Highlights
-
•
First report on Arginine deiminase production from Pseudomonas putida using RSM.
-
•
Optimum conditions for ADI production were established in shake flasks.
-
•
ADI production was assessed in a 14 L bioreactor under the optimized conditions.
-
•
Repressor effect of aeration on ADI production was observed in 14 L bioreactor.
-
•
Substantial improvement of 4.5-folds in ADI titre was achieved.
Keywords: Arginine deiminase, Enzyme production, Fermentation, Enzyme bioreactors, Pseudomonas putida KT2440, Submerged culture
Abstract
Statistically designed experiments were used to optimize the production of arginine deiminase (ADI) by Pseudomonas putida KT2440 in batch culture. A Plackett-Burman design involving eleven factors showed that ADI production was most influenced by the initial pH and the initial concentrations of glucose and yeast extract. A central composite experimental design showed that the optimal values of these factors were 8.0, 10 g/L and 12.5 g/L, respectively. The other components of the optimal culture medium were bacto peptone 7.5 g/L, Triton X–100 0.30% (v/v), and arginine 3 g/L, for a culture temperature of 25 °C. Compared with the basal medium, the ADI activity in the optimized medium had nearly 4.5-fold increase (4.31 U/mL). The optimized medium was then used for a further study of ADI production in a 14 L stirred tank bioreactor. The agitation speed and the aeration rates were varied to determine suitable values of these variables.
1. Introduction
Arginine deiminase (ADI) (EC 3.5.3.6) is a prokaryotic enzyme originally isolated from a Mycoplasma. ADI catalyses an irreversible deimination of the guanidine group of l-arginine to citrulline and ammonia [1]. ADI and various drugs based on it are potential treatments for certain cancers. PEGylated ADI (e.g. ADI-PEG20) has shown promising outcomes in the treatment of human malignancies [2]. ADI-PEG20, the PEGylated recombinant Mycoplasmal arginine deiminase, has been designated as an orphan drug by US Food and Drug Administration for the treatment of hepatocellular carcinomas and malignant melanomas. The European Agency for the Evaluation of Medicinal Products (EMEA) has also recognized ADI-PEG20 as an orphan drug for the treatment of hepatocellular carcinomas. Considerable interest exists in the use of ADI-mediated arginine deprivation therapy for various cancers [3], [4], [5] and ADI-PEG20 is in clinical trials for the treatment of certain cancers [6], [7].
In view of its anti-tumor potential, ADI from various bacterial genera has been characterized [8]. Biochemical and cellular responses to ADI are a focus of increasing attention [9]. Despite a strong interest in clinical use of ADI and its derivatives, very few studies have focused on the optimization of production of ADI to increase its titer in bacterial fermentation processes [10], [11], [12]. The present study attempts to address the fermentative production of ADI. Pseudomonas putida efficiently converts arginine to citrulline and has the highest capability of ADI production. Kakimoto et al. [10] have screened a large number (83 strains of bacteria, 31 strains of yeasts, 15 strains of molds and 15 strains of actinomycetes) of organisms and have found Pseudomonas putida as the best organism for ADI production. However, most of the studies till date are mainly focused upon Mycoplasmal arginine deiminase. Quite the opposite, far fewer studies have focused on the process optimization aspects of improving ADI yield from Pseudomonas sp.
Production of enzymes is profoundly affected by the composition of the fermentation medium and the culture conditions. Multiple nutritional and environmental factors interact to influence enzyme production. Therefore, statistical methods of fermentation optimization which take into account the interactive effects of the factors are superior to the conventional optimization approach in which a single factor is varied at any one time [13], [14].
This work reports on a two-step sequential optimization of production of ADI by batch culture of Pseudomonas putida KT2440. Initially, a Plackett-Burman screening strategy was used to identify the most influential factors affecting ADI production. In a second step, the optimum levels of the screened factors were identified using a central composite experiment design. Finally, ADI production was further assessed in a 14 L bioreactor under the optimized environmental and nutritional conditions.
2. Materials and methods
2.1. Chemicals
All chemicals were purchased from Hi-Media Laboratories (Mumbai, India). All reagents and solvents used were of analytical grade.
2.2. Microorganism and cultivation conditions
Pseudomonas putida KT2440 was a kind gift from Professor Manfred Zinn, Laboratory for Biomaterials, Empa-Swiss Federal Laboratories for Materials Science and Technology, Switzerland. The stock cultures were maintained on Luria Bertani (LB) medium and stored at −80 °C. Starter cultures were grown for 12 h at 37 °C in a 50 mL flask by suspending a loopful of the stock in 10 mL medium of the following composition (g/L): yeast extract 5, tryptone 10 and sodium chloride 10. After 12 h, 1 mL culture was transferred to a 250 mL flask containing 50 mL medium of the composition specified in the statistical design matrix.
2.3. Optimization of process parameters
The environmental and nutritional parameters were optimized to maximize the production of ADI and the biomass concentration. The following statistical approach was used. The initial screening of the variables potentially affecting ADI production was carried out by Plackett-Burman design. The selected factors were subjected to response surface methodology to determine their optimum values. All the submerged batch cultivations were performed in triplicate in shake flasks. The results are presented as the average values of three trials.
2.3.1. Plackett-Burman design
The Plackett-Burman design has been frequently used for the screening of process variables to improve production of enzymes. This design is based on a first-order model with no interaction among the factors [15]. To maximize growth and ADI production, the variable factors and their effective ranges were selected based on preliminary experiments and the literature data [10], [12], [16], [17]. For screening purpose, the following eleven variables were used: glucose, fructose, tryptone, bacteriological peptone, yeast extract, meat extract, Triton X–100, Tween-80, arginine, pH and temperature. Each of these factors was evaluated at two levels: −1 for a low level and +1 for a high level. Table 1 provides the ranges of the variables examined in the experimental design.
Table 1.
Range of values of the various factors used in Plackett–Burman design.
| Factor | Low actual (−) | High actual (+) |
|---|---|---|
| Glucose (g/L) | 5 | 20 |
| Fructose (g/L) | 5 | 20 |
| Tryptone (g/L) | 2.5 | 7.5 |
| Bacteriological peptone (g/L) | 2.5 | 7.5 |
| Yeast extract (g/L) | 1 | 4 |
| Meat extract (g/L) | 1 | 4 |
| Triton X–100 (% v/v) | 0.1 | 0.3 |
| Tween-80 (% v/v) | 0.1 | 0.3 |
| Arginine (g/L) | 1.0 | 5.0 |
| pH | 5 | 8 |
| Temperature (°C) | 25 | 37 |
The responses (ADI activity, biomass concentration) were measured in a total of 12 experiments designed using the software Design Expert (Stat-Ease Inc, USA; trial version 9). All experiments were carried out in 250 mL Erlenmeyer flasks containing 50 mL of the medium specified by the experimental design. All flasks were incubated at 200 rpm (Kühner incubator shaker; www.kuhner.com) at the specified temperature, and the responses were measured at 24, 30 and 36 h. These responses were subjected to a regression analysis to identify the most significant factors affecting the production of ADI. The optimal values of these selected factors were then determined using a central composite experiment design.
2.3.2. Response surface methodology
The response surface methodology was used to examine the dependence of the responses on the experimental factors [15]. The three selected factors, identified through screening experiments for use in the central composite design, were the initial concentration of glucose, the initial concentration of yeast extract, and pH. The ranges of values for the relevant factors are shown in Table 2.
Table 2.
Concentration ranges of the factors used in the central composite design.
| Factor | Actual levels of coded
factors |
||
|---|---|---|---|
| −1 | 0 | +1 | |
| Glucose (g/L) | 10 | 30 | 50 |
| Yeast extract (g/L) | 2.5 | 7.5 | 12.5 |
| pH | 5 | 6.5 | 8 |
To determine the optimum values of the selected three factors, a set of 20 experiments was generated using a 23 full-factorial central composite design with six replicates at the centre point. A second order polynomial model was used to fit the responses to the values of the factors. The levels of the factors in each experiment were as specified by the design software. The experiments were carried out in shake flasks as specified above. The following values were fixed at the levels shown: bacto peptone 7.5 g/L; Triton X–100 0.30% (v/v); arginine 3 g/L; the inoculum size 2.0% v/v; temperature 25 °C; and agitation rate 200 rpm. All experiments were in triplicate. Although, P-value of arginine was more significant than glucose, however, due to the negative β-coefficient value of arginine, it was used as a constant factor (3 g/L) in the optimization experiments. It may be mentioned that negative β-coefficient value depicts the detrimental effect on the response. Samples were withdrawn at 24, 30 and 36 h and used to measure the ADI activity and the biomass concentration. The dependent variables, or the responses, were the average ADI activity and the biomass concentration. The response data were fitted to a second order polynomial to generate the contour plots.
2.3.3. Validation of the model
An experiment was performed in triplicate, to verify the statistical model at the optimal values of the three factors (initial glucose concentration, initial yeast extract concentration, initial pH). The observed values of the ADI activity and the biomass concentration at 30 h of fermentation were compared with the model predicted values.
2.4. Production of ADI in a stirred-tank bioreactor
The bacterium was grown aseptically in a 14 L stirred-tank bioreactor (BioFlo 310; New Brunswick Scientific, USA) with a working volume of 7 L. The culture medium and the bioreactor were autoclaved (121 °C, 20 min) and cooled to 25 °C. The medium contained the optimized levels of glucose and yeast extract and the initial pH after autoclaving was adjusted to identified optimal value. The pH was measured in situ, but not controlled. The inoculum volume was always 700 mL, or 10% of the initial volume of the culture medium. The temperature was controlled at 25 °C. The initial concentrations of bacto peptone and arginine were always 7.5 and 3 g/L, respectively. The initial concentration of Triton X–100 was 0.30% (v/v), unless specified otherwise.
Agitation was provided by a Rushton turbine type agitator at the specified values in different experiments. Filter sterilized air was sparged through the culture at specified values for different runs. Foaming was controlled by automatic addition of sterile polypropylene glycol (Sigma-Aldrich; catalog no. 20,2339) as needed. Fermentations were run for 36 h. The broth was sampled every 4 h to measure biomass concentration, ADI activity and residual glucose concentration.
The inoculum used for the bioreactor studies had been prepared as follows. 100 μL stock suspension was transferred to 100 mL LB medium in a 500 mL shake flask and incubated overnight at 25 °C. A 25 mL portion of this overnight culture was transferred to a 1 L shake flask containing 225 mL of the above specified production medium and incubated at 25 °C, 200 rpm, for 12 h. A 700 mL portion of this culture from multiple shake flasks was used to inoculate the bioreactor.
2.5. Analytical methods
2.5.1. ADI activity
The ADI activity was determined using a modified method previously described by Ni et al. [18]. Briefly, the cell pellet from 1 mL culture broth was recovered by centrifugation at 7000g for 10 min. The cells were washed by resuspending it in 1 mL 50 mM phosphate buffer. The washed cells were recovered as above and suspended in 1 mL of cetyltrimethyl ammonium bromide (CTAB) (1 mg/mL) for 10 min. The treated cells were collected by centrifugation. The cells were then resuspended in 1 mL mixture containing 30 mM L-arginine and 200 mM sodium phosphate buffer (pH 6.0). The suspended cells were incubated at 37 °C for 30 min. This suspension was centrifuged (7000g, 5 min) to remove the cells. A 100 μL clear supernatant thus obtained was used to measure the ADI activity. The ADI activity was determined by measuring the production of l-citrulline from l-arginine. One unit of ADI activity was defined as the amount of enzyme that converted 1 μmol l-arginine to 1 μmol citrulline per min at 37 °C.
The amount of citrulline produced from l-arginine was estimated using the diacetyl monoxime thiosemicarbazide (DAM) method [19]. For this, a calibration curve for citrulline was produced by diluting a 3 mM stock solution of l-citrulline to various concentrations (0.25–3.00 mM). A 100 μL portion of the diluted l-citrulline solution was mixed with 3 mL of a chromogenic solution. The mixture was incubated at 95 °C for 5 min, cooled, and the spectrophotometric absorbance was measured at 530 nm against a blank of distilled water treated the same way as the l-citrulline standard solution. The measured absorbance was plotted against the known concentration of l-citrulline to obtain a calibration curve.
The chromogenic solution was prepared just before use by adding 5 mg thiosemicarbazide to 50 mL diacetyl monoxime solution (500 mg diacetyl monoxime was dissolved in 100 mL distilled water) and mixing in 100 mL acid-ferric solution. The latter had been made by adding 25 mL concentrated sulfuric acid and 20 mL concentrated phosphoric acid to 55 mL distilled water, cooling to room temperature, and dissolving 25 mg of ferric chloride.
2.5.2. Biomass concentration
The bacterial biomass in the fermentation broth was quantified by measuring optical density at 600 nm. Thus, a broth sample was suitably diluted with the fresh medium and the optical density was measured against a blank of the fresh medium. The measured optical density was converted to a dry cell weight concentration using a calibration curve. The calibration curve was made by measuring the optical density of a serially diluted broth sample having a precisely known biomass dry weight concentration. The latter was determined by recovering the cells from the broth by centrifugation at 7000g for 10 min in a 1.5 mL micro-centrifuge tube, washing twice by resuspending in distilled water, and drying the recovered cell pellet in an oven at 90 °C for 24 h.
2.5.3. Glucose concentration
Residual glucose concentration in the culture supernatant was determined using the 3,5-dinitro salicylic acid (DNS) method [20]. Briefly, 0.5 mL cell free culture supernatant was made up to 1.0 mL with distilled water and this solution was mixed with 1.5 mL 3,5-dinitro salicylic acid solution. The mixture was boiled for 5 min and made up to 5.0 mL with distilled water. The absorbance of this solution was read at 540 nm against a blank of distilled water treated the same way as the sample. The absorbance value was converted to a glucose concentration using a standard curve.
3. Results and discussion
3.1. Evaluation of the factors affecting ADI production
In screening experiments conducted according to a Plackett–Burman design, the response Y (i.e. the ADI activity in broth) was correlated with the values of the independent factors (Xi), as follows:
| (1) |
where, β0 is the model intercept and βi is the variable estimate.
Eleven factors were assessed each at two levels of high and low (Table 1). The factors were: pH, temperature, and concentrations of glucose, fructose, tryptone, bacteriological peptone, yeast extract, meat extract, Triton X–100, Tween-80 and arginine.
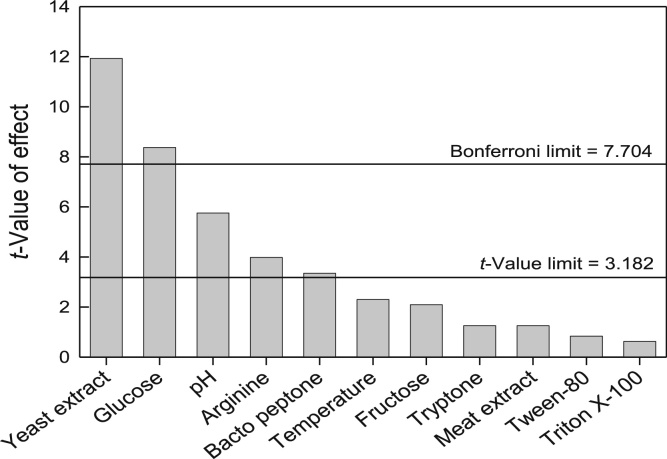
The results are shown in Table 3 for the 12 different trials. The response, or the ADI titer, varied widely (2.83–4.15 U/mL) for the different runs. The highest ADI activity (4.15 U/mL) occurred in run 6 while the lowest activity (2.83 U/mL) occurred in run 2 (Table 3). The relative effects of the factors on the ADI titer are summarized in the Pareto chart in Fig. 1. The bars in Fig. 1 show the magnitude of the influence of each factor on the response. The three most influential factors affecting the response were the concentrations of yeast extract and glucose and the pH (Fig. 1). In addition, the concentrations of arginine and bacteriological peptone had a significant influence on the ADI titer. All the other factors within the ranges tested, had no significant impact on the ADI titer.
Table 3.
The Placket–Burman experimental design matrix for the screening experiments.
| Run | Glucose (g/L) | Fructose(g/L) | Tryptone(g/L) | Bacto peptone (g/L) | Yeast extract (g/L) | Meat extract (g/L) | Triton X-100(% v/v) | Tween 80 (% v/v) | Arginine (g/L) | pH | Temperature (°C) | Biomass
(g/L) |
ADI activity
(U/mL) |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimental | Predicted | Experimental | Predicted | ||||||||||||
| 1 | 20 | 20 | 7.5 | 2.5 | 1 | 1 | 0.3 | 0.1 | 5 | 8 | 25 | 4.36 | 4.51 | 3.28 | 3.35 |
| 2 | 5 | 5 | 2.5 | 7.5 | 1 | 4 | 0.3 | 0.1 | 5 | 8 | 37 | 4.60 | 4.49 | 2.83 | 2.84 |
| 3 | 20 | 5 | 7.5 | 7.5 | 1 | 4 | 0.3 | 0.3 | 1 | 5 | 25 | 6.81 | 6.69 | 3.24 | 3.20 |
| 4 | 5 | 20 | 7.5 | 7.5 | 1 | 1 | 0.1 | 0.3 | 1 | 8 | 37 | 4.76 | 4.87 | 2.87 | 2.87 |
| 5 | 20 | 20 | 2.5 | 2.5 | 1 | 4 | 0.1 | 0.3 | 5 | 5 | 37 | 4.31 | 4.16 | 3.09 | 3.02 |
| 6 | 20 | 5 | 2.5 | 2.5 | 4 | 1 | 0.3 | 0.3 | 1 | 8 | 37 | 4.30 | 4.37 | 4.15 | 4.18 |
| 7 | 5 | 20 | 7.5 | 2.5 | 4 | 4 | 0.3 | 0.1 | 1 | 5 | 37 | 5.08 | 5.01 | 3.32 | 3.33 |
| 8 | 20 | 20 | 2.5 | 7.5 | 4 | 4 | 0.1 | 0.1 | 1 | 8 | 25 | 6.25 | 6.15 | 4.04 | 4.02 |
| 9 | 20 | 5 | 7.5 | 7.5 | 4 | 1 | 0.1 | 0.1 | 5 | 5 | 37 | 5.26 | 5.42 | 3.44 | 3.47 |
| 10 | 5 | 20 | 2.5 | 7.5 | 4 | 1 | 0.3 | 0.3 | 5 | 5 | 25 | 6.35 | 6.42 | 3.11 | 3.14 |
| 11 | 5 | 5 | 2.5 | 2.5 | 1 | 1 | 0.3 | 0.1 | 1 | 5 | 25 | 6.76 | 6.88 | 2.99 | 3.03 |
| 12 | 5 | 5 | 7.5 | 2.5 | 4 | 4 | 0.1 | 0.3 | 5 | 8 | 25 | 6.09 | 5.97 | 3.70 | 3.63 |
Fig. 1.
Relative effect of the different factors on ADI titer.
In other studies, arginine has been reported as an inducer of the ADI activity, particularly in Lactobacilli strains [16], [21], [22]. This induction effect of arginine has been observed also in Pseudomonas putida ATCC 4359 [10] and Pseudomonas plecoglossicida CGMCC2039 [12]. These observations are consistent with arginine being identified as an influential factor for ADI production in the screening experiments of this study.
ANOVA of screening experiments is shown in Table 4. The correlation coefficient (R) of the model (Eq. (1)) was 0.994, indicating a good agreement between the experimental data and the model-predicted values. The determination coefficient (R2) value was 0.989, indicating that nearly 99% of the variation in ADI titer could be attributed to the experimental factors. As the concentrations of yeast extract, concentration of glucose and the pH were the most influential factors, with confidence levels of 99.87%, 96.4% and 98.98%, respectively, they were selected for optimization by the central composite design.
Table 4.
Statistical analysis of Plackett–Burman design for ADI production.
| Variable | SS | DF | MS | F-ratio | P-value | β-coefficient |
|---|---|---|---|---|---|---|
| Glucose (g/L) | 0.49 | 1 | 0.49 | 69.49 | 0.036* | 0.20 |
| Fructose (g/L) | 0.033 | 1 | 0.033 | 4.70 | 0.1187 | −0.052 |
| Tryptone (g/L) | 0.012 | 1 | 0.012 | 1.66 | 0.2886 | −0.031 |
| Peptone (g/L) | 0.081 | 1 | 0.081 | 11.57 | 0.0424 | −0.082 |
| Yeast extract (g/L) | 1.00 | 1 | 1.00 | 142.35 | 0.0013* | 0.29 |
| Arginine (% w/v) | 0.12 | 1 | 0.12 | 16.57 | 0.0267 | −0.098 |
| pH | 0.24 | 1 | 0.24 | 33.62 | 0.0102* | 0.14 |
| Temperature (°C) | 0.037 | 1 | 0.037 | 5.33 | 0.1042 | −0.056 |
SS − Sum of squares.
DF − Degrees of freedom.
MS − Mean sum of squares.
3.2. Central composite design
The levels of the three variables, or factors, identified via screening experiments to have the strongest influence on ADI titre, were optimized through a central composite experiment design. The relevant factors were the initial concentrations of glucose and yeast extract and the culture pH. Each of these factors was assessed at three levels. In addition, the medium contained the following other components at the values (g/L) shown: bacto peptone 7.5 g/L, arginine 3 g/L, and Triton X–100 0.30% (v/v). The inoculum constituted 2% (v/v) of the culture volume. The agitation rate was 200 rpm and the temperature was maintained at 25 °C. A total of 20 runs were carried out with the values of the factors as shown in Table 5. The predicted and measured responses of the experiments are also shown in Table 5 and they were in good agreement.
Table 5.
Experimental setup for central composite design matrix.
| Run | Glucose (g/L) | Yeast extract (g/L) | pH | ADI activity
(U/mL) |
|
|---|---|---|---|---|---|
| Experimental | Predicted | ||||
| 1 | 30 | 7.5 | 6.5 | 3.42 | 3.13 |
| 2 | 30 | 7.5 | 5 | 3.61 | 3.11 |
| 3 | 30 | 7.5 | 6.5 | 2.70 | 3.13 |
| 4 | 30 | 7.5 | 6.5 | 2.51 | 3.13 |
| 5 | 10 | 2.5 | 5 | 2.59 | 2.53 |
| 6 | 10 | 12.5 | 8 | 4.26 | 4.61 |
| 7 | 50 | 2.5 | 5 | 1.74 | 1.64 |
| 8 | 10 | 2.5 | 8 | 2.27 | 2.55 |
| 9 | 50 | 2.5 | 8 | 1.44 | 1.67 |
| 10 | 50 | 12.5 | 5 | 3.61 | 3.70 |
| 11 | 30 | 7.5 | 8 | 3.77 | 3.14 |
| 12 | 30 | 2.5 | 6.5 | 2.41 | 2.10 |
| 13 | 30 | 7.5 | 6.5 | 2.98 | 3.13 |
| 14 | 30 | 12.5 | 6.5 | 4.37 | 4.16 |
| 15 | 50 | 7.5 | 6.5 | 2.14 | 2.69 |
| 16 | 30 | 7.5 | 6.5 | 2.92 | 3.13 |
| 17 | 10 | 7.5 | 6.5 | 4.17 | 3.57 |
| 18 | 50 | 12.5 | 8 | 4.23 | 3.73 |
| 19 | 10 | 12.5 | 5 | 4.28 | 4.59 |
| 20 | 30 | 7.5 | 6.5 | 3.15 | 3.13 |
The response measured at 30 h, was correlated with the values of the factors using the following equation:
| (2) |
In the above equation, Y1 is the response (or ADI titer, U/mL); A (g/L) is the concentration of glucose; B (g/L) is the concentration of yeast extract; and C is the pH.
The ADI titer did not change much after 30 h of fermentation. The maximum ADI titer of 4.37 U/mL was measured in run 14 and the corresponding predicted titer was 4.16 U/mL, or within ±5% of the measured value (Table 5). The ANOVA of the model (Eq. (2)) is shown in Table 6. The F-value, calculated as the ratio between the lack-of-fit mean square and pure error mean square, is a statistically valid measure of how well the factors describe the variation in the data about its mean. The greater the F-value from unity, the more certain is that the factors explain adequately the variation in the data about its mean and the estimated values are real [15]. The F-value for the present model (i.e. Eq. (2)) was 6.68 (Table 6), suggesting that the model was significant and the probability of the F-value of the model being due to experimental noise was only 0.29% (Table 6). The interactive effects were not significant.
Table 6.
Analysis of variance for response surface quadratic model.
| Source | SS | DF | MS | F- value | P-value Prob > F |
|---|---|---|---|---|---|
| Model | 13.21 | 9 | 1.47 | 6.68 | 0.0029 |
| Glucose (A) | 1.95 | 1 | 1.95 | 9.12 | 0.0129 |
| Yeast extract (B) | 10.61 | 1 | 10.61 | 49.74 | <0.0001 |
| pH (C) | 1.900 × 10−3 | 1 | 1.900 × 10−3 | 8.904 × 10−3 | 0.9267 |
| AB | 0.12 | 1 | 0.12 | 0.56 | 0.4727 |
| AC | 0.054 | 1 | 0.054 | 0.25 | 0.6252 |
| BC | 0.19 | 1 | 0.19 | 0.89 | 0.3686 |
| A2 | 0.19 | 1 | 0.19 | 0.87 | 0.3724 |
| B2 | 1.759 × 10−3 | 1 | 1.759 × 10−3 | 8.245 × 10−3 | 0.9294 |
| C2 | 0.21 | 1 | 0.21 | 0.96 | 0.3496 |
| Residual | 2.13 | 10 | 0.21 | ||
| Lack-of-fit | 1.61 | 5 | 0.32 | 3.11 | 0.1194 |
| Pure error | 0.52 | 5 | 0.10 | ||
| Cor. Total | 15.34 | 19 |
R2 = 0.8609; adjusted R2 = 0.7358; predicted R2 = 0.2640; adequate precision = 9.446; coefficient of variation (%) = 14.76.
The determination coefficient (R2) of the model (Eq. (2)) was 0.86; therefore, the model could explain 86% of the variation in the predicted responses of the ADI titer. The ‘adequate precision value’, an index of the signal-to-noise ratio, of Eq. (2) was 9.446 (Table 6), and this also suggested that the model can be used to navigate the design space. An adequate precision value of greater than 4 is a prerequisite for a model to be considered a good fit to the data. In view of Table 6, the concentration of yeast extract had the strongest influence on the ADI titer.
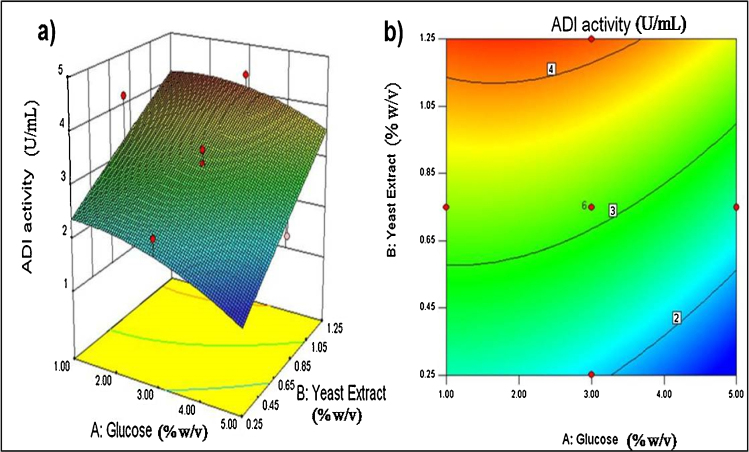
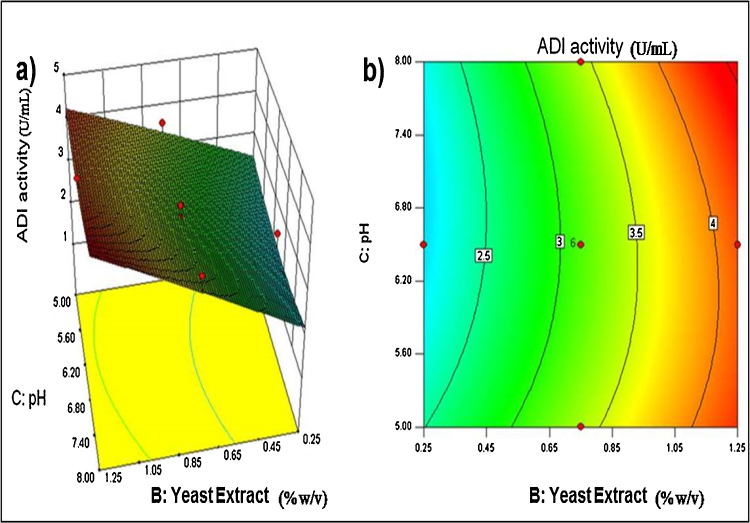
The response surface and contour plots generated using Eq. (2) are shown in Fig. 2, Fig. 3. Fig. 2 shows the effect of concentrations of glucose and yeast extract on ADI titer at a fixed pH 6.5. Fig. 3 shows the effect of the pH and yeast extract concentration on ADI titer at a fixed glucose concentration of 30 g/L. In other studies with various microorganisms [10], [12], [16], [17], peptone and tryptone have been reported as the best nitrogen sources for producing ADI. In the present work, the screening experiments revealed a significant effect of both yeast extract and bacto peptone on the production of the ADI. A linear increase in ADI titer with increasing concentration of yeast extract is seen in Fig. 3. Yeast extract has also been used in the production of ADI by various microorganisms including Pseudomonas putida [23], Streptococcus faecalis, Pseudomonas plecoglossicida CGMCC2039 [12], Enterococcus faecium [16], Pseudomonas sp. [17], Pseudomonas aeruginosa PAO [24], and different strains of the genera Micrococcus, Sarcina, Staphylococcus, Lactobacillus, Leuconostoc, Streptococcus and Mycobacterium [10]. Supplementation of yeast extract along with peptone to the medium used for the cultivation of Pseudomonas putida ATCC 4359 has been demonstrated to significantly improve ADI production up-to 9.2 U/mL [10]. Pseudomonas plecoglossicida CGMCC2039 cultivated in a 3 L stirred tank bioreactor in a complex medium containing yeast extract yielded a maximum ADI titer of 1.6 U/mL [12].
Fig. 2.
Response surface plot (a) and contour plot (b) showing the interactive effect of glucose and yeast extract concentration on the ADI titre at a fixed initial pH of 6.5.
Fig. 3.
Response surface plot (a) and contour plot (b) showing the interactive effect of pH and yeast extract concentration on the ADI titer at a fixed initial glucose concentration of 30 g/L.
The ADI titer was enhanced by increasing the concentration of glucose up to 30 g/L, but not higher (Fig. 2). This was likely because of a substrate inhibitory effect of glucose. The ADI titer increased linearly with increasing concentration of yeast extract right up to the maximum concentration tested (Fig. 2). The pH had a minimal effect on ADI titer at the pH levels tested, but a somewhat alkaline pH appeared to favour ADI production. Although PB design identified pH as an influential factor, CCD experiments did not show significant P-value for pH. It should be noted that PB design is based on the first-order model with no interaction among the factors, while CCD outcomes are more relevant regarding the interactive effects of the variables on responses. Non-consideration of the interactive effects between independent factors might be the plausible reason for the discrepancy in P-values as shown by PB design and CCD experiments.
In other work, the production of ADI has been reported to be affected by the type and concentration of sugars used in the culture medium [10], [16], [25], although the magnitude of the effect depends strongly on the microbial species. Glucose is a known repressor of the deiminase pathway in Pseudomonas aeruginosa [25]. In Pseudomonas putida ATCC 4359, an initial glucose concentration in excess of 20 g/L has been found to reduce the production of ADI [10]. Much lower concentrations have produced similar effect in other bacteria. For example, in Lactobacillus sake [26] and Lactobacillus sanfranciscensis CB1 [27], the ADI induction was suppressed when the initial glucose concentration exceeded 0.6 mM (∼0.1 g/L) and 54 mM (∼9.7 g/L), respectively. A maximum ADI titer of 2.17 U/mL was observed when a Pseudomonas sp. was grown on glucose [17]. In P. plecoglossicida CGMCC2039, use of glucose as a carbon source limited the peak ADI titre to only 1.6 U/mL [12]. In the present work, the contour plots (Fig. 2) revealed an initial glucose concentration in the range of 10 to 30 g/L as being satisfactory for maximizing the ADI production.
3.3. Validation of the statistical model
The statistical model (Eq. (2)) was validated at the combination of the optimum values of the three factors. The optimum point, i.e. the point of maximum ADI titer, was determined using the point prediction feature of the software. The optimal values calculated for the factors were: a glucose concentration of 10 g/L, a yeast extract concentration of 12.5 g/L and a pH of 8.0. A further shake flask experiment was carried out in triplicate, at the optimal combination of the factors and the ADI titer was measured at 30 h. The ADI titer of was found to be 4.31 ± 0.1 U/mL. This compared closely with the model predicted titer of 4.61 U/mL. The measured data and the predicted values agreed within ±7% of the measured value, thus validating the model.
Compared with the non-optimized conditions (i.e. a medium containing 10 g/L tryptone, 5 g/L yeast extract, 10 g/L sodium chloride; 1% (v/v) inoculum; an initial pH of 7.5; and an incubation temperature of 37 °C), the ADI titer increased from 0.96 to 4.31 U/mL under the optimized condition. This was a substantial 4.5-fold increase in the ADI titer.
3.4. Production of ADI in bioreactor
The culture conditions optimized in shake flasks were used to produce ADI in a 14 L stirred tank bioreactor. In a preliminary fermentation run, the aeration rate and the agitation speed were fixed at 0.36 vvm and 200 rpm, respectively. Although the aeration rate was relatively low, acute foaming occurred immediately after inoculation. Furthermore, the foaming could not be controlled despite the addition of large quantities of polypropylene glycol as an antifoaming agent. The intense foaming was attributed to the presence of the surfactant Triton X–100 in the culture medium. Therefore, in all the subsequent experiments in bioreactors, Triton X–100 was omitted from the medium. The aeration rate was set to 0.5 vvm, still relatively low for a small bioreactor, and the agitation speed was increased to 300 rpm to ensure a good oxygen supply.
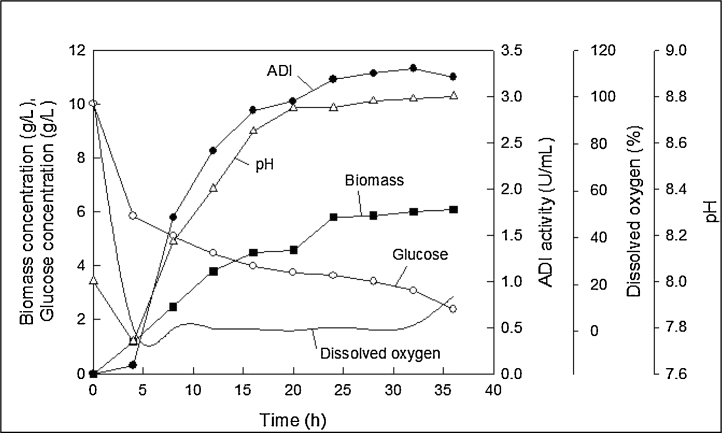
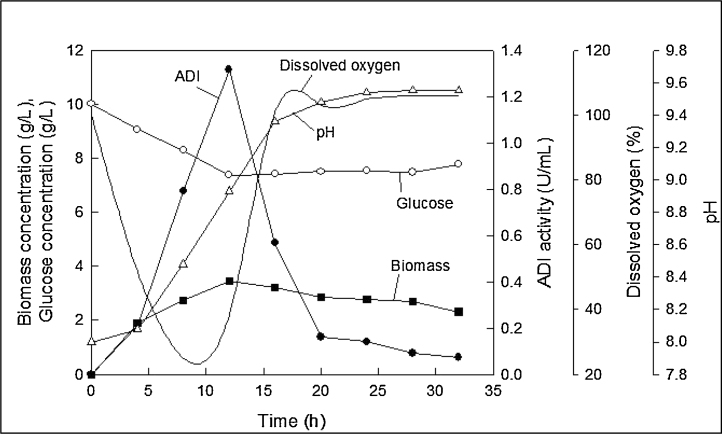
The fermentation profiles are shown in Fig. 4. The cells grew rapidly for the first 12 h and then gradually entered the stationary phase. A maximum biomass concentration of 6.1 g/L was attained after 36 h of fermentation. The pH dropped rapidly within about 3 h of inoculation, although the magnitude of the drop was small (Fig. 4). Subsequently, the pH rose and the rise almost in parallel with the biomass growth. The final pH was around 8.8 after 36 h. This rise in pH was attributed to the consumption of arginine and concomitant production of ammonia. The ADI titer increased in parallel with the biomass concentration, suggesting the enzyme production to be growth-associated. The maximum ADI titre of 3.3 U/mL was attained around 32 h (Fig. 4). The fermentation was likely to be oxygen limited, as during much of the growth phase the dissolved oxygen concentration was nearly zero (Fig. 4). Thus, all the oxygen being provided via aeration was being consumed and there was no accumulation in the culture broth (Fig. 4).
Fig. 4.
Fermentation profiles at an agitation speed of 300 rpm and an aeration rate of 0.5 vvm in a stirred tank bioreactor.
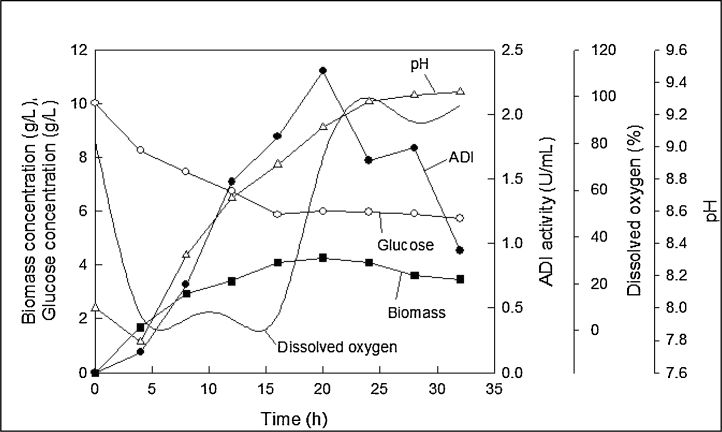
In view of the oxygen limitation of this fermentation (Fig. 4), a further fermentation was carried out at a higher agitation speed of 450 rpm in combination with an increased aeration rate of 1 vvm (7 L/min). All the other conditions were maintained as before. The results of this fermentation are shown in Fig. 5. The dissolved oxygen concentration declined rapidly during the phase of rapid growth and subsequently recovered as the growth slowed because of the consumption of nutrients. However, the dissolved oxygen concentration never declined to below 20% of the air saturation value (Fig. 5) and, therefore, there was always sufficient oxygen available. The biomass concentration and the ADI titre peaked at 12 h (Fig. 5), but were both quite low relative to the case of the earlier fermentation (Fig. 4). The peak ADI titre at 12 h was only 1.31 U/mL. The pH profile of the fermentation (Fig. 5) was essentially the same as discussed earlier for Fig. 4. The final pH at 36 h was nearly 9.3.
Fig. 5.
Fermentation profiles at an agitation speed of 450 rpm and an aeration rate of 1 vvm in a stirred tank bioreactor.
As intense agitation and aeration did not improve the biomass concentration and the ADI titer, despite an excess of oxygen (Fig. 5), a further fermentation (Fig. 6) was carried out with the agitation speed set to 350 rpm and the aeration rate set to 0.75 vvm. In this fermentation, the dissolved oxygen concentration did fall to nearly zero (Fig. 6), but the duration of this oxygen limited period was much shorter than in the profiles shown in Fig. 4. The peak biomass concentration and the ADI titer were 4.3 g/L and 2.3 U/mL, respectively. To our surprise, despite a low initial concentration of glucose of only 10 g/L, glucose was not totally consumed in any of the fermentations (Fig. 4, Fig. 5, Fig. 6). The oxygen limited fermentation (Fig. 4), nearly 76% of the glucose was consumed, but in the oxygen-sufficient fermentation (Fig. 5), the consumption was only 23%. At intermediate levels of oxygen supply (Fig. 6), nearly 43% of the initial glucose was consumed. Incomplete glucose consumption despite an excess of nitrogen source, was likely a consequence of the increasing pH interfering with the cell growth.
Fig. 6.
Fermentation profiles at an agitation speed of 350 rpm and an aeration rate of 0.75 vvm in a stirred tank bioreactor.
Studies in the bioreactor showed that excessive agitation and aeration adversely affected the ADI titer in Pseudomonas putida KT2440. In Pseudomonas aeruginosa a limitation of oxygen has been previously shown to induce ADI production [25]. The transcriptional regulator ANR (for anaerobic regulation of arginine catabolism and nitrate reduction) has been shown to mediate ADI induction by oxygen limitation [28], [29], [30]. Aeration has been found to repress the ADI in other bacteria such as Streptococcus sanguis and Streptococcus rattus [31].
4. Conclusion
The optimal conditions for the production of ADI in a batch shake flask fermentation were as follows: an initial pH of 8.0; 25 °C; an agitation rate of 200 rpm; an inoculum size of 2% (v/v) of the initial culture volume; and the following concentrations of the components in the medium—glucose 10 g/L, yeast extract 12.5 g/L, peptone 7.5 g/L, arginine 3 g/L, and Triton X–100 0.30% (v/v). With these conditions, the peak titer of ADI was 4.31 U/mL. Optimization of the fermentation conditions enhanced the ADI titre by 4.5-fold relative to the nonoptimzied base case. Bioreactor fermentations did not improve the ADI titer relative to the best-case shake flasks apparently because of the repressory effect of oxygen on the production of ADI.
Acknowledgments
MP and GP gratefully acknowledge Department of Biotechnology (DBT), New Delhi, India for the award of Senior Research Fellowship. Authors are thankful to Mr. Mukesh Kumar for the technical assistance provided to carry out bioreactor runs. We are also thankful to Dr. Jayeeta Bhaumik for editorial assistance.
References
- 1.Shirai H., Blundell T.L., Mizuguchi K. A novel superfamily of enzymes that catalyse the modification of guanidine groups. Trends Biochem. Sci. 2001;26:465–468. doi: 10.1016/s0968-0004(01)01906-5. [DOI] [PubMed] [Google Scholar]
- 2.Chiaviello A., Paciello I., Veneziani B.M., Palumbo G., Aloj S.M. Cells derived from normal or cancer breast tissue exhibit different growth properties when deprived of arginine. Med. Oncol. 2012;29:2543–2551. doi: 10.1007/s12032-011-0130-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L., Liu M., Jamil S., Han R., Xu G., Ni Y. PEGylation and pharmacological characterization of a potential anti-tumor drug, an engineered arginine deiminase originated from Pseudomonas plecoglossicida. Cancer Lett. 2015;357:346–354. doi: 10.1016/j.canlet.2014.11.042. [DOI] [PubMed] [Google Scholar]
- 4.Fayura L.R., Boretsk Y.R., Pynyaha Y.V., Wheatley D.N., Sibirny A.A. Improved method for expression andisolation of the Mycoplasma hominis arginine deiminase fromthe recombinant strain of Escherichia coli. J. Biotechnol. 2013;167:420–426. doi: 10.1016/j.jbiotec.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 5.Ahn K.-Y., Lee B., Han K.-Y., Song J.-A., Lee J. Synthesis of Mycoplasma arginine deiminase in E. coli using stress-responsive proteins. Enzyme Microbiol. Technol. 2014;63:46–49. doi: 10.1016/j.enzmictec.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Ott P.A., Carvajal R.D., Pandit-Taskar N., Jungbluth A.A., Hoffman E.W., Wu B.W., Bomalaski J.S., Venhaus R., Pan L.D., Old L.J. Phase I/II study of pegylated arginine deiminase (ADI-PEG 20) in patients with advanced melanoma. Invest. New Drugs. 2013;31:425–434. doi: 10.1007/s10637-012-9862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Synakiewicz A., Stachowicz-Stencel T., Adamkiewicz-Drozynska E. The role of arginine and the modified arginine deiminase enzyme ADI-PEG 20 in cancer therapy with special emphasis on phase I/II clinical trials. Expert Opin. Investig. Drugs. 2014;23:1–13. doi: 10.1517/13543784.2014.934808. [DOI] [PubMed] [Google Scholar]
- 8.Ni Y., Schwaneberg U., Sun Z.H. Arginine deiminase, a potential anti-tumor drug. Cancer Lett. 2008;261:1–11. doi: 10.1016/j.canlet.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 9.Szlosarek P.W., Luong P., Phillips M.M., Baccarini M., Ellis S., Szyszko T., Sheaff M.T., Avril N. Metabolic response to pegylated arginine deiminase in mesothelioma with promoter methylation of argininosuccinate synthetase. J. Clin. Oncol. 2013;31:e111–e113. doi: 10.1200/JCO.2012.42.1784. [DOI] [PubMed] [Google Scholar]
- 10.Kakimoto T., Shibatani T., Nishimura N., Chibata I. Enzymatic production of l-citrulline by Pseudomonas putida. Appl. Microbiol. 1971;22:992–999. doi: 10.1128/am.22.6.992-999.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaur B., Kaur R. Statistical screening of media components for the production of arginine deiminase by Weissella confuse GR7. Int. J. Food Ferment. Technol. 2012;2:81–89. [Google Scholar]
- 12.Liu Y.-M., Sun Z.-H., Ni Y., Zheng P., Liu Y.-P., Meng F.-J. Isolation and identification of an arginine deiminase producing strain Pseudomonas plecoglossicida CGMCC2039. World J. Microbiol. Biotechnol. 2008;24:2213–2219. [Google Scholar]
- 13.Ghevariya C.M., Bhatt J.K., Dave B.P. Enhanced chrysene degradation by halotolerant Achromobacter xylosoxidans using response surface methodology. Bioresour. Technol. 2011;102:9668–9674. doi: 10.1016/j.biortech.2011.07.069. [DOI] [PubMed] [Google Scholar]
- 14.Dubey S., Singh A., Banerjee U.C. Response surface methodology of nitrilase production by recombinant Escherichia coli. Braz. J. Microbiol. 2011;42:1085–1092. doi: 10.1590/S1517-838220110003000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar R., Mahajan S., Kumar A., Singh D. Identification of variables and value optimization for optimum lipase production by Bacillus pumilis RK31 using statistical methodology. New Biotechnol. 2011;28:65–71. doi: 10.1016/j.nbt.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Kaur B., Kaur R. Application of response surface methodology for optimizing arginine deiminase production medium for Enterococcus faecium sp. GR7. Sci. World J. 2013 doi: 10.1155/2013/892587. (article ID 892587.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng F.-J., Liu Y.-M., Sun Z.-H., Ni Y., Zheng P. Preparation of arginine deiminase by Pseudomonas sp. Chin. J. Pharm. 2008 http://en.cnki.com.cn/Article_en/CJFDTOTAL-ZHOU200807014.htm (Abstract only) [Google Scholar]
- 18.Ni Y., Liu Y., Schwaneberg U., Zhu L., Li N., Li L., Sun Z. Rapid evolution of arginine deiminase for improved anti-tumor activity. Appl. Microbiol. Biotechnol. 2011;90:193–201. doi: 10.1007/s00253-010-3051-z. [DOI] [PubMed] [Google Scholar]
- 19.Boyde T.R.C., Rahmatullah M. Optimization of conditions for the calorimetric determination of citrulline using diacetyl monoxime. Anal. Biochem. 1980;107:424–431. doi: 10.1016/0003-2697(80)90404-2. [DOI] [PubMed] [Google Scholar]
- 20.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–427. [Google Scholar]
- 21.Yu J.J., Oh S.H. Isolation and characterization of lactic acid bacteria strains with ornithine producing capacity from natural sea salt. J. Microbiol. 2010;48:467–472. doi: 10.1007/s12275-010-0204-9. [DOI] [PubMed] [Google Scholar]
- 22.de Orduňa R.M., Liu S.Q., Patchett M.L., Pilone G.J. Ethyl carbamate precursor citrulline formation from arginine degradation by malolactic wine lactic acid bacteria. FEMS Microbiol. Lett. 2000;183:31–35. doi: 10.1111/j.1574-6968.2000.tb08929.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto K., Sato T., Tosa T., Chibata I. Continuous production of l-citrulline by immobilized Pseudomonas putida cells. Biotechnol. Bioeng. 1974;16:1589–1599. doi: 10.1002/bit.260161203. [DOI] [PubMed] [Google Scholar]
- 24.Mercenier A., Stalon V., Simon J.-P., Haas D. Mapping of the arginine deiminase gene in Pseudomonas aeruginosa. J. Bacteriol. 1982;149:787–788. doi: 10.1128/jb.149.2.787-788.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercenier A., Simon J.-P., Wauven C.V., Haas D., Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J. Bacteriol. 1980;144:159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montel M.-C., Champomier M.-C. Arginine catabolism in Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 1987;53:2683–2685. doi: 10.1128/aem.53.11.2683-2685.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelis M.D., Mariotti L., Rossi J., Servili M., Fox P.F., Rollán G., Gobbetti M. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 2002;68:6193–6201. doi: 10.1128/AEM.68.12.6193-6201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C.-D., Winteler H., Abdelal A., Haas D. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 1999;181:2459–2464. doi: 10.1128/jb.181.8.2459-2464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galimand M., Gamper M., Zimmermann A., Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamper M., Zimmermann A., Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 1991;173:4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burne R.A., Parsons D.T., Marquis R.E. Environmental variables affecting arginine deiminase expression in oral Streptococci. In: Dunny P.P., Cleary G.M., McKay L.L., editors. Genetics and Molecular Biology of Streptococci, Lactococci and Enterococci. American Society for Microbiology; Washington, DC: 1991. pp. 276–280. [Google Scholar]