Abstract
A substantial proportion of patients with posterior polymorphous corneal dystrophy (PPCD) lack a molecular diagnosis. We evaluated 14 unrelated probands who had a clinical diagnosis of PPCD who were previously determined to be negative for mutations in ZEB1 by direct sequencing. A combination of techniques was used including whole-exome sequencing (WES), single-nucleotide polymorphism (SNP) array copy number variation (CNV) analysis, quantitative real-time PCR, and long-range PCR. Segregation of potentially pathogenic changes with disease was confirmed, where possible, in family members. A putative run of homozygosity on chromosome 10 was identified by WES in a three-generation PPCD family, suggestive of a heterozygous deletion. SNP array genotyping followed by long-range PCR and direct sequencing to define the breakpoints confirmed the presence of a large deletion that encompassed multiple genes, including ZEB1. Identification of a heterozygous deletion spanning ZEB1 prompted us to further investigate potential CNVs at this locus in the remaining probands, leading to detection of two additional heterozygous ZEB1 gene deletions. This study demonstrates that ZEB1 mutations account for a larger proportion of PPCD than previously estimated, and supports the hypothesis that haploinsufficiency of ZEB1 is the underlying molecular mechanism of disease for PPCD3.
INTRODUCTION
Posterior polymorphous corneal dystrophy (PPCD) is an autosomal dominant disorder with characteristic changes of the corneal endothelium and Descemet membrane. Clinically, abnormalities of the posterior corneal layers present as geographical and vesicular lesions, bands, and opacities. Corneal endothelial cell dysfunction can lead to corneal oedema, secondary glaucoma, and loss of vision.1 Endothelial cells from affected corneas demonstrate characteristics of epithelial cells including the ability to proliferate, leading to focal formation of multilayered areas, and multilamination of Descemet membrane.2, 3, 4
PPCD is genetically heterogeneous with three identified loci; PPCD1 (OMIM #122000) on chromosome 20p with a currently undefined causative gene,5, 6, 7 PPCD2 (OMIM #609140) associated with mutations in COL8A2,8 and PPCD3 (OMIM #609141) caused by mutations in ZEB1.9 To date, over 30 heterozygous nonsense and frameshifting mutations in ZEB1 have been identified.9, 10, 11, 12, 13, 14, 15, 16, 17
There are regional variations in the contribution of these PPCD subtypes, and the proportion attributed to ZEB1 mutations (PPCD3) has been reported to be 9% in New Zealand, 20% in the Czech Republic, 30% in the United Kingdom and Canada, and 34% in the United States.9, 10, 11, 12, 13, 14, 15, 16, 17 In contrast to other PPCD subtypes, PPCD3 is often associated with corneal steepening.16, 18 Systemic features such as inguinal hernias have also been described.9, 10, 16 There is a substantial variability in disease onset and presentation even among affected members of the same family.19 PPCD3 can arise as a result of de novo mutations, and some individuals with a pathogenic mutation in ZEB1 do not exhibit clinical signs of the disease indicating either incomplete, or age-related, penetrance.9, 14, 16, 19
ZEB1 encodes a zinc finger transcription factor (zinc finger E-box binding homeobox 1) that has a role in many cellular processes, including epithelial–mesenchymal transition (EMT) that involves the transformation of adhesive, non-mobile, epithelial cells into cells with a mesenchymal phenotype that have the ability to migrate.20, 21 ZEB1 induces EMT by suppressing the expression of epithelial-specific factors such as E-cadherin.22 EMT is crucial in early embryogenesis for delamination of the neural crest from the neural tube and for defining the ectodermal–mesodermal boundary.23 ZEB1 is also thought to have a role in neural crest cell migration and the subsequent development of derivative structures, such as the corneal endothelium.20, 24
PPCD often presents in small nuclear families or as an isolated case such that linkage to a specific genomic region is not conclusive, or is confounded by incomplete penetrance in some cases, so a substantial proportion of patients currently lack a molecular diagnosis.9, 10, 11, 12, 14, 15, 16, 17 In this study we demonstrate that, in some of these cases, PPCD can be attributed to heterozygous deletions at the ZEB1 locus, providing insights into the molecular diagnosis of previously unexplained PPCD cases and verifying haploinsufficiency as the mechanism of disease for PPCD3.
METHODS
Study subjects and clinical examination
The study followed the tenets of the Declaration of Helsinki, was prospectively reviewed and approved by local research ethics boards and all patients provided informed consent. Blood samples were collected and genomic DNA extracted from peripheral lymphocytes using conventional methodologies. We included a total of 14 PPCD probands (Table 1) and selected family members. A diagnosis of PPCD was based on established clinical criteria documented previously in detail.1, 16 For inclusion in this analysis, the presence of ZEB1 coding and/or splice-site mutations by direct sequencing had previously been excluded.11, 16, 17 All probands identified to be positive for ZEB1 locus deletions in the current study were also negative for mutations in COL8A2, either by Sanger sequencing or by whole-exome sequencing (WES) (unpublished data).11
Table 1. Summary of posterior polymorphous corneal dystrophy (PPCD) probands included in this study.
| No | Family ID | Gender/age (years) | Ethnic origin | Family history | Methods used for analysis of CNVs in the ZEB1 genomic region |
|---|---|---|---|---|---|
| 1 | C15 | M/34 | Caucasian | Y | Illumina HumanOmniExpress BeadChip qRT-PCR |
| 2 | C17 | M/32 | Caucasian | Y | WES Illumina HumanCytoSNP-12 v2.1 BeadChip qRT-PCR |
| 3 | C21 | M/27 | Caucasian | N | qRT-PCR |
| 4 | C23 | M/48 | Caucasian | N | Illumina HumanOmniExpress BeadChip qRT-PCR |
| 5 | C26 | F/54 | Caucasian | Y | Illumina HumanOmniExpress BeadChip qRT-PCR |
| 6 | C27 | F/77 | Caucasian | N | Illumina HumanOmniExpress BeadChip qRT-PCR |
| 7 | C29 | F/48 | Caucasian | N | qRT-PCR |
| 8 | B8 | M/26 | Caucasian | N | qRT-PCR Illumina HumanCytoSNP-12 v2.1 BeadChip |
| 9 | NZ6 | M/37 | Caucasian | Y | qRT-PCR |
| 10 | NZ54 | M/57 | Caucasian | N | qRT-PCR |
| 11 | NZ220 | F/18 | Caucasian | Y | qRT-PCR |
| 12 | NZ222 | M/10 | Maori | N | qRT-PCR |
| 13 | NZ226 | F/80 | Caucasian | N | qRT-PCR |
| 14 | NZ308 | M/39 | Tongan | N | qRT-PCR |
Abbreviations: B, British; C, Czech; CNVs, copy number variations; F, female; M, male; NZ, New Zealander; qRT-PCR, quantitative real-time PCR; WES, whole-exome sequencing.
All probands were previously determined to be negative for ZEB1 coding and/or splice-site mutations. A positive family history indicates that at least one other family member has been examined ophthalmologically and diagnosed with PPCD. Negative family history does not entirely exclude the presence of the disease in other family members as not all first-degree relatives could be examined.
Whole-exome sequencing
In family C17, WES was performed on DNA samples from three affected individuals (II:2, III:1, and IV:1) (Figure 1a) using a TruSeq exome enrichment kit (Illumina, Inc., San Diego, CA, USA) in accordance with the manufacturer's recommendations, with subsequent analysis on an Illumina HiSeq 2000. Reads were aligned to the GRCh37/hg19 human reference sequence using Novoalign (Novocraft, www.novocraft.com) version 2.05. The ANNOVAR tool was used to annotate single-nucleotide polymorphisms (SNPs) and small insertions/deletions.25
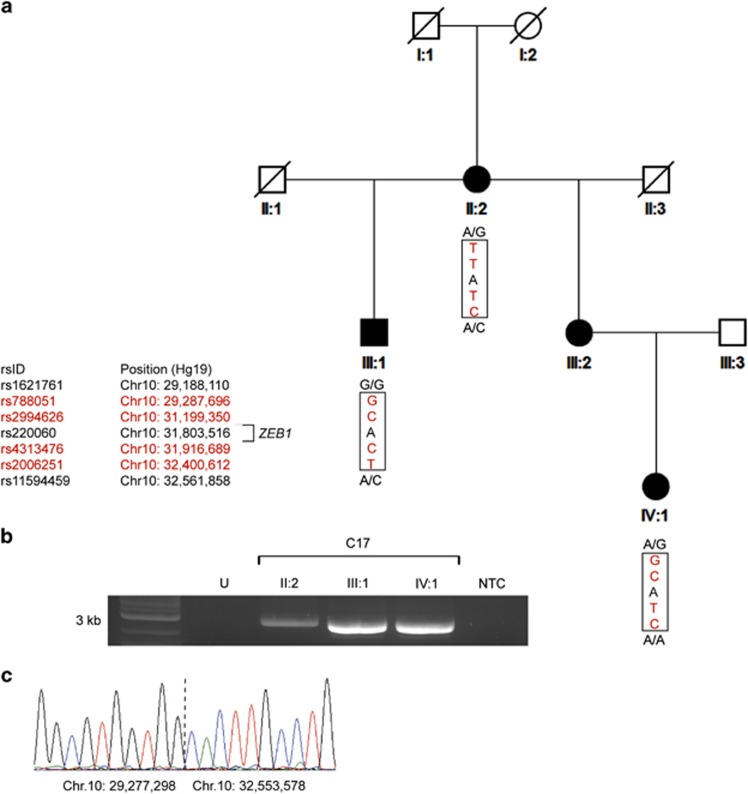
Figure 1.
Detection of a deletion encompassing the entire ZEB1 gene in a family (C17) with posterior polymorphous corneal dystrophy 3. (a) WES revealed a run of homozygous SNPs spanning the ZEB1 locus in three affected individuals. Informative SNPs in red show what appears to be inconsistent inheritance of alleles within the pedigree. The region predicted to be deleted, on the basis of these data, is shown within the boxed area with flanking heterozygous SNPs demonstrating the presence of two alleles. (b) Long-range PCR using primers that flank the deleted region produced an ~3-kb amplimer in affected individuals II:2, III:1, IV:1, but not in an unaffected (U) control sample or non-template control (NTC). (c) Sanger sequencing across the deletion breakpoints in proband II:2 revealed a deletion spanning chr10.hg19:g.29,277,299_32,553,577del. A full colour version of this figure is available at the European Journal of Human Genetics journal online.
SNP array analysis
Individuals III:1 and IV:1 from family C17 (Figure 1a) and proband B8 were screened for copy number variations (CNVs) using the HumanCytoSNP-12 v2.1 BeadChip (Illumina) with ~300 000 markers that provide a mean genome-wide spacing of 10 kb. Four additional probands of Czech origin (C15, C23, C26, and C27) were analysed using the HumanOmniExpress BeadChip (Illumina) containing ~730 000 markers with a mean genome-wide spacing of 4 kb. Labelling and hybridization were performed according to the recommended Infinium HD assay Ultra protocol from Illumina, and scanning was performed with an Illumina iScan System. GenomeStudio Software v2011.1 (Illumina) was used for genotype calling and quality control. Results were analysed with the CNV Partition 2.4.4 algorithm plug-in within the GenomeStudio (Illumina) software and manually inspected in the Illumina Chromosome Browser (display option of the GenomeStudio).
Defining chromosomal deletion breakpoints
GoTaq Long (Promega, Southampton, UK) enzyme was utilized for all long-range PCR assays. Using co-ordinates derived from SNP array genotyping data, the following primers were designed to span the predicted deletion breakpoint in proband C27; ZEB1_C27_F: AAGAAATCCTGGGGCCTGAA and ZEB1_C27_R: TGGGTGACGGATTTTGCAAA. To refine the breakpoint in family C17, conventional PCR and Sanger sequencing were used to genotype polymorphic SNPs surrounding the predicted breakpoints identified by WES and SNP array genotyping. The following primers were designed to amplify across the predicted breakpoint: ZEB1_C17_F: ACGACACAAAGCCTCCTGTA and ZEB1_C17_R: ACTTCAGGGTATTTGGCCTGA. The identity of each amplimer generated by long-range PCR was confirmed by direct sequencing (primer details and conditions available on request).
Quantitative real-time PCR
Two pairs of quantitative real-time PCR (qRT-PCR) primers were designed to target the 5' and 3' ends of the ZEB1 transcript. At the 5' end of the gene ZEB1_CNV_5'F: ACACGTACATTTCGGACCGA and ZEB1_CNV_5'R: GGGGAGCACGAGAGAGTTTT primers were used to amplify a 157-bp amplimer within intron 1. At the 3' end, primers ZEB1_CNV_3'F: AAGGTCTGGCTCTAGTCACC and ZEB1_CNV_3'R: CAGGAGGCTGAGATGAAAGGA were used to amplify a 112-bp region of intron 8 (exons and introns numbered in reference to NG_017048.1). For both qRT-PCR reactions, TWIST1 was used as a reference gene, amplified with TWIST1_CNV_F: ACTGGCCTGCAAAACCATAG and TWIST1_CNV_R: TGCATTTTACCATGGGTCCT primers. All qRT-PCR reactions were performed using labTAQ Green Hi Rox (LabTech, Aylesbury, UK) on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Data analysis was performed with the relative standard curve method, using unaffected control DNA. Each qRT-PCR assay was performed in triplicate. Interpolated gDNA levels of ZEB1 were normalized to those of TWIST1 to produce a ZEB1/TWIST1 ratio.
The identified mutations have been submitted to the ZEB1 specific Leiden Open Variation Database (www.lovd.nl/ZEB1) with individual IDs #00046902 (C17), #00046903 (C27), and #00046952 (B8).
RESULTS
ZEB1 haploinsufficiency in a three-generation PPCD family
Family C17 comprises four affected individuals in three generations (Figure 1a). Analysis of the WES data for affected individuals (II:2, III:1, and IV:1) confirmed that there were no mutations in the coding region or splice sites of the PPCD-associated genes ZEB1 and COL8A2.11 Furthermore, no shared rare (minor allele frequency ≤1% in NHLBI ESP Release Version: v.0.0.25 (http://evs.gs.washington.edu/EVS/) and 1000 Genomes dataset release version 14 (http://www.1000genomes.org/)) variants were identified in any other corneal dystrophy-associated genes.26 However, analysis of the WES data revealed that there was a putative region of homozygosity on chromosome 10p12.1-p11.22 encompassing the ZEB1 gene. Inconsistencies in the segregation of haplotypes for related individuals in this family were observed in the putative homozygous region, suggesting that there may be a heterozygous deletion spanning the entire ZEB1 locus (Figure 1a).
SNP genotyping array analysis of two affected individuals (III:1 and IV:1) revealed a large deletion spanning the region on chr10p12.1-p11.22 that encompassed the ZEB1 gene (Figure 2a). Further haplotype analysis, using a combination of the SNP array data and the WES data, enabled us to refine the co-ordinates of the deletion breakpoints. Long-range PCR was then used to amplify across the deletion breakpoints (Figure 1b). Sanger sequencing of the amplified product confirmed that a large heterozygous deletion (~3.3 Mb; chr10.hg19:g.29,277,299_32,553,577del) was present in all three affected individuals (Figure 1c). The deletion spans the entire ZEB1 gene and an additional nine protein coding transcripts (Figure 3). According to the database of genomic variants, no deletion encompassing the ZEB1 gene has been identified in the control population (http://projects.tcag.ca/variation/; accessed January 2015).
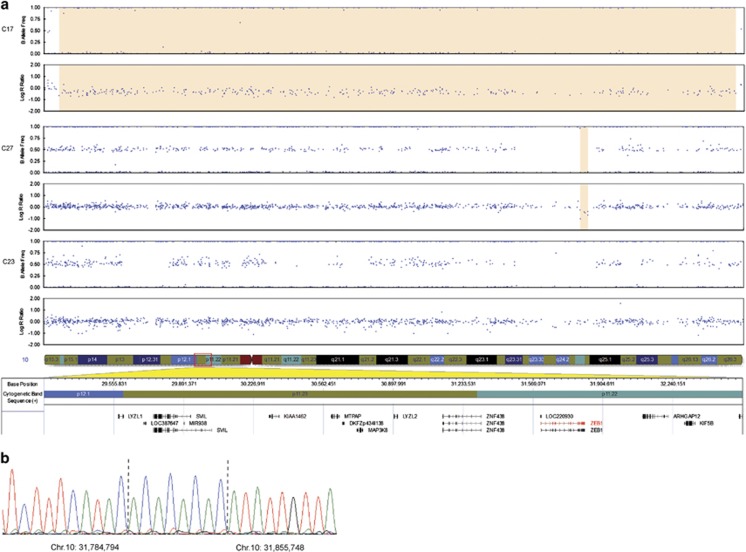
Figure 2.
SNP array CNV analysis of the ZEB1 region in probands with PPCD. (a) A heterozygous ~3.3-Mb deletion was detected in individual III:1 from family C17 (top two traces, highlighted region) and a putative deletion in the ZEB1 genomic region in proband C27 (third and fourth traces, highlighted region). No CNV was identified in proband C23. Blue dots represent individual array probes. The horizontal axis indicates genomic position of array probes and the vertical axis indicates the allele frequency (top trace for each individual) or log R ratio (bottom trace for each individual). The genomic region is shown below the SNP array data. (b) Long-range PCR across the deletion breakpoints and subsequent Sanger sequencing (proband C27) revealed chr10.hg19:g.31,784,795_31,855,747delinsACACACAC. The inserted sequence is shown between the dotted lines. A full colour version of this figure is available at the European Journal of Human Genetics journal online.
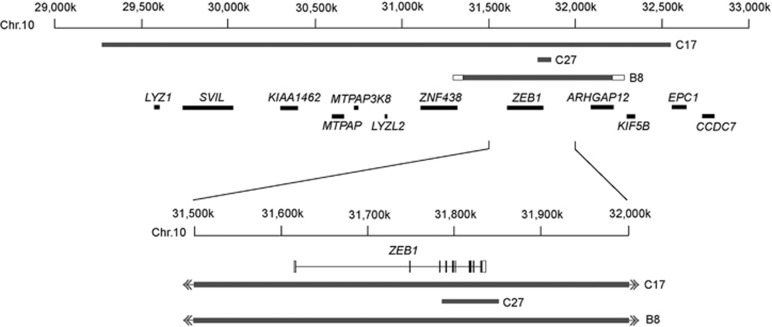
Figure 3.
Schematic representation of three ZEB1 deletions causing posterior polymorphous corneal dystrophy 3. The deletions identified in C17, C27, and B8 do not share any common breakpoints. A large deletion spanning ~3.3 Mb was identified in family C17. This mutation encompasses the entire ZEB1 gene and nine other protein coding genes. C27 had an ~71-kb deletion, which spans the 3' end of ZEB1 from intron 3 and a region downstream containing no additional genes. Proband B8 has a 0.8–1 Mb deletion encompassing the entire ZEB1 gene and at least partially encompasses the flanking gene ARHGAP12. All three deletions differ in size and position and do not share common breakpoints (grey bar: deleted region, white bar: unknown region).
ZEB1 CNV analysis by SNP array genotyping
To test whether other probands previously identified as negative for ZEB1 mutations might also harbour deletions encompassing ZEB1, we performed CNV analysis using SNP genotyping arrays in four unrelated and molecularly unsolved PPCD probands (C15, C23, C26, and C27: Table 1). The SNP array data were suggestive of a deletion encompassing the 3' end of the ZEB1 gene in one proband (C27), while normal copy number was observed at this locus for the remaining three: C23 (Figure 2a), C15, and C26 (data not shown). To confirm the presence of the deletion in proband C27, primers were designed to span the potential deletion breakpoints. The resulting amplimer was Sanger sequenced and revealed the presence of a large deletion with an 8-bp insertion replacing the deleted region (chr10.hg19:g.31,784,795_31,855,747delinsACACACAC). The deletion spans ~71 kb and encompasses exons 4–9 of the ZEB1 gene, as well as the entire 3' untranslated region and ~37 kb downstream (Figure 3).
ZEB1 copy number analysis by qRT-PCR in a molecularly unsolved PPCD cohort
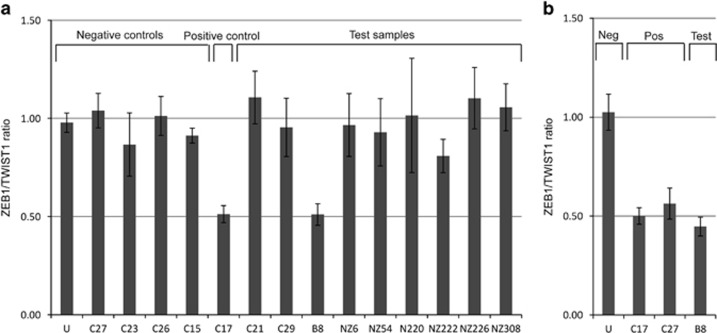
Given that heterozygous full or partial ZEB1 gene deletions were identified as a cause of PPCD3, we designed a ZEB1 qRT-PCR copy number assay to analyse a further nine molecularly unsolved PPCD probands (Table 1). Primers were designed to target the 5' (intron 1) and 3' ends (intron 8) of the ZEB1 gene to allow for detection of partial deletions, as previously observed in proband C27. Genomic DNA levels of ZEB1 were normalized to a reference gene, TWIST1, with an assumed copy number of 2 on the basis that haploinsufficiency of TWIST1 is highly unlikely in control subjects as it causes Saethre–Chotzen syndrome (OMIM #101400).27 The proband from family C17 (Figure 1a; III:1) was used as a positive control for both the 5' and 3' qRT-PCR assays, whereas proband C27 acted as a negative control for the 5' assay and a positive control for the 3' assay. Samples C15, C23, and C26, previously identified as negative for ZEB1 CNVs by SNP array, were included as negative controls for the 5' assay. The 5' assay accurately determined ZEB1 copy number for all positive and negative controls and was used to screen for the presence of a CNV in the remaining unsolved PPCD cohort (Figure 4a). This assay revealed a deletion of the 5' region of the ZEB1 gene in proband B8, whereas the remaining eight samples screened had two copies of the gene. For the 3' qRT-PCR assay the data were outside the expected range for some control samples; therefore, the assay was not sufficiently robust to use as a screening tool. However, the 3' qRT-PCR assay successfully confirmed the heterozygous 3' ZEB1 deletions in probands C17 and C27, and suggested that the heterozygous 5' deletion identified in proband B8 (Figure 4a) extended to the 3' end of the gene (Figure 4b). Using the primers and conditions optimized for amplification across the deletion breakpoints in probands C17 and C27, no PCR product was obtained for sample B8, indicating that the deletion breakpoints in this individual differ from the other two deletions identified. Subsequently, SNP array analysis in proband B8 confirmed the presence of a heterozygous deletion, chr10.hg19:g.(31,298,325_31,350,494)_(32,214,122_32,285,574) del, encompassing the entire ZEB1 gene (Figure 3; Supplementary File 1).
Figure 4.
qRT-PCR screening of a molecularly unsolved PPCD cohort for ZEB1 CNV. ZEB1/TWIST1 ratio ±standard deviation is shown. (a) ZEB1 primers targeted to the 5‘ end of the gene (intron 1). Positive and negative controls include samples that were also analysed by SNP array and an unaffected (U) control DNA sample. Positive control (patient sample C17 with a confirmed deletion of the 5‘ end of the gene) and test sample for patient B8 have a ZEB1/TWIST1 ratio of ~0.5, equivalent to one copy of ZEB1. (b) ZEB1 primers targeted to the 3' end of the gene (intron 8). Positive controls (patient samples C17 and C27) and test sample for patient B8 have a ZEB1/TWIST1 ratio of ~0.5, equivalent to one copy of ZEB1.
DISCUSSION
In a significant proportion of PPCD cases, Sanger sequencing of the COL8A2 and ZEB1 genes does not result in the identification of a pathogenic change, suggesting that there are as-yet undiscovered genes or alternative disease mechanisms.9, 10, 11, 12, 14, 15, 16, 17 In this study, we show that heterozygous deletions of the ZEB1 gene cause PPCD3, verifying that haploinsufficiency of ZEB1 is the mechanism of disease.
The three deletion events that we found in this study differed both in size and in position, with no common deletion breakpoints. We identified a 36-bp region of microhomology for the large deletion in family C17 that originates from either the proximal or distal breakpoint. These data indicate that this microhomology influenced the extent of the deletion.28 The deleted region (chr10.hg19:g.29,277,299_32,553,577del) encompassed ZEB1 and nine flanking genes. Currently, there is only one annotated gene within the deleted region that is known to cause a Mendelian disorder; mutations in MTPAP are associated with recessive spastic ataxia 4 (OMIM #613672).29 The four individuals with this deletion had no apparent systemic disorder, suggesting that haploinsufficiency of the other genes in the region does not cause a clinically distinct phenotype. For the deletion identified in patient C27, no microhomology was observed at the breakpoints; however, a repeat sequence (4xAC) was found to be inserted in the deleted region (Figure 2b). SNP array analysis confirmed that the deletion identified in proband B8 differed from that of families C17 and C27.
The absence of a ZEB1 gene deletion in the remaining 11 probands indicates that PPCD in these patients could be caused by a small CNV not detected by the methods described, by a mutation in COL8A2, by the as-yet unidentified gene on chromosome 20p, or due to another genetic mechanism.
It is hypothesized that PPCD3 manifests as a result of mesenchymal to epithelial dysregulation during prenatal corneal development.30 Interestingly, overexpression of ZEB1 in cancers has been shown to trigger EMT by ZEB1-mediated repression of epithelial-specific genes.31 Conversely, enhanced expression of epithelial-specific genes in this pathway, including E-cadherin (CDH1) and COL4A3, has been demonstrated in embryonic null and adult haploinsufficient ZEB1 mice.20, 30 Importantly, the mice display abnormal corneal endothelial and keratocyte proliferation, as well as abnormal posterior corneal morphology, mimicking the PPCD3 phenotype.30 Interestingly, the corneal endothelium from PPCD3 patients exhibits increased expression of markers for an epithelial cell phenotype, such as keratins 7 and 19.2, 3 Patients with PPCD also have multilayering of the corneal endothelium that is probably due to uncontrolled growth of a dividing endothelium.2, 3, 4
The proportion of PPCD cases attributed to mutations in ZEB1 has been reported to be between 9 and 34%.9, 10, 11, 12, 14, 15, 16, 17 Our results demonstrate that the number of cases with PPCD3 is underestimated, because standard screening techniques do not detect deletions spanning such large regions. We have therefore developed an assay for the rapid and reliable detection of ZEB1 deletions using qRT-PCR. The assay targeting the 5' end of the ZEB1 gene was reproducibly robust, and corroborated the data obtained using different techniques for five PPCD samples. Furthermore, the deletion in proband B8, identified using this assay, was subsequently confirmed with SNP array CNV analysis. This assay can be used as an initial screening method for molecularly unsolved PPCD cases. The integration of a variety of approaches for the detection of ZEB1 mutations, such as those described here, or development of an appropriate multiplex ligation-dependent probe amplification assay should be applied to achieve a molecular diagnosis for PPCD.
Acknowledgments
We thank all patients and their families for participating in this research. PL was supported by research grant GACR P301/12/P591, UNCE 204011, and PRVOUK-P24/LF1/3 programs of the Charles University in Prague. VS was supported by BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund and by grant NT13116-4/2012 from the Ministry of Health of the Czech Republic. AJH, SJT, AED, and CJE were supported by National Institute for Health Research (NIHR) Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, Moorfields Special Trustees, Moorfields Eye Charity, The Lanvern Foundation, Fight for Sight and Rosetrees Trust, ALV by Save Sight Society of New Zealand and School of Medicine Foundation and LD by SVV UK 260148/2015.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Cibis GW, Krachmer JA, Phelps CD, Weingeist TA: The clinical spectrum of posterior polymorphous dystrophy. Arch Ophthalmol 1977; 95: 1529–1537. [DOI] [PubMed] [Google Scholar]
- Moroi SE, Gokhale PA, Schteingart MT et al: Clinicopathologic correlation and genetic analysis in a case of posterior polymorphous corneal dystrophy. Am J Ophthalmol 2003; 135: 461–470. [DOI] [PubMed] [Google Scholar]
- Jirsova K, Merjava S, Martincova R et al: Immunohistochemical characterization of cytokeratins in the abnormal corneal endothelium of posterior polymorphous corneal dystrophy patients. Exp Eye Res 2007; 84: 680–686. [DOI] [PubMed] [Google Scholar]
- Merjava S, Malinova E, Liskova P et al: Recurrence of posterior polymorphous corneal dystrophy is caused by the overgrowth of the original diseased host endothelium. Histochem Cell Biol 2011; 136: 93–101. [DOI] [PubMed] [Google Scholar]
- Gwilliam R, Liskova P, Filipec M et al: Posterior polymorphous corneal dystrophy in Czech families maps to chromosome 20 and excludes the VSX1 gene. Invest Ophthalmol Vis Sci 2005; 46: 4480–4484. [DOI] [PubMed] [Google Scholar]
- Hosseini SM, Herd S, Vincent AL, Heon E: Genetic analysis of chromosome 20-related posterior polymorphous corneal dystrophy: genetic heterogeneity and exclusion of three candidate genes. Mol Vis 2008; 14: 71–80. [PMC free article] [PubMed] [Google Scholar]
- Yellore VS, Papp JC, Sobel E et al: Replication and refinement of linkage of posterior polymorphous corneal dystrophy to the posterior polymorphous corneal dystrophy 1 locus on chromosome 20. Genet Med 2007; 9: 228–234. [DOI] [PubMed] [Google Scholar]
- Biswas S, Munier FL, Yardley J et al: Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet 2001; 10: 2415–2423. [DOI] [PubMed] [Google Scholar]
- Krafchak CM, Pawar H, Moroi SE et al: Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet 2005; 77: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldave AJ, Yellore VS, Yu F et al: Posterior polymorphous corneal dystrophy is associated with TCF8 gene mutations and abdominal hernia. Am J Med Genet A 2007; 143A: 2549–2556. [DOI] [PubMed] [Google Scholar]
- Liskova P, Tuft SJ, Gwilliam R et al: Novel mutations in the ZEB1 gene identified in Czech and British patients with posterior polymorphous corneal dystrophy. Hum Mutat 2007; 28: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AL, Niederer RL, Richards A, Karolyi B, Patel DV, McGhee CN: Phenotypic characterisation and ZEB1 mutational analysis in posterior polymorphous corneal dystrophy in a New Zealand population. Mol Vis 2009; 15: 2544–2553. [PMC free article] [PubMed] [Google Scholar]
- Nguyen DQ, Hosseini M, Billingsley G, Heon E, Churchill AJ: Clinical phenotype of posterior polymorphous corneal dystrophy in a family with a novel ZEB1 mutation. Acta Ophthalmol 2010; 88: 695–699. [DOI] [PubMed] [Google Scholar]
- Bakhtiari P, Frausto RF, Roldan AN, Wang C, Yu F, Aldave AJ: Exclusion of pathogenic promoter region variants and identification of novel nonsense mutations in the zinc finger E-box binding homeobox 1 gene in posterior polymorphous corneal dystrophy. Mol Vis 2013; 19: 575–580. [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Dash DP, Muszynska D et al: Mutational spectrum of the ZEB1 gene in corneal dystrophies supports a genotype-phenotype correlation. Invest Ophthalmol Vis Sci 2013; 54: 3215–3223. [DOI] [PubMed] [Google Scholar]
- Liskova P, Palos M, Hardcastle AJ, Vincent AL: Further genetic and clinical insights of posterior polymorphous corneal dystrophy 3. JAMA Ophthalmol 2013; 131: 1296–1303. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Liskova P, Dudakova L et al: Identification of six novel mutations in ZEB1 and description of the associated phenotypes in patients with posterior polymorphous corneal dystrophy 3. Ann Hum Genet 2015; 79: 1–9. [DOI] [PubMed] [Google Scholar]
- Aldave AJ, Ann LB, Frausto RF, Nguyen CK, Yu F, Raber IM: Classification of posterior polymorphous corneal dystrophy as a corneal ectatic disorder following confirmation of associated significant corneal steepening. JAMA Ophthalmol 2013; 131: 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskova P, Filipec M, Merjava S, Jirsova K, Tuft SJ: Variable ocular phenotypes of posterior polymorphous corneal dystrophy caused by mutations in the ZEB1 gene. Ophthalmic Genet 2010; 31: 230–234. [DOI] [PubMed] [Google Scholar]
- Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC: Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 2008; 135: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne G, Sayan AE, Tulchinsky E: ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell Cycle 2010; 9: 886–891. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME: The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA: The early steps of neural crest development. Mech Dev 2001; 105: 27–35. [DOI] [PubMed] [Google Scholar]
- Darling DS, Stearman RP, Qi Y, Qiu MS, Feller JP: Expression of Zfhep/deltaEF1 protein in palate, neural progenitors, and differentiated neurons. Gene Expr Patterns 2003; 3: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H: ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JS, Moller HU, Aldave AJ et al: IC3D classification of corneal dystrophies–edition 2. Cornea 2015; 34: 117–159. [DOI] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED et al: Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet 1997; 15: 36–41. [DOI] [PubMed] [Google Scholar]
- McVey M, Lee SE: MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet 2008; 24: 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby AH, Patel H, Chioza BA et al: Defective mitochondrial mRNA maturation is associated with spastic ataxia. Am J Hum Genet 2010; 87: 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Peng X, Tan J, Darling DS, Kaplan HJ, Dean DC: Zeb1 mutant mice as a model of posterior corneal dystrophy. Invest Ophthalmol Vis Sci 2008; 49: 1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC et al: The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11: 1487–1495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.