although the precise function has not been elucidated yet, the UFM1 cascade seems to be involved in cellular homeostasis, influencing cell division, growth and endoplasmatic reticulum function.42 UFM1 is highly expressed in the pancreatic islets of Langerhans and has a role in the development of type 2 diabetes. Another study showed possible involvement in the development of ischemic heart disease. In this study, chronic inflammation in mice led to a strong upregulation of UFM1 in cardiomyocytes.43 As VWF:Ag levels also have been associated with an increased risk of ischemic heart disease, this is an interesting finding. However, UFM1 has not yet been linked to VWF directly yet and is a novel association needing replication.
Four of the identified loci for low VWF:Ag levels (ie, ABO, VWF, STXBP5 and STAB2) have previously shown to be involved in the regulation of VWF:Ag levels in general.44 The other identified new genetic loci for continuous VWF:Ag levels (ie, SCARA5, STX2, TC2N and CLEC4M) were not associated with low VWF levels.
UFM1 is a novel genetic locus associated with low VWF levels that was not associated with the continuous VWF:Ag levels. Rs17057285:A>C, the SNP with the highest P-value that marks this locus, has a very small minor allele frequency of ~0.5%. Therefore, this finding should be interpreted with care.
In today's clinical practice, it is hard to distinguish between physiologically low VWF levels and VWF:Ag levels due to VWD, because both VWF levels and bleeding symptoms are highly variable and occur frequently in the general population.45 Until recently, it was believed that low VWF:Ag levels and VWD are caused by variations in the VWF gene only. However, now it has been shown that 35% of type 1 (partial quantitative deficiency of VWF) VWD patients have no apparent VWF variations.10, 11 This suggests that genetic variations in genes other than VWF may lead to low VWF:Ag levels, also in patients diagnosed as having VWD.13 Indeed, our current findings confirm this hypothesis that next to ABO blood group and VWF, other genetic loci are involved in the occurrence of low VWF levels.
In the current study, we have not included a replication cohort. Generally, it has been recommended to include all cohorts in the discovery panel to maximize statistical power, rather than use some of the cohorts for replication. In addition, the identified genetic loci comprise extremely small P-values and were previously discovered in the meta-analysis using VWF:Ag as a continuous measure. For these reasons, it is very unlikely that our findings are false positive or came out by chance.
In conclusion, we identified five genetic loci that are associated with low VWF levels: ABO, VWF, STXBP5, STAB2 and UFM1. Our findings confirm the hypothesis that genes other than VWF lead to low VWF:Ag levels. Further research is warranted in order to elucidate whether these genetic loci also contribute to the incidence of bleeding symptoms and VWD.
Acknowledgments
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute Contract U01HG004402; and National Institutes of Health Contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
We acknowledge the use of phenotype and genotype data from the B58C DNA collection, funded by Medical Research Council Grant G0000934 and the Wellcome Trust Grant 068545/Z/02 (http://www.b58cgene.sgul.ac.uk/).
Genotyping of the Lothian Birth Cohorts 1921 and 1936 were supported by the UK's Biotechnology and Biological Sciences Research Council (BBSRC). Phenotype collection in the Lothian Birth Cohort 1921 was supported by The Chief Scientist Office of the Scottish Government (ETM/55). Phenotype collection in the Lothian Birth Cohort 1936 was supported by Research Into Ageing (continues as part of Age UK's The Disconnected Mind project). The work was undertaken in The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Funding from the UK's BBSRC, EPSRC, ESRC and MRC is gratefully acknowledged.
PROSPER is supported by the Scottish Executive Chief Scientist Office, Health Services Research Committee grant number CZG/4/306 and the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NOW 911-03-016).
The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Netherlands Heart Foundation (DHF-2007B159); the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO; 175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was further supported by the Netherlands Genomics Initiative (NGI)/NWO Project No. 050-060-810 and NWO/ZonMw Grant No. 918-76-619 and NWO/ZonMw Grant No. 918-76-619. Dr Dehghan is supported by NWO Grant (veni, 916.12.154) and the EUR Fellowship.
PREVEND genetics is supported by the Dutch Kidney Foundation (Grant E033), the EU project grant GENECURE (FP-6 LSHM CT 2006 037697), the National Institutes of Health (Grant 2R01LM010098), The Netherlands Organization for Health Research and Development (NWO-Groot Grant 175.010.2007.006, NWO VENI Grant 916.761.70, ZonMw Grant 90.700.441) and the Dutch Inter-University Cardiology Institute Netherlands (ICIN).
The VIS study in the Croatian island of Vis was supported through grants from the Medical Research Council UK and Ministry of Science, Education and Sport of the Republic of Croatia (No. 108-1080315-0302) and the European Union framework program 6 EUROSPAN project (Contract No. LSHG-CT-2006-018947). ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society and the European Union framework program 6 EUROSPAN project (Contract No. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh.
Dr Psaty serves on a DSMB for a clinical trial of a device funded by Zoll LifeCor. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Ruggeri ZM: Old concepts and new developments in the study of platelet aggregation. J Clin Invest 2000; 105: 699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri ZM, Ware J: von Willebrand factor. FASEB J 1993; 7: 308–316. [DOI] [PubMed] [Google Scholar]
- Nichols WL, Hultin MB, James AH et al: von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA). Haemophilia 2008; 14: 171–232. [DOI] [PubMed] [Google Scholar]
- Bladbjerg EM, de Maat MP, Christensen K, Bathum L, Jespersen J, Hjelmborg J: Genetic influence on thrombotic risk markers in the elderly – a Danish twin study. J Thromb Haemost 2006; 4: 599–607. [DOI] [PubMed] [Google Scholar]
- Martinelli I: von Willebrand factor and factor VIII as risk factors for arterial and venous thrombosis. Semin Hematol 2005; 42: 49–55. [DOI] [PubMed] [Google Scholar]
- Koster T, Blann AD, Briet E, Vandenbroucke JP, Rosendaal FR: Role of clotting factor VIII in effect of von Willebrand factor on occurrence of deep-vein thrombosis. Lancet 1995; 345: 152–155. [DOI] [PubMed] [Google Scholar]
- Spiel AO, Gilbert JC, Jilma B: von Willebrand factor in cardiovascular disease: focus on acute coronary syndromes. Circulation 2008; 117: 1449–1459. [DOI] [PubMed] [Google Scholar]
- Smith A, Patterson C, Yarnell J, Rumley A, Ben-Shlomo Y, Lowe G: Which hemostatic markers add to the predictive value of conventional risk factors for coronary heart disease and ischemic stroke? The Caerphilly Study. Circulation 2005; 112: 3080–3087. [DOI] [PubMed] [Google Scholar]
- Sadler JE, Budde U, Eikenboom JC et al: Update on the pathophysiology and classification of von Willebrand disease: a report of the Subcommittee on von Willebrand Factor. J Thromb Haemost 2006; 4: 2103–2114. [DOI] [PubMed] [Google Scholar]
- Goodeve A, Eikenboom J, Castaman G et al: Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD). Blood 2007; 109: 112–121. [DOI] [PubMed] [Google Scholar]
- James PD, Notley C, Hegadorn C et al: The mutational spectrum of type 1 von Willebrand disease: results from a Canadian cohort study. Blood 2007; 109: 145–154. [DOI] [PubMed] [Google Scholar]
- Sadler JE: Low von Willebrand factor: sometimes a risk factor and sometimes a disease. Hematol Am Soc Hematol Educ Program 2009; 1: 106–112. [DOI] [PubMed] [Google Scholar]
- Sanders YV, Van der Bom JG, Isaacs A et al: CLEC4M and STXBP5 gene variations contribute to von Willebrand factor level variation in von Willebrand disease. J Thromb Haemost 2015; 13: 956–966. [DOI] [PubMed] [Google Scholar]
- Psaty BM, O'Donnell CJ, Gudnason V et al: Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009; 2: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman A, Breteler MM, van Duijn CM et al: The Rotterdam Study: objectives and design update. Eur J Epidemiol 2007; 22: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ARIC Investigators.: The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol 1989; 129: 687–702. [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE Jr: Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health 1951; 41: 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan DP, Rudnicka AR, Power C et al: Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int J Epidemiol 2007; 36: 522–531. [DOI] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R et al: Runs of homozygosity in European populations. Am J Hum Genet 2008; 83: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Kathiresan S, Lin JP, Tofler GH, O'Donnell CJ: Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med Genet 2007; 8 (Suppl 1): S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlihan LM, Davies G, Tenesa A et al: Common variants of large effect in F12, KNG1, and HRG are associated with activated partial thromboplastin time. Am J Hum Genet 2010; 86: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudan I, Campbell H, Rudan P: Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Coll Antropol 1999; 23: 531–546. [PubMed] [Google Scholar]
- Hillege HL, Fidler V, Diercks GF et al: Urinary albumin excretion predicts cardiovascular and noncardiovascular mortality in general population. Circulation 2002; 106: 1777–1782. [DOI] [PubMed] [Google Scholar]
- Trompet S, de Craen AJ, Postmus I et al: Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med Genet 2011; 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J, Blauw GJ, Murphy MB et al: Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet 2002; 360: 1623–1630. [DOI] [PubMed] [Google Scholar]
- Pare G, Chasman DI, Kellogg M et al: Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet 2008; 4: e1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D: Testing for association on the X chromosome. Biostatistics 2008; 9: 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P: A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Servin B, Stephens M: Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 2007; 3: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ding J, Abecasis GR: Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006; 79: S2290. [Google Scholar]
- Devlin B, Roeder K, Wasserman L: Genomic control, a new approach to genetic-based association studies. Theor Popul Biol 2001; 60: 155–166. [DOI] [PubMed] [Google Scholar]
- Gallinaro L, Cattini MG, Sztukowska M et al: A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood 2008; 111: 3540–3545. [DOI] [PubMed] [Google Scholar]
- Van Schie MC, Wieberdink RG, Koudstaal PJ et al: Genetic determinants of von Willebrand factor plasma levels and the risk of stroke: the Rotterdam Study. J Thromb Haemost 2012; 10: 550–556. [DOI] [PubMed] [Google Scholar]
- Desch KC, Ozel AB, Siemieniak D et al: Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc Natl Acad Sci USA 2013; 110: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie MC, de Maat MP, Isaacs A et al: Variation in the von Willebrand factor gene is associated with von Willebrand factor levels and with the risk for cardiovascular disease. Blood 2011; 117: 1393–1399. [DOI] [PubMed] [Google Scholar]
- van Schie MC, van Loon JE, de Maat MP, Leebeek FW: Genetic determinants of von Willebrand factor levels and activity in relation to the risk of cardiovascular disease: a review. J Thromb Haemost 2011; 9: 899–908. [DOI] [PubMed] [Google Scholar]
- Campos M, Sun W, Yu F et al: Genetic determinants of plasma von Willebrand factor antigen levels: a target gene SNP and haplotype analysis of ARIC cohort. Blood 2011; 117: 5224–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein CJ, Morrell CN, Yamakuchi M: Regulation of Weibel–Palade body exocytosis. Trends Cardiovasc Med 2005; 15: 302–308. [DOI] [PubMed] [Google Scholar]
- van Loon JE, Leebeek FW, Deckers JW et al: Effect of genetic variations in syntaxin-binding protein-5 and syntaxin-2 on von Willebrand factor concentration and cardiovascular risk. Circ Cardiovasc Genet 2010; 3: 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EN, Weigel PH: The ligand-binding profile of HARE: hyaluronan and chondroitin sulfates A, C, and D bind to overlapping sites distinct from the sites for heparin, acetylated low-density lipoprotein, dermatan sulfate, and CS-E. Glycobiology 2008; 18: 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K et al: A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 2004; 23: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Liebau E: The ufm1 cascade. Cells 2014; 3: 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE: Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol 2006; 291: H1411–H1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NL, Chen MH, Dehghan A et al: Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation 2010; 121: 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon JE, Sanders YV, de Wee EM, Kruip MJ, de Maat MP, Leebeek FW: Effect of genetic variation in STXBP5 and STX2 on von Willebrand factor and bleeding phenotype in type 1 von Willebrand disease patients. PLoS One 2012; 7: e40624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.