Abstract
Hereditary cerebellar ataxias and hereditary spastic paraplegias are clinically and genetically heterogeneous and often overlapping neurological disorders. Mutations in SPG7 cause the autosomal recessive spastic paraplegia type 7 (SPG7), but recent studies indicate that they are also one of the most common causes of recessive cerebellar ataxia. In Quebec, a significant number of patients affected with cerebellar ataxia and spasticity remain without a molecular diagnosis. We performed whole-exome sequencing in three French Canadian (FC) patients affected with spastic ataxia and uncovered compound heterozygous variants in SPG7 in all three. Sanger sequencing of SPG7 exons and exon/intron boundaries was used to screen additional patients. In total, we identified recessive variants in SPG7 in 22 FC patients belonging to 12 families (38.7% of the families screened), including two novel variants. The p.(Ala510Val) variant was the most common in our cohort. Cerebellar features, including ataxia, were more pronounced than spasticity in this cohort. These results strongly suggest that variants affecting the function of SPG7 are the fourth most common form of recessive ataxia in FC patients. Thus, we propose that SPG7 mutations explain a significant proportion of FC spastic ataxia cases and that this gene should be considered in unresolved patients.
Introduction
Hereditary cerebellar ataxias and hereditary spastic paraplegias (HSPs) are clinically and genetically heterogeneous and often overlapping neurological disorders. HSPs are characterized by a predominant progressive spasticity and weakness in the lower limbs due to degeneration of the corticospinal tracts,1 whereas the main feature of cerebellar ataxias is progressive cerebellar degeneration leading to impaired balance, gait and speech.2 Both HSP and hereditary ataxias can be associated with other neurological and non-neurological features, resulting in complex phenotypes with frequent intra- and inter-familial variability. Significantly, cerebellar ataxias are very often associated with pyramidal involvement leading to >50% of recessive ataxias manifesting as spastic ataxias.3 Because of the individual rarity and genetic heterogeneity of these conditions, their molecular diagnosis remains challenging and time-consuming.
Mutations in the gene SPG7 were the first identified genetic cause of autosomal recessive HSP in 1998 (MIM602783).4 Since then, a significant number of causative mutations were found in several HSP cohorts from different populations.5, 6, 7, 8, 9, 10, 11, 12, 13, 14 SPG7 can be characterized by a pure or complex HSP phenotype. Increasingly, reports documented that cerebellar ataxia and cerebellar atrophy on magnetic resonance imaging (MRI) are the most frequent additional features in complex SPG7 cases.6, 7, 9, 12, 15 In a study of a large Dutch cohort, cerebellar ataxia was found in 57% of cases and it was even the dominating clinical symptom in some of these patients.6 In addition, a recent report suggests that SPG7 mutations are a frequent cause of adult-onset undiagnosed cerebellar ataxia in patients of British descent.16 The sequencing of SPG7 in next-generation sequencing panels has further identified cases of spastic ataxia carrying compound heterozygous variants, supporting that SPG7 may be one of the most common forms of recessive ataxias worldwide.8 Thus, a growing number of studies indicate that SPG7 should be considered in the differential diagnosis of recessive cerebellar ataxia.6, 16, 17
In Quebec, Friedreich ataxia (MIM229300), autosomal recessive spastic ataxia of Charlevoix–Saguenay (MIM270550) and autosomal recessive spinocerebellar ataxia type 8 (MIM610743) account for the majority of autosomal recessive cerebellar ataxia cases and, except for the latter one, they usually present as spastic ataxias.18 However, a large number of French Canadian (FC) cerebellar ataxia cases remain unresolved, many of which have associated spasticity and often milder adult-onset phenotypes. Only recently were three SPG7 FC cases reported from the province of Ontario.19 The relative prevalence of SPG7 in the FC population is unknown.
As for many other rare diseases, the implementation of whole-exome sequencing (WES) in research and in clinical settings has greatly accelerated the identification of disease-causing genes in the field of ataxias. Here we report on the identification of causative SPG7 variants in 22 unresolved FC spastic ataxia cases belonging to 12 families, making it the fourth most common recessive ataxia in this population.
Subjects and methods
Subjects
Patients presenting with cerebellar ataxia, spasticity and a family history suggestive of autosomal recessive or sporadic inheritance were seen at several neuromuscular clinics in the province of Quebec and Eastern Ontario between 2002 and 2014. Mutations in SACS and FRDA were ruled out in the majority of cases. All participating family members signed an informed consent form approved by the institutional ethics committee of the Centre de Recherche du Centre Hospitalier de l'Université de Montréal (CRCHUM) or the Children's Hospital of Eastern Ontario.
Molecular analyses
Genomic DNA was extracted from peripheral blood cells using standard methods. WES was performed on individuals 1, 5 and 9 using the SureSelect exome capture kit v.5 (Agilent Technologies, Santa Clara, CA, USA). The libraries were sequenced on an Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) with paired-end 100-bp reads at the McGill University and Genome Quebec Innovation Center (Montreal, Quebec, Canada). Sequences were aligned to the human reference genome (UCSC hg19) using the BWA (Burrows–Wheeler Aligner) algorithm, variant calling was performed using SAMtools20 and annotation was done using ANNOVAR21 and custom scripts, as previously described. For Sanger sequencing, PCR was used to amplify selected individual exons and intron–exon boundaries of SPG7. PCR products were sent to McGill University and Genome Quebec Innovation Center for sequencing, using a 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Mutation detection analysis was performed using SeqMan v.4.03 (DNASTAR Inc., Madison, WI, USA) and 4Peaks (A. Griekspoor and Tom Groothuis, mekentosj.com).
Patient 22 was analyzed using the HSP Sanger panel at the Hospital for Sick Children (Toronto, Ontario, Canada), as previously described.19 Variants identified were submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
For complementary DNA (cDNA) sequencing, skin fibroblasts were derived from individual 19 and a healthy control and were grown according to standard protocols. Total RNA was extracted using Trizol reagent (Ambion, Foster City, CA, USA) and reverse transcribed into cDNA using the Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Exons 7 and 8 of SPG7 were amplified by PCR from the cDNA and sequenced. Sequencing and variant detection analysis were performed as described above.
Results
Exome sequencing
A group of nine unrelated patients with unresolved autosomal recessive spastic ataxia were sent for WES. As there was some phenotypic heterogeneity within the cohort regarding age of onset, severity and associated neurological symptoms, it was expected that causative variants in different genes would be uncovered. We searched for homozygous or compound heterozygous variants with a minor allele frequency <3% in the 1000 Genomes and Exome Variant Server (EVS) databases. As expected, we identified distinct candidate genes in several individuals, confirming the genetic heterogeneity within our cohort. Nevertheless, in three patients (1, 5 and 9), we uncovered compound heterozygous variants in SPG7 (NM_003119, NG_00808.2.1; Table 1, Supplementary Table S1 and S2). The three cases carried the most frequent previously reported variant c.1529C>T (p.(Ala510Val)) on one allele.15 To assess the possibility that the p.(Ala510Val) variant arose from a single event, we looked at genotypes of known SNPs surrounding this shared variant. Single-nucleotide polymorphisms were selected if they were reported in dbSNP138, had a mapping quality >50 and coverage higher than × 10. Carriers of this variant shared a common 1.33-Mb haplotype (rs1107678-rs7196459), suggesting that p.(Ala510Val) derives from a single ancestral event (Supplementary Table S3).
Table 1. Clinical features and variants identified in French Canadian SPG7 cases.
| Individual ID | Family ID | Mutations | Age of onset | Age at exam | Urinary urgency | CPEO | Optic atrophy | Nystagmus | Dysarthria | Spasticity | Hyper-reflexia | Babinski | Ataxia | Dysmetria | Proximal weakness | Age documented proximal weakness | Cerebellar atrophy on MRI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | Ex 11c.1529C>T | 20 | 50 | − | − | − | + | ++ | ++ | + | + | ++ | + | − | ++ | |
| 2 | p.(Ala510Val) | 55 | 67 | − | − | − | − | + | + | + | + | + | − | LE | 67 | ++ | |
| 3 | Ex 17c.2249C>T | 40 | 57 | + | − | − | + | − | ++ | + | + | + | − | − | + | ||
| 4 | p.(Pro750Leu) | 32 | 44 | + | − | − | + | + | + | + | + | + | + | − | − | ||
| 5 | B | Ex 11c.1529C>T | 15 | 58 | + | − | − | ++ | ++ | ++ | + | ++ | + | − | ++ | ||
| 6 | p.(Ala510Val) | 43 | 66 | + | − | − | ++ | + | ++ | + | − | ++ | + | − | ++ | ||
| 7 | Ex 13c.1715C>T | 50 | 74 | + | − | − | − | ++ | ++ | + | − | ++ | + | − | ++ | ||
| 8 | p.(Ala572Val) | 45 | 68 | + | − | − | − | + | ++ | + | + | ++ | + | − | + | ||
| 9 | C | Ex 11c.1529C>T p.(Ala510Val) Ex 8c.988−1G>A | 25 | 43 | − | − | − | + | ++ | ++ | + | + | ++ | ++ | − | + | |
| 10 | D | Ex 11c.1529C>T p.(Ala510Val) Ex 13c.1715C>T p.(Ala572Val) | 28 | 61 | − | − | − | + | +++ | +++ | + | + | ++ | + | UE/LE | 61 | ++ |
| 11 | E | Ex 8c.1045G>A p.(Gly349Ser) Ex 17c.2249C>T | 32 | 48 | + | − | − | − | + | ++ | + | + | ++ | + | LE | 48 | − |
| 12 | p.(Pro750Leu) | 40 | 48 | − | − | − | + | ++ | + | + | + | ++ | + | − | NA | ||
| 13 | F | Ex 8c.1045G>A p.(Gly349Ser) Ex 17c.2249C>T p.(Pro750Leu) | 48 | 51 | + | − | − | + | + | + | + | + | ++ | + | − | + | |
| 14 | G | Ex 11c.1529C>T p.(Ala510Val) Ex 2c.233T>A p.(Leu78Ter) | 30 | 51 | + | − | − | + | ++ | ++ | + | + | ++ | ++ | − | 58 | ++ |
| 15 | H | Ex 11c.1529C>T p.(Ala510Val) (hmz) | 20 | 29 | − | − | − | + | + | + | + | + | ++ | + | − | − | |
| 16 | I | Ex 8c.988−1G>A Ex 17c.1715C>T | 25 | 45 | − | − | − | − | ++ | ++ | + | + | ++ | + | − | ++ | |
| 17 | p.(Ala572Val) | 37 | 43 | − | − | − | − | + | + | + | + | + | + | − | ++ | ||
| 18 | J | Ex 2c.233T>A p.(Leu78Ter) Ex 8c.988−1G>A | 25 | 33 | + | − | − | − | + | ++ | + | + | + | + | − | 41 | − |
| 19 | K | Ex 8c.988−1G>A | 40 | 45 | + | − | − | + | ++ | + | + | − | ++ | + | − | 54 | NA |
| 20 | (hmz) | 35 | 39 | + | − | − | + | + | ++ | + | + | + | + | − | NA | ||
| 21 | 39 | 44 | + | − | − | + | − | ++ | + | + | + | − | − | 52 | + | ||
| 22 | M | Ex 4c.473_474del p.(Leu158GlnfsTer30) Ex 8c.988−1G>A | 30 | 50 | + | − | + | + | ++ | ++ | + | + | + | + | − | NA | − |
Abbreviations: +, mild; ++ moderate; +++, severe; Ex, exon; CPEO, chronic external ophtalmoplegia; LE, lower extremities; MRI, magnetic resonance imaging; UE, upper extremities.
Exons are numbered according to reference NG_00808.2.1.
Novel variants are indicated in bold characters.
In addition, patients 1 and 5 each carried a different previously reported pathogenic missense variant on the second allele (c.2249C>T (p.(Pro750Leu)) and c.1715C>T (p.(Ala572Val)), respectively).5, 7 In patient 9, we found a novel intronic variant c.988-1G>A located one base pair before exon 8. This variant was absent from the 1000 Genomes and EVS databases and present at a frequency of 1.66e-05 in the Exome Aggregation Consortium (ExAC). In addition, it was found at a frequency of 2/2000 in our in-house exome database. Furthermore, it was predicted to be disease causing due to the loss of an acceptor splice site by MutationTaster. Sanger sequencing validated the presence of the corresponding variants in the three patients and co-segregation with disease status was confirmed in families A and B.
Identification of SPG7 variants in additional spastic ataxia patients
To identify additional patients with SPG7 variants, we screened 21 additional unrelated patients from our cohort of unresolved cases. Because of the genetic homogeneity observed in FC, we focused our analysis on SPG7 exons in which we had previously identified variants in patients. Strikingly, we uncovered rare variants in eight additional unrelated patients (38.1%, 8/21). We confirmed the presence of the identified variants in four affected relatives, which brought the total to 12 cases belonging to eight families (Table 1). In parallel, we also identified variants in SPG7 in subject 22 using a Sanger sequencing HSP panel. This included five missense variants that were previously reported to affect the protein function,5, 7, 10, 15, 22 and two novel variants (Table 1). In fact, five families (eight individuals) carried the novel splice site variant c.988-1G>A described above, making it the second most common variant in our cohort. Furthermore, patient 22 carried a novel variant (c.473_474del, p.(Leu158GlnfsTer30)), which is predicted to induce a frameshift and a premature stop codon at position 187 of SPG7. We confirmed co-segregation of all variants with the disease status in family members for which DNA was available. Thus, we uncovered rare homozygous or compound heterozygous variants in SPG7 in 38.7% of the families screened (12/31).
cDNA sequencing
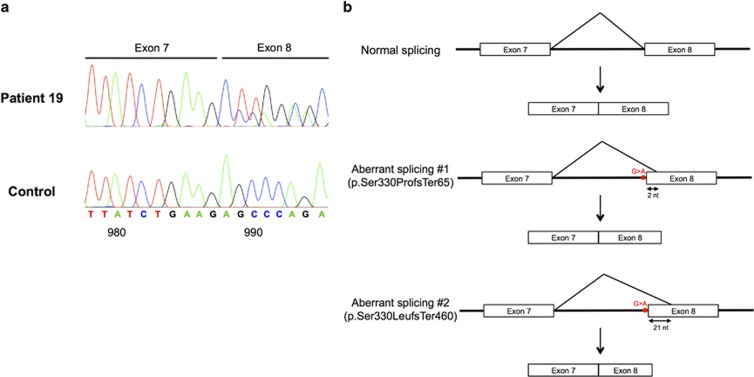
To confirm the pathogenicity of the splice site variant c.988-1G>A, we sequenced exons 7 and 8 of the cDNA of patient 19, who was homozygous for this variant. The results confirm that the substitution of G to A causes the loss of the acceptor splice site and leads to the use of two alternative cryptic acceptor splice sites within exon 8 (Figure 1). This leads to frameshifts and premature stop codons that likely produce two truncated SPG7 proteins (p.Ser330ProfsTer65 and p.Ser330LeufsTer460). Thus, these results strongly suggest that the SPG7 variant c.988-1G>A is indeed a mutation because it affects the function of the SPG7 protein.
Figure 1.
cDNA analysis of the splice site variant c.988-1G>A. (a) Complementary DNA sequence chromatograms are shown for a non-affected control and patient 19 who is homozygous for the c.988-1G>A variant. The G to A substitution leads to the loss of the acceptor splice site and the use of two alternate cryptic acceptor sites located within exon 8, causing the deletion of 2 and 21 nucleotides from the beginning of exon 8, respectively, as well as a frameshift and premature stop codon. (b) Schematic representation of normal splicing of SPG7 (upper panel) and aberrant splicing as seen in case 19 (middle and lower panel).
Clinical features of patients with SPG7 variants
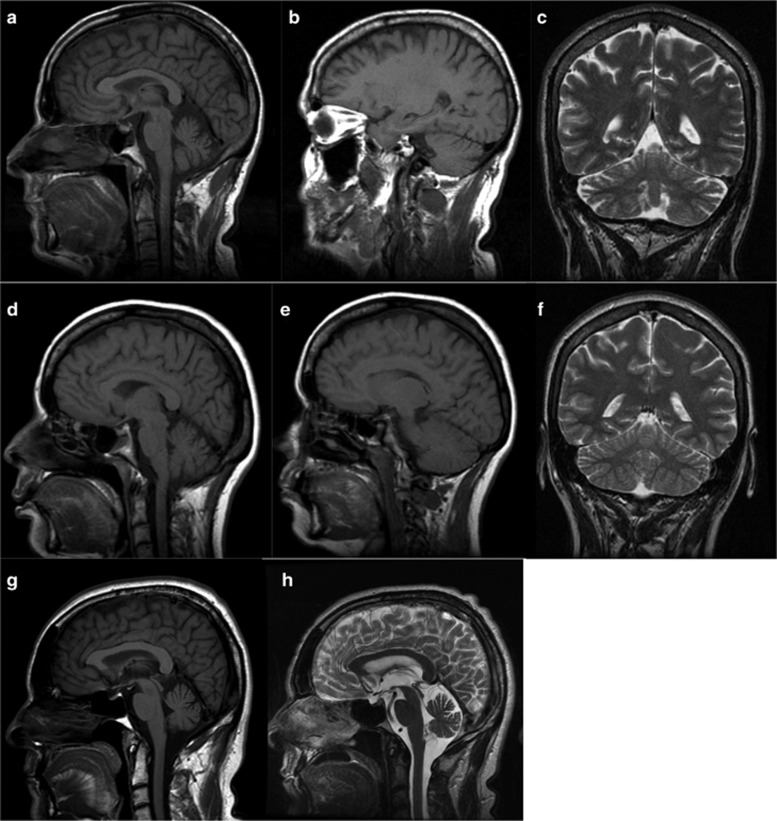
Despite the allelic heterogeneity, this FC cohort supports a more homogeneous core phenotype where spasticity and ataxia are both a constant feature (Table 1). The fact that 100% of cases presented with ataxic features explains why SPG7 was not screened initially in these patients. This cohort confirms the variable but generally adult age of onset (mean 34.2, 15–55) and the phenotypic intra-familial variability.15 Urinary urgency was a very common symptom that required medical treatment in many cases (14/22; 63.6%). Compared with other series, there was never any chronic external ophthalmoplegia documented even in the older cases, though clearly some pursuit difficulties above abnormal saccades and nystagmus (15/22; 68.1%) seem to appear with age.23 Only one patient demonstrated optic atrophy as has been reported in other cases.23, 24 Although proximal lower limb weakness was a rare finding at initial evaluation (3/22; 13.6%), it clearly develops with time (mean age 54.4, 7/21; 33.3%). Ambulatory loss appears to be exceedingly rare in patients. MRI data were available for 16 patients. Various degrees of cerebellar atrophy were present in the majority of them (14/16; 87.5%) and it was associated with mild supratentorial atrophy in a few cases (Table 1, Figure 2). Intra-familial variability was also observed on MRI, as cerebellar atrophy was moderate in the more ataxic siblings, but absent or milder in others with less ataxia (Table 1, Figure 2). Follow-up MRI was available for one case presenting mild cerebellar atrophy over a period of 7 years (age 51–58); subject 13 did not show marked progression of cerebellar atrophy despite clinical progression of his ataxia without loss of independent ambulation (Figure 2). However, more serial MRI data would be necessary to establish a correlation between the increasing ataxia and the cerebellar atrophy. The evolution of the gait difficulty in patients is clearly due to both the progression of the ataxia and the spasticity. Most patients were followed in a rehabilitation clinic on a yearly basis. Lioresal in doses of 10–90 mg per day in divided doses were given to many of the more spastic patients. In a few rare cases, Botox injections in the lower extremities were given. Urinary urgency was medically treated in the >50% of cases presenting this symptom.
Figure 2.
Brain MRI of FC cases with SPG7 variants. (a–f) Sagittal T1-weighted (a, b, d, e) and coronal T2-weighted (c, f) images show moderate atrophy of the cerebellar vermis and hemispheres in subject 2 at age 57 (a–c), whereas cerebellar atrophy is milder in subject 3 at age 51 (d–f). Sagittal T1- (g) and T2- (h) weighted images show mild cerebellar atrophy on the initial MRI of subject 13 at age 51 (g) without significant progression at the follow-up MRI at age 58.
Discussion
We report the identification of causative variants in SPG7 in 22 FC patients from 12 families affected with autosomal recessive spastic ataxia. This is only the second report of SPG7 mutations in FC patients,19 and the first report of a large cohort. These results strongly suggest that homozygous or compound heterozygous SPG7 variants explain a significant proportion of FC spastic ataxia cases (38.7% of families in our cohort). Sanger sequencing detected a slightly higher frequency of SPG7 cases (9/22, 40.9%) than WES (3/9, 33.3%). This may be due to a larger phenotypic heterogeneity in the patients sent for WES compared with the ones screened by Sanger sequencing. SPG7 should be considered in spastic ataxia patients lacking a genetic diagnosis. The SPG7 c.1529C>T (p.(Ala510Val)) variant was present in six families (12 patients), including one homozygote, making it the most frequent variant identified in our cohort. It was also found to be the most common SPG7 variant in several other populations.9, 15, 16, 25 Whereas it was first thought to be a benign variant, its pathogenicity was later demonstrated15, 22 and its presence in our FC spastic ataxia cohort supports this claim. In addition, this variant was present at a frequency slightly above 1% in our in-house exome database. This is significantly higher than public databases (1000 Genomes, EVS and ExAc), but is comparable to what has been reported in the literature for British, Spanish, Italian and German populations.9, 10, 15, 22 A threshold of one percent is frequently used for the filtering of recessive variants. However, ‘not so rare' variants have been identified as causing recessive diseases and this should be taken into account when searching for disease variants altering protein function. Stringent filtering parameters might prevent the uncovering of disease variants, as would have been the case in our SPG7 cohort. In addition to this more common variant, we identified two previously unreported SPG7 variants, including a novel splice site variant present in five families (eight patients), making it the second most frequent variant in our cohort. Despite the two more common variants in our cohort, the identification of seven distinct SPG7 variants supports the growing documentation that there is allelic heterogeneity in the FC founder population, even for rare diseases.26 Additional patients are currently being investigated for SPG7 mutations in a clinical setting, which may lead to the identification of more variants in the FC population. Furthermore, considering the recurrence of several variants in our cohort and the relatively high frequency of the p.(Ala510Val) variant in the FC population, our results highlight the importance of genetic counseling for SPG7 variant carriers.
Variants in three of the patients were identified through WES, again illustrating the efficiency of exome sequencing for the diagnosis of inherited ataxias, as it identified the genetic cause in patients for whom SPG7 would not have been considered based solely on clinical symptoms. This study, as well as more recent publications on SPG7 mutation carriers, supports that cerebellar ataxia is a very frequent feature.5, 6, 7, 9, 15 When present in milder adult-onset cases with spasticity, it suggests this diagnosis if not associated with cognitive deficit or peripheral neuropathy. In fact, cerebellar features, including ataxia, are more pronounced than spasticity in our cohort. This agrees with more recent literature suggesting that cerebellar ataxia can be the dominating clinical symptom in SPG7 and that SPG7 should be considered in the differential diagnosis of inherited cerebellar ataxia. In conclusion, we show for the first time that mutations in SPG7 are an important cause of autosomal recessive spastic ataxia in the FC population.
Acknowledgments
We thank the patients and their relatives who accepted to partake in this study. This project was financially supported by the Fondation Monaco. This work was selected for study by the Care4Rare (Enhanced Care for Rare Genetic Diseases in Canada) Consortium Gene Discovery Steering Committee: Kym Boycott (lead; University of Ottawa), Alex MacKenzie (co-lead; University of Ottawa), Jacek Majewski (McGill University), Michael Brudno (University of Toronto), Dennis Bulman (University of Ottawa), and David Dyment (University of Ottawa) and was funded in part by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec and the Children's Hospital of Eastern Ontario Foundation. The authors wish to acknowledge the contribution of the high-throughput sequencing platform of the McGill University and Génome Québec Innovation Centre, Montréal, Canada. KC and RLP received a Doctoral Award from the Fonds de recherche du Québec - Santé (FRQS). MT received a post-doctoral award from the Réseau de Médecine Génétique Appliquée and FRQS.
Web resources
EVS: https://evs.gs.washington.edu/EVS/. ExAC: Cambridge, MA, USA (http://exac.broadinstitute.org; March 2015). MutationTaster: www.mutationtaster.org
Accession codes
Gene: SPG7. Variants/accession numbers: c.1529C>T, p.(Ala510Val)/SCV000245719; c.2249C>T, p.(Pro750Leu)/SCV000245720; c.1715C>T, p.(Ala572Val)/SCV000245721; c.988-1G>A/SCV000245722; c.1045G>A, p.(Gly349Ser)/SCV000245723; c.233T>A, p.(Leu78Ter)/SCV000245724; c.473_474del, p.(Leu158QfsTer30)/SCV000245726.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Noreau A, Dion PA, Rouleau GA: Molecular aspects of hereditary spastic paraplegia. Exp Cell Res 2014; 325: 18–26. [DOI] [PubMed] [Google Scholar]
- Hersheson J, Haworth A, Houlden H: The inherited ataxias: genetic heterogeneity, mutation databases, and future directions in research and clinical diagnostics. Hum Mutat 2012; 33: 1324–1332. [DOI] [PubMed] [Google Scholar]
- Ruano L, Melo C, Silva MC, Coutinho P: The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology 2014; 42: 174–183. [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S et al: Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 1998; 93: 973–983. [DOI] [PubMed] [Google Scholar]
- Wilkinson PA, Crosby AH, Turner C et al: A clinical, genetic and biochemical study of SPG7 mutations in hereditary spastic paraplegia. Brain 2004; 127 (Pt 5): 973–980. [DOI] [PubMed] [Google Scholar]
- van Gassen KL, van der Heijden CD, de Bot ST et al: Genotype-phenotype correlations in spastic paraplegia type 7: a study in a large Dutch cohort. Brain 2012; 135 (Pt 10): 2994–3004. [DOI] [PubMed] [Google Scholar]
- Klebe S, Depienne C, Gerber S et al: Spastic paraplegia gene 7 in patients with spasticity and/or optic neuropathy. Brain 2012; 135 (Pt 10): 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KR, Blair NF, Vandebona H et al: Targeted next generation sequencing in SPAST-negative hereditary spastic paraplegia. J Neurol 2013; 260: 2516–2522. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ferrero E, Coto E, Beetz C et al: SPG7 mutational screening in spastic paraplegia patients supports a dominant effect for some mutations and a pathogenic role for p.A510V. Clin Genet 2013; 83: 257–262. [DOI] [PubMed] [Google Scholar]
- Arnoldi A, Tonelli A, Crippa F et al: A clinical, genetic, and biochemical characterization of SPG7 mutations in a large cohort of patients with hereditary spastic paraplegia. Hum Mutat 2008; 29: 522–531. [DOI] [PubMed] [Google Scholar]
- Doi H, Ohba C, Tsurusaki Y et al: Identification of a novel homozygous SPG7 mutation in a Japanese patient with spastic ataxia: making an efficient diagnosis using exome sequencing for autosomal recessive cerebellar ataxia and spastic paraplegia. Intern Med 2013; 52: 1629–1633. [DOI] [PubMed] [Google Scholar]
- Orsucci D, Petrucci L, Ienco EC et al: Hereditary spastic paraparesis in adults. A clinical and genetic perspective from Tuscany. Clin Neurol Neurosurg 2014; 120: 14–19. [DOI] [PubMed] [Google Scholar]
- Racis L, Tessa A, Di Fabio R et al: The high prevalence of hereditary spastic paraplegia in Sardinia, insular Italy. J Neurol 2014; 261: 52–59. [DOI] [PubMed] [Google Scholar]
- Elleuch N, Depienne C, Benomar A et al: Mutation analysis of the paraplegin gene (SPG7) in patients with hereditary spastic paraplegia. Neurology 2006; 66: 654–659. [DOI] [PubMed] [Google Scholar]
- Roxburgh RH, Marquis-Nicholson R, Ashton F et al: The p.Ala510Val mutation in the SPG7 (paraplegin) gene is the most common mutation causing adult onset neurogenetic disease in patients of British ancestry. J Neurol 2013; 260: 1286–1294. [DOI] [PubMed] [Google Scholar]
- Pfeffer G, Pyle A, Griffin H et al: SPG7 mutations are a common cause of undiagnosed ataxia. Neurology 2015; 84: 1174–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Warrenburg BP, van Gaalen J, Boesch S et al: EFNS/ENS Consensus on the diagnosis and management of chronic ataxias in adulthood. Eur J Neurol 2014; 21: 552–562. [DOI] [PubMed] [Google Scholar]
- Dupre N, Gros-Louis F, Bouchard JP, Noreau A, Roueau GA. SYNE1-Related autosomal recessive cerebellar ataxia. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH et al (eds): GeneReviews(R). University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Yoon G, Baskin B, Tarnopolsky M et al: Autosomal recessive hereditary spastic paraplegia-clinical and genetic characteristics of a well-defined cohort. Neurogenetics 2013; 14: 181–188. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H: ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn F, Pantakani K, Shoukier M, Langer T, Mannan AU: Functional evaluation of paraplegin mutations by a yeast complementation assay. Hum Mutat 2010; 31: 617–621. [DOI] [PubMed] [Google Scholar]
- Pfeffer G, Gorman GS, Griffin H et al: Mutations in the SPG7 gene cause chronic progressive external ophthalmoplegia through disordered mitochondrial DNA maintenance. Brain 2014; 137 (Pt 5): 1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotulli C, Leonardi L, Tessa A et al: Early-onset optic neuropathy as initial clinical presentation in SPG7. J Neurol 2014; 261: 1820–1821. [DOI] [PubMed] [Google Scholar]
- Brugman F, Scheffer H, Wokke JH et al: Paraplegin mutations in sporadic adult-onset upper motor neuron syndromes. Neurology 2008; 71: 1500–1505. [DOI] [PubMed] [Google Scholar]
- Thiffault I, Dicaire MJ, Tetreault M et al: Diversity of ARSACS mutations in French-Canadians. Can J Neurol Sci 2013; 40: 61–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.