Abstract
Alveolar epithelial cell (AEC) dysfunction underlies the pathogenesis of pulmonary fibrosis in Hermansky-Pudlak syndrome (HPS) and other genetic syndromes associated with interstitial lung disease; however, mechanisms linking AEC dysfunction and fibrotic remodeling are incompletely understood. Since increased macrophage recruitment precedes pulmonary fibrosis in HPS, we investigated whether crosstalk between AECs and macrophages determines fibrotic susceptibility. We found that AECs from HPS mice produce excessive MCP-1, which was associated with increased macrophages in the lungs of unchallenged HPS mice. Blocking MCP-1/CCR2 signaling in HPS mice with genetic deficiency of CCR2 or targeted deletion of MCP-1 in AECs normalized macrophage recruitment, decreased AEC apoptosis, and reduced lung fibrosis in these mice following treatment with low-dose bleomycin. We observed increased TGF-β production by HPS macrophages, which was eliminated by CCR2 deletion. Selective deletion of TGF-β in myeloid cells or of TGF-β signaling in AECs through deletion of TGFBR2 protected HPS mice from AEC apoptosis and bleomycin-induced fibrosis. Together, these data reveal a feedback loop in which increased MCP-1 production by dysfunctional AECs results in recruitment and activation of lung macrophages that produce TGF-β, thus amplifying the fibrotic cascade through AEC apoptosis and stimulation of fibrotic remodeling.
Interactions between alveolar epithelial cells and macrophages determine fibrotic susceptibility in Hermansky-Pudlak syndrome through a MCP-1/CCR2/TGF-β feedback cycle.
Introduction
Hermansky-Pudlak syndrome (HPS), the most penetrant of the genetic pulmonary fibrosis syndromes, provides a compelling paradigm for studying the cellular pathogenesis of pulmonary fibrosis. Recessive mutations in ubiquitously expressed HPS genes result in dysfunction of key hetero-oligomeric intracellular protein trafficking complexes, leading to clinical manifestations that include oculocutaneous albinism and a bleeding diathesis (1). To date, 10 genetic loci have been associated with HPS in humans, and pulmonary fibrosis has been reported in several of these subtypes, including HPS1 and HPS2 (1–5). Naturally occurring mutations in HPS mice reliably model important features of the human disease, including susceptibility to profibrotic stimuli, which is restricted to the same HPS subtypes that are associated with pulmonary fibrosis in humans (6–8). We have demonstrated that intrinsic defects in the alveolar epithelium due to HPS mutations underlie the fibrotic susceptibility in HPS mice. Specifically, epithelial-specific transgenic correction of the β1 subunit of adaptor protein 3 (AP-3) in HPS2 mice protected the animals from bleomycin-induced alveolar epithelial cell (AEC) apoptosis and fibrosis (7). However, the mechanisms by which dysfunctional AECs facilitate lung fibrosis following injury remain incompletely defined.
Compelling data from HPS patients demonstrate that lung inflammation precedes pulmonary fibrosis (9). HPS patients with preserved lung function have increased numbers of macrophages in bronchoalveolar lavage (BAL), and macrophage alveolitis has been described in lung histology from HPS patients (9). Although alveolar macrophages have been implicated in lung fibrosis, macrophages may alternately promote fibrosis or resolution at different times after injury (10–18). We postulated that aberrant signaling between AECs and lung macrophages contributes to the marked sensitivity to low-dose bleomycin in susceptible HPS mice and provides a mechanistic link between AEC dysfunction and subsequent development of lung fibrosis. Our studies show that increased production of monocyte chemotactic protein-1 (MCP-1; also known as C-C motif ligand 2 [CCL2]) by AECs from HPS mice regulates macrophage recruitment and TGF-β production via CCR2. Macrophage-derived TGF-β then drives a feedback cycle promoting AEC apoptosis and further MCP-1 production. These studies provide evidence for epithelial-macrophage interactions that impact development of lung fibrosis.
Results
HPS AECs produce excess MCP-1.
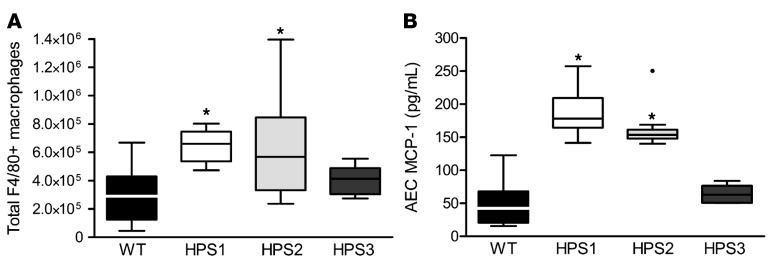
By flow cytometry, we found that numbers of F4/80+ macrophages were increased in the lungs of unchallenged HPS1 and HPS2 mice compared with WT controls and HPS3 mice (which do not have increased susceptibility to bleomycin-induced fibrosis) (Figure 1A), suggesting that increased numbers of macrophages are associated with the propensity to develop fibrosis in HPS. In contrast, the numbers of dendritic cells, granulocytes, and CD4+ and CD8+ T lymphocytes were similar in the lungs of HPS and WT mice (data not shown). We next examined potential mechanisms for excess recruitment of macrophages in the lungs of HPS mice. We isolated primary type II AECs from unchallenged HPS and WT mice and evaluated production of monocyte/macrophage chemoattractants as well as other cytokines to identify factors that were elevated in conditioned media from AECs from both HPS1 and HPS2, but not fibrosis-resistant HPS3, mice. Primary type II AECs from unchallenged HPS1 and HPS2 mice secreted excess MCP-1 compared with WT and HPS3 AECs (Figure 1B). Levels of MCP-1 were reduced in a HPS2 transgenic model with AEC-specific correction of AP-3 expression (7). In contrast, similar levels of other monocyte/macrophage chemoattractants, including MCP-3, MCP-5, granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage CSF (M-CSF), RANTES, and MIP1α, were produced by WT and HPS AECs (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.88947DS1). Together, these findings from HPS mice were consistent with those of prior reports demonstrating increased concentration of MCP-1 and increased macrophages in BAL from HPS patients (9) and suggested that epithelial-derived MCP-1 could be responsible for recruitment of macrophages to the lungs in HPS.
Figure 1. Increased MCP-1 production by type II alveolar epithelial cells correlates with excess macrophage accumulation in the lungs of HPS1 and HPS2 mice.
(A) F4/80+ macrophages from lungs of unchallenged mice were quantitated by flow cytometry with gating on CD45+ viable cells. Data are presented as box-and-whisker Tukey plots; n = 14 WT, n = 6 HPS1, n = 9 HPS2, and n = 5 HPS3. Comparison between groups was conducted by ANOVA, *P < 0.01 vs. WT and HPS3 mice. (B) MCP-1 was measured by ELISA in cell culture media from type II alveolar epithelial cells (AECs) from unchallenged mice after 24 hours in culture; n = 15 WT, n = 6 HPS1, n = 9 HPS2, and n = 6 HPS3. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.001 vs. WT and HPS3.
The MCP-1/CCR2 pathway regulates macrophage recruitment and fibrotic susceptibility in HPS mice.
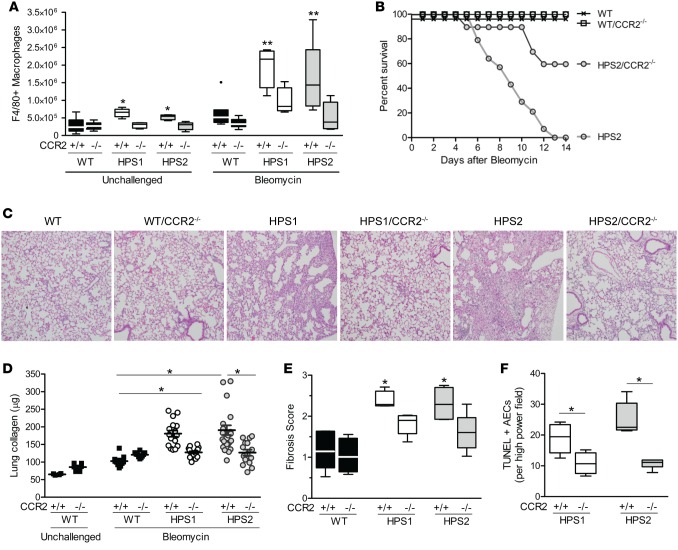
To determine whether MCP-1 contributes to macrophage recruitment and fibrosis in HPS mice in vivo, we crossed mice that were globally deficient for the MCP-1 receptor, CCR2 (CCR2–/–, C57BL/6J background), with HPS1 or HPS2 mice. The increased recruitment of F4/80+ myeloid cells observed in the lungs of unchallenged HPS1 and HPS2 mice was blocked in both HPS1/CCR2–/– and HPS2/CCR2–/– mice (Figure 2A). Further examination of myeloid subpopulations in HPS mice indicated that the subset of interstitial macrophages (identified by F4/80lo, CD11clo, CD11b+, Gr1–) was selectively increased in unchallenged HPS mice compared with WT controls, while no differences were observed in the numbers of alveolar macrophages (F4/80+, CD11c+, CD11b–, Gr1–) or monocytes (F4/80lo, CD11c–, CD11b+, Gr1lo) (Supplemental Figure 2). CCR2 deficiency reduced the number of interstitial macrophages in unchallenged HPS mice to levels similar to those of WT controls (with or without CCR2 deletion). In contrast, CCR2 deletion did not affect the population of alveolar macrophages and similarly reduced monocytes in the lungs of HPS and WT strains.
Figure 2. CCR2 regulates increased recruitment of macrophages and exaggerated fibrotic susceptibility in HPS mice.
HPS1 or HPS2 mutant mice were bred to homozygosity with global CCR2-deficient (CCR2–/–) mice. (A) Numbers of macrophages in unchallenged and bleomycin-treated mice (day 7) were quantified by flow cytometry using the F4/80 marker. Data are presented as box-and-whisker Tukey plots. For unchallenged groups, n = 13 WT, n = 9 WT/CCR2–/–, n = 6 HPS1, n = 4 HPS1/CCR2–/–, n = 4 HPS2, and n = 10 HPS2/CCR2–/–. For bleomycin-challenged groups, n = 7 WT and WT/CCR2–/–, n = 4 HPS1 and HPS1/CCR2–/–, n = 10 HPS2, and n = 6 HPS2/CCR2–/–. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.05 vs. unchallenged WT and CCR2–/– groups, **P < 0.05 vs. bleomycin-challenged WT and CCR2–/– groups. (B) Survival after IT bleomycin; n = 8/group WT and WT/CCR2–/–, n = 14 HPS2, and n = 10 HPS2/CCR2–/–. P < 0.05 for HPS2/CCR2–/– vs. other groups by log-rank test. (C) Histology of lung sections from mice 7 days after bleomycin challenge. Representative H&E images; original magnification, ×10. (D) Lung collagen content of left lung quantified by Sircol assay in unchallenged mice or 7 days after bleomycin challenge (mean ± SEM). For unchallenged groups, n = 4 WT and n = 6 WT/CCR2–/–. For bleomycin-challenged groups, n = 11 WT, n = 10 WT/CCR2–/–, n = 17 HPS1, n = 19 HPS1/CCR2–/–, n = 24 HPS2, and n = 15 HPS2/CCR2–/–. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.001 for HPS2/CCR2+/+ and HPS1/CCR2+/+ vs. WT and CCR2–/– groups. (E) Modified Ashcroft fibrosis score from trichrome-stained lung sections. Data are presented as box-and-whisker Tukey plots; n = 4/group, except n = 5 HPS1/CCR2–/– and HPS2, and n = 6 HPS2/CCR2–/–, with comparisons between groups conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. WT and CCR2–/– groups. (F) TUNEL+ alveolar epithelial cells (AECs) in lung sections from mice 24 hours after bleomycin challenge; n = 6 HPS1, n = 5 HPS2, n = 10 HPS1/CCR2–/–, and n = 7 HPS2/CCR2–/–. Comparison with CCR2–/– groups was assessed using Mann-Whitney U analysis, *P < 0.01.
In CCR2-deficient HPS mice, we observed a reduction in F4/80+ macrophages after intratracheal (IT) bleomycin treatment compared with the matched HPS strain (Figure 2A). In addition, CCR2 deficiency was associated with significantly improved survival in response to bleomycin treatment in HPS2 mice (Figure 2B), which exhibit accelerated fibrotic susceptibility and high mortality following low-dose bleomycin challenge (6, 7). Consistent with mortality data, CCR2-deficient HPS mice exhibited a marked reduction in fibrosis compared with HPS mice at 7 days after bleomycin treatment, as measured by collagen content assay and histologic assessment of lung sections (Figure 2, C–E). In contrast, CCR2 deficiency did not alter the mild fibrotic response of WT mice to low-dose bleomycin. Since AEC apoptosis has been linked to subsequent fibrosis in bleomycin-treated mice (6, 7), we quantified TUNEL+ AECs and found that apoptotic AECs were reduced in CCR2-deficient HPS1 and HPS2 mice compared with HPS1 and HPS2 mice at 24 hours after bleomycin challenge (Figure 2F).
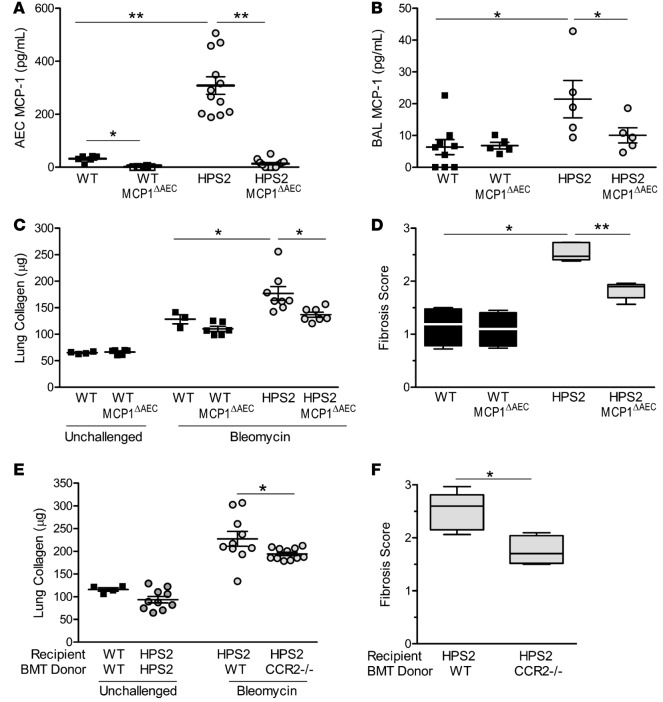
Since multiple cell types can produce MCP-1 and express CCR2, we performed experiments to directly test whether the lung epithelium was the relevant cellular source of MCP-1 in HPS mice in vivo. For these studies, we used the SPC.Cre promoter to generate a model in which MCP-1 was deleted specifically in epithelial cells (MCP1ΔAEC), resulting in reduced MCP-1 in the BAL of HPS mice (Figure 3, A and B). Following bleomycin treatment, HPS2/MCP1ΔAEC mice had reduced lung collagen content and improved fibrosis scores compared with HPS2 mice, thus supporting the idea that the epithelial MCP-1/myeloid CCR2 axis regulates fibrosis in HPS mice (Figure 3, C and D). This conclusion was further supported by bone marrow transplantation experiments using CCR2–/– and WT mice as donors and HPS2 mice as recipients. Compared with HPS mice that were transplanted with WT marrow, HPS mice that received CCR2–/– marrow had significantly reduced lung collagen content and fibrosis scores after bleomycin challenge (Figure 3, E and F). Taken together, these studies demonstrate that epithelial MCP-1 recruits macrophages to the lungs via CCR2 signaling and that these cells play an important role in mediating the exaggerated fibrotic susceptibility of HPS mice.
Figure 3. Epithelial MCP-1 drives fibrotic susceptibility in HPS2 mice via CCR2 signaling in myeloid cells.
(A–D) MCP-1 was deleted in the lung epithelium of HPS mice by crossing HPS2 mice with SPC.Cre/MCP1 floxed mice (HPS2/MCP1ΔAEC mice). (A) MCP-1 production determined by ELISA from alveolar epithelial cells (AECs) isolated from unchallenged mice (mean ± SEM); n = 6 for WT groups and n = 12 for HPS groups. Comparisons between groups were conducted by ANOVA with Tukey post-test, *P < 0.05 vs. WT, **P < 0.001 vs. others. (B) Bronchoalveolar lavage MCP-1 levels 24 hours after bleomycin challenge (mean ± SEM); n = 9 WT and n = 5 each for WT/MCP1 ΔAEC, HPS2, and HPS2/MCP1ΔAEC. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. other groups. (C) Lung collagen content of left lung quantitated in age-matched unchallenged mice or 7 days after bleomycin (mean ± SEM). For unchallenged groups, n = 4 WT and n = 6 WT/MCP1ΔAEC. For bleomycin-challenged groups, n = 6 WT/MCP1ΔAEC, n = 3 WT/SPC.Cre– littermate controls, n = 7 HPS2/MCP1ΔAEC, and n = 8 HPS2 SPC.Cre– littermate controls. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.05 vs. other bleomycin-challenged groups. (D) Fibrosis score. Data are presented as box-and-whisker Tukey plots; n = 4 for WT groups, n = 5 for HPS groups, and n = 3 SPC.Cre– controls. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. WT, **P < 0.05 vs. HPS2/MCP1ΔAEC. (E and F) Mice underwent whole body irradiation followed by transplantation of whole marrow from WT, HPS2, or WT/CCR2–/– mice. After 60 days, transplanted mice were challenged with bleomycin. (E) Lung collagen content of the left lung quantitated in unchallenged transplant controls or 7 days after bleomycin treatment (mean ± SEM). For unchallenged groups, n = 4 WT and n = 10 HPS2. For bleomycin-challenged groups, n = 10 recipients of WT marrow and n = 11 recipients of CCR2–/– marrow. Comparisons between bleomycin-challenged groups were conducted using Mann-Whitney U analysis, *P < 0.01. (F) Fibrosis score. Data are presented as box-and-whisker Tukey plots; n = 6 per group. Comparisons were conducted using Mann-Whitney U analysis, *P < 0.05.
HPS macrophages express increased M2 markers after bleomycin treatment.
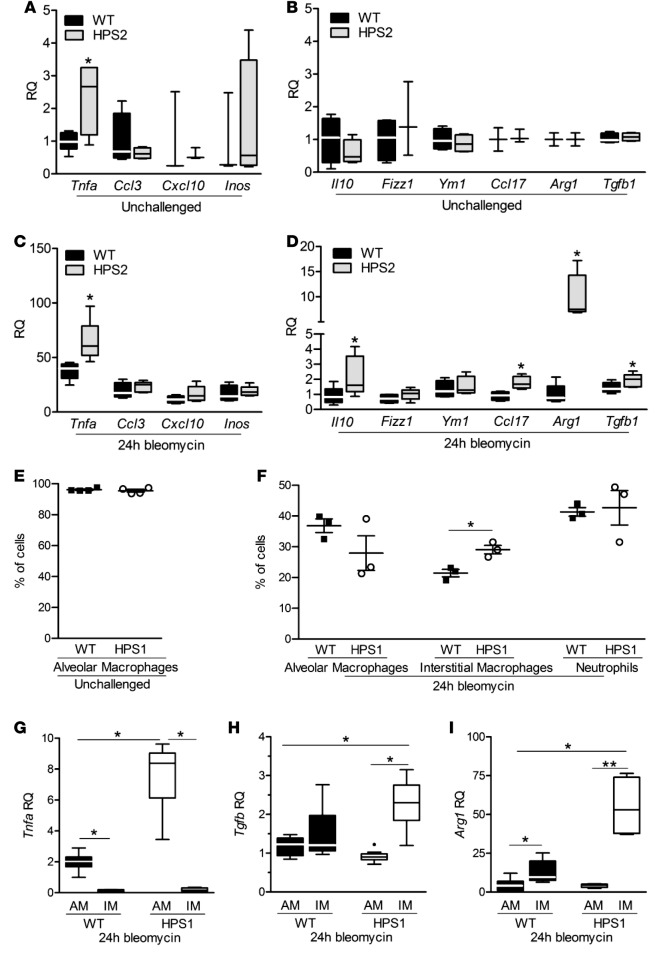
We next aimed to determine how macrophages were contributing to lung fibrosis in HPS mice. We isolated macrophages from the BAL of HPS and WT mice and used quantitative PCR to examine gene expression profiles. Macrophages in BAL from unchallenged HPS mice had a gene expression profile similar to that of cells from WT mice, except for increased expression of Tnfa (Figure 4, A and B), a finding consistent with our prior reports (8). At 24 hours after bleomycin treatment, M1 markers were similarly increased in macrophages isolated from BAL of HPS and WT mice; however, we identified increased expression of the M2 markers Il10, Ccl17, Arg1, and Tgfb in HPS macrophages relative to WT cells (Figure 4, C and D). Macrophages from HPS1 mice exhibited similar expression profiles as cells from HPS2 mice (data not shown). To determine which subgroup of macrophages was responsible for increased M2 marker expression after bleomycin treatment, we examined macrophage subpopulations present in the airways of WT and HPS mice. While almost all BAL cells from unchallenged HPS and WT mice had cell surface markers consistent with alveolar macrophages, increased numbers of interstitial macrophages and neutrophils were present in the BAL from both HPS and WT mice at 24 hours after bleomycin treatment (Figure 4, E and F). More interstitial macrophages were present in BAL from HPS mice compared with WT mice after bleomycin treatment, and these cells expressed increased levels of M2 markers Tgfb and Arg1 compared with cells from WT mice. In contrast, Tnfa expression was highest in alveolar macrophages from HPS mice (Figure 4, G–I). Collectively, these data indicate that increased numbers of interstitial macrophages are recruited to the lungs of HPS mice followed bleomycin injury and these cells are skewed toward production of M2 macrophage markers, including TGF-β.
Figure 4. Interstitial macrophages from HPS mice produce increased M2 markers after bleomycin treatment.
(A–D) Quantitative PCR was performed for a panel of M1 (A and C) and M2 (B and D) markers from WT and HPS macrophages isolated from bronchoalveolar lavage (BAL) by adherence to cell culture plates. BAL macrophages were recovered from unchallenged mice (A and B) and mice at 24 hours after bleomycin treatment (C and D). Gene expression was normalized to GAPDH. Data are presented as box-and-whisker Tukey plots; n = 3-5/group. Comparison between WT and HPS for each gene was assessed using Mann-Whitney U analysis, *P < 0.05. (E and F) Evaluation of cell populations in the airways of WT and HPS1 mice. BAL cells were recovered from unchallenged mice (E) and mice at 24 hours after bleomycin treatment (F). Flow cytometry was performed on BAL cells with cell types defined as follows: alveolar macrophages, F4/80+, CD11c+, CD11b–, Gr1–; interstitial macrophages, F4/80lo, CD11clo, CD11b+, Gr1–; and neutrophils, F4/80–, CD11b+, Gr1+; n = 4/group for unchallenged mice and n = 3/bleomycin-treated group. *P < 0.05 by Mann-Whitney U analysis. (G–I) Comparison of gene expression from alveolar macrophages (AM) and interstitial macrophages (IM). Quantitative PCR was performed for Tnfa (G), Tgfb (H), and Arg1 (I), normalized to Act, with results presented in comparison to flow-sorted WT unchallenged alveolar macrophages. Data are presented as box-and-whisker Tukey plots; n = 6/group. Comparison between macrophage subtypes were performed using Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.05, **P < 0.001.
Excess TGF-β produced by HPS macrophages is controlled by lung epithelium.
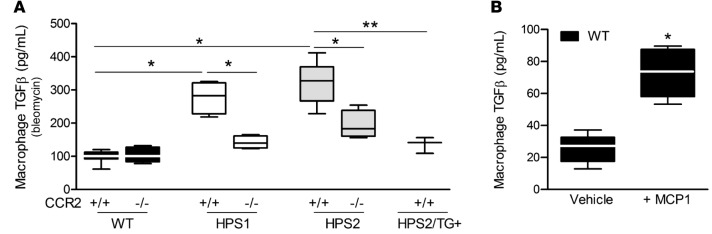
Given the increased TGF-β expression by HPS lung macrophages (6) and the well-established role of TGF-β in fibrosis, we wondered whether CCR2 signaling was regulating TGF-β levels in HPS macrophages. Consistent with the data presented above, macrophages isolated from BAL of IT bleomycin-treated HPS mice by adherence to cell culture plates produced increased TGF-β compared with WT macrophages in a manner that was diminished by CCR2 deficiency (Figure 5A). Utilizing our epithelial-specific corrected HPS2 transgenic model (7), we showed that the lung epithelium regulates increased TGF-β production by HPS macrophages. Further studies showed that MCP-1 induced TGF-β production by WT macrophages in vitro (Figure 5B), supporting a direct effect of MCP-1 on macrophage activity in addition to its chemoattractant properties. Taken together, these data demonstrate that the lung epithelium regulates TGF-β production by HPS macrophages through the MCP-1/CCR2 axis.
Figure 5. The MCP-1/CCR2 axis regulates macrophage TGF-β1 production.
(A) Macrophage TGF-β1 production. Mice were challenged with IT bleomycin, and bronchoalveolar lavage (BAL) was performed 24 hours later. Macrophages were separated by adherence to cell culture plates and then cultured for 24 hours in vitro. Total TGF-β1 was quantitated in the cell media by ELISA. Data are presented as box-and-whisker Tukey plots; n = 7 WT and HPS2/CCR2–/–, n = 6 WT/CCR2–/–, n = 5 HPS2, and n = 4/group for HPS1, HPS1/CCR2–/–, and HPS2/TG+. Comparisons were conducted by 2-way ANOVA with Bonferroni’s post-hoc test, *P < 0.05 vs. WT and CCR2–/–, **P < 0.05 vs. HPS2/TG+. (B) MCP-1 augments TGF-β production by WT macrophages. Macrophages were isolated by BAL from unchallenged WT mice and cultured with recombinant MCP-1 (10 ng/ml) or vehicle for 60 minutes. Total TGF-β1 was quantitated in the cell media by ELISA. Data are presented as box-and-whisker Tukey plots; n = 6/group. Comparisons between groups were conducted by Mann-Whitney U analysis, *P < 0.05.
Macrophage TGF-β contributes to the excess fibrotic response in HPS mice through TGF-β receptor signaling in AECs.
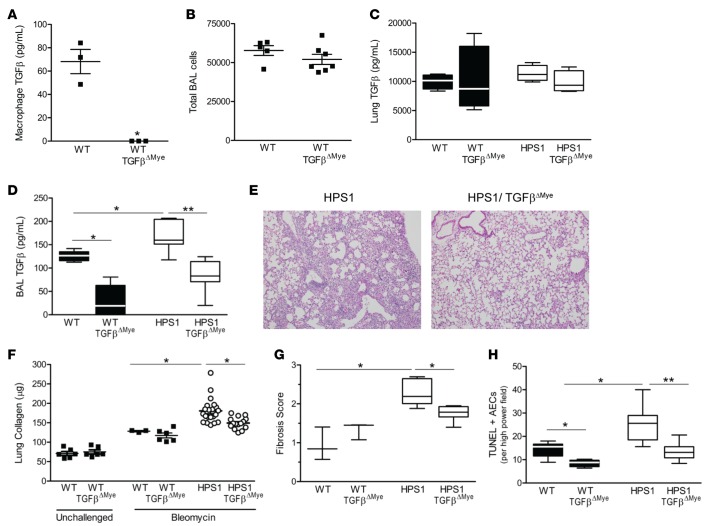
In order to directly test the contribution of macrophage TGF-β to fibrotic susceptibility, we generated mice with selective deletion of TGF-β1 in myeloid cells (TGF-βΔMye) by breeding mice expressing Cre recombinase under control of the lysozyme M promoter (LysM.Cre) with TGFB1f/f mice. We confirmed the absence of measurable TGF-β1 production from macrophages in this model (Figure 6A). No significant differences were observed in total BAL cell counts (Figure 6B) or in lung histology (data not shown) in these mice compared with age-matched WT controls. Although total lung TGF-β1 levels were not significantly different (Figure 6C), TGF-βΔMye mice showed a marked reduction in TGF-β levels in BAL, indicating that macrophages are largely responsible for producing TGF-β present in the airspaces (Figure 6D). We then generated HPS1/TGF-βΔMye mice for study in comparison to LysM.Cre– littermate controls and showed that these mice had decreased total TGF-β in BAL compared with HPS1 mice (Figure 6D). Following bleomycin treatment, biochemical and histologic evidence of fibrosis was reduced in HPS1/TGF-βΔMye mice compared with HPS1 mice (Figure 6, E–G). In addition, the number of TUNEL+ AECs was also diminished in lung sections from HPS1/TGF-βΔMye mice compared with HPS1 mice (Figure 6H). To confirm findings in the HPS2 model, we performed complementary experiments utilizing bone marrow transplant chimeras. In these studies, irradiated HPS1 or HPS2 mice were recipients of bone marrow from either TGF-βΔMye or WT control mice. When bleomycin challenge was performed 60 days after marrow transplantation, a similar reduction in fibrotic endpoints was observed in both the HPS1 and HPS2 recipients of marrow from myeloid TGF-β–deficient mice compared with those transplanted with WT marrow (Supplemental Figure 3). Together, these data demonstrate that macrophages play an important role in HPS pulmonary fibrosis through local production of TGF-β.
Figure 6. Macrophage TGF-β contributes to fibrotic susceptibility in HPS1 mice.
(A–C) Mice deficient in myeloid TGF-β1 (LysM.Cre/TGFb1f/f; denoted WT/TGF-βΔMye) were studied in comparison to LysM.Cre– littermate controls (LysM.Cre–/TGFb1f/f; denoted WT). (A) Macrophages were isolated by bronchoalveolar lavage (BAL), and total TGF-β1 was measured in the cell culture media by ELISA after 24 hours in culture (mean ± SEM); n = 3/group; *P < 0.05 by Mann-Whitney U analysis. (B) BAL cell counts from unchallenged mice; n = 5 WT and n = 7 WT/ TGF-βΔMye, P = NS by Mann-Whitney U analysis. (C) Total TGF-β1 in lung homogenates from unchallenged mice. Data are presented as box-and-whisker Tukey plots; n = 4/group, P = NS by Mann-Whitney U analysis. (D–H) TGF-βΔMye mice were bred onto the HPS1 background to generate HPS1/TGF-βΔMye mice or HPS1/Cre– littermate controls (denoted HPS1). (D) Total TGF-β1 was determined by ELISA in BAL fluid from unchallenged mice; n = 15 WT/TGF-βΔMye, n = 8 HPS1/TGF-βΔMye, n = 7 WT controls, and n = 11 HPS1 controls. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. WT, **P < 0.05 vs. HPS1/TGF-βΔMye. (E) Representative H&E histologic images (original magnification, ×10) of lung sections at 7 days after IT bleomycin. (F) Lung collagen content of the left lung in unchallenged mice and at 7 days after bleomycin (mean ± SEM). For unchallenged groups, n = 6. For bleomycin-challenged groups, n = 3 WT, n = 6 WT/TGF-βΔMye, n = 21 HPS1, and n = 15 HPS1/TGF-βΔMye mice. Comparisons between bleomycin-challenged groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. other bleomycin-challenged groups. (G) Fibrosis scoring on trichrome-stained lung tissue (mean ± SEM); n = 3/group for WT and n = 6/group for HPS1, *P < 0.05. (H) TUNEL+ alveolar epithelial cells in lung sections from mice 24 hours after bleomycin challenge (mean ± SEM); n = 10 WT/LysM.Cre– (WT) and n = 12 for other groups. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. WT, **P < 0.05 vs. HPS1/ TGF-βΔMye.
To better examine the role of myeloid TGF-β in lung fibrosis in WT mice, we performed experiments in which we challenged TGF-βΔMye or WT mice with a higher dose of IT bleomycin (0.05 units), which produces substantial fibrosis at 14 days. Deletion of myeloid TGF-β reduced lung collagen content and histologic fibrosis scores in TGF-βΔMye mice compared with WT littermate controls (Supplemental Figure 4). These data suggest that the mechanisms by which macrophages contribute to fibrosis in HPS may provide generalizable insights into common pathways mediating lung fibrosis.
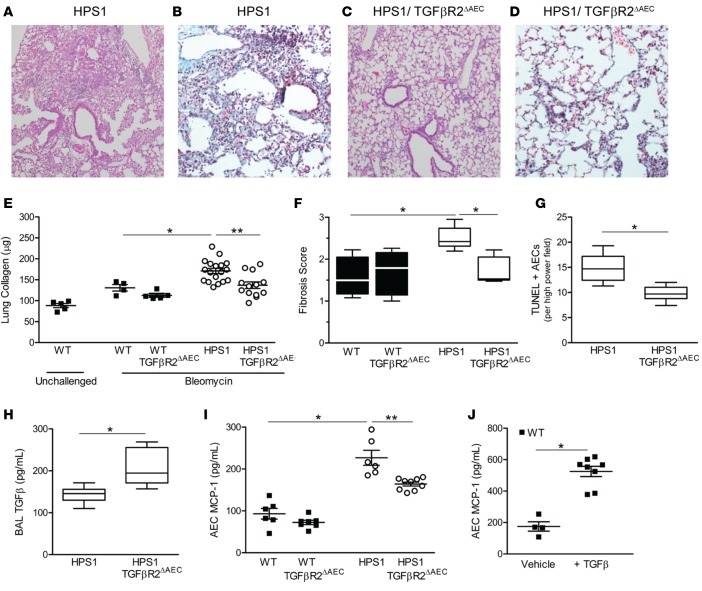
Deficiency of TGFBR2 disrupts the TGF-β signaling pathway, as binding of TGF-β to TGFBR2 at the cell surface results in recruitment of TGFBR1, which forms a heterodimer, with subsequent phosphorylation of TGFBR1 by the kinase activity of TGFBR2. To determine how increased TGF-β levels in the airways could contribute to fibrosis, HPS1 mutant mice were crossed with SPC.Cre/TGFBR2f/f mice (19). Isolated type II AECs from HPS1/TGFBR2ΔAEC mice showed minimal pSMAD2 signaling in response to TGF-β stimulation as compared with HPS1 controls, thus confirming effective reduction of TGF-β signaling (Supplemental Figure 5). BAL cell counts and lung histology were similar in 8- to 10-week-old HPS1/TGFBR2ΔAEC mice compared with littermate controls (data not shown). Following low-dose bleomycin treatment, HPS1/TGFBR2ΔAEC mice were protected from lung fibrosis compared with HPS1 mice (Figure 7, A–F). Further, compared with HPS1 mice, we found a significant reduction in numbers of TUNEL+ AECs in the lungs of HPS1/TGFBR2ΔAEC mice (Figure 7G). The protection from fibrosis and AEC apoptosis occurred despite a significant increase in total TGF-β levels in the BAL fluid of mice with epithelial TGFBR2 deficiency (Figure 7H). In addition, AEC MCP-1 production was reduced, but not eliminated, in HPS1/TGFBR2ΔAEC mice compared with HPS1 mice (Figure 7I), and exogenous TGF-β enhanced MCP-1 production from WT AECs (Figure 7J). Together, these findings indicate that epithelial dysfunction in HPS stimulates TGF-β production in macrophages via the MCP-1/CCR2 signaling axis, amplified by a TGF-β–driven feedback loop promoting AEC apoptosis and further MCP-1 production, resulting in pulmonary fibrosis.
Figure 7. Epithelial-specific deletion of TGFBR2 attenuates lung fibrosis in HPS1 mice.
HPS1 mice were bred to homozygosity with SPC.Cre+/TGFBR2f/f mice (denoted HPS1/TGFBR2ΔAEC) and studied in comparison to HPS1/SPC.Cre– littermate controls (denoted HPS1). (A–D) Representative histologic images of lung sections from mice at 7 days after bleomycin. (A and C) H&E images (original magnification, ×10). (B and D) Trichrome images (original magnification, ×20). (E) Lung collagen content of left lung in unchallenged mice or at 7 days after bleomycin (mean ± SEM); n = 5 WT unchallenged, n = 4 WT bleomycin, n = 5 WT/TGFBR2 ΔAEC; n = 13 HPS1/TGFBR2ΔAEC; and n = 18 HPS1. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.01 vs. unchallenged groups and WT bleomycin groups, **P < 0.05 vs. HPS1/ TGFBR2ΔAEC. (F) Fibrosis scoring on trichrome-stained lung tissue. Data are presented as box-and-whisker Tukey plots; n = 6 WT/TGFBR2ΔAEC, n = 5 HPS1/ TGFBR2ΔAEC, n = 4 WT/Cre– littermate controls, and n = 6 HPS1 controls, *P < 0.05 vs. all other groups. (G) TUNEL+ alveolar epithelial cells (AECs) in lung sections from mice 24 hours after bleomycin challenge; n = 10 HPS1/TGFBR2ΔAEC and n = 6 HPS1. Comparison between groups was assessed using Mann-Whitney U analysis, *P < 0.01. (H) Total TGF-β1 in BAL from unchallenged mice quantitated by ELISA; n = 14 HPS1/TGFBR2ΔAEC and n = 13 Cre– controls; Mann-Whitney U analysis, *P < 0.001. (I) MCP-1 production from type II AECs isolated from unchallenged mice and cultured for 24 hours; n = 9 WT/TGFBR2 ΔAEC and HPS1/TGFBR2ΔAEC and n = 6/group for WT and SPC.Cre– littermate controls. Comparisons between groups were conducted by Kruskal-Wallis test with Dunn’s multiple comparisons post-test, *P < 0.05 vs. WT, **P < 0.01 vs. HPS1/TGFBR2ΔAEC. (J) MCP-1 production from WT type II AECs after exposure to TGF-β. WT AECs were cultured for 24 hours in the presence of 20 ng/ml TGF-β or vehicle control, and MCP-1 levels were measured in the cell culture media by ELISA (mean ± SEM); n = 4 WT + vehicle and n = 8 WT + TGF-β, *P < 0.001.
Discussion
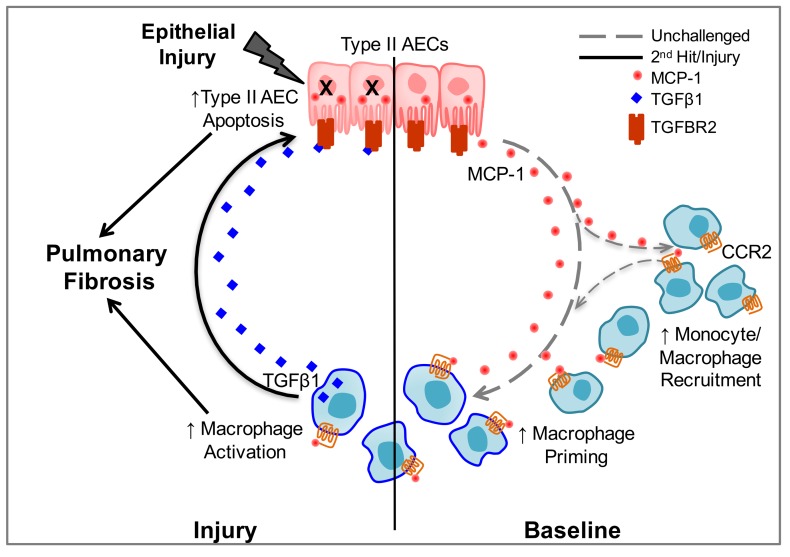
These studies demonstrate that cross-talk between AECs and macrophages determines fibrotic susceptibility in HPS, thus illuminating a mechanism by which AEC dysfunction underlies the pathogenesis of pulmonary fibrosis in HPS. We found that increased macrophage influx into the lungs of unchallenged HPS mice was associated with excessive MCP-1 production from AECs. Blocking MCP-1/CCR2 signaling eliminated increased macrophage recruitment and attenuated the excess fibrotic response to low-dose bleomycin in HPS mice. Further, we demonstrated that the MCP-1/CCR2 axis regulated excess TGF-β production by HPS macrophages and that selective deletion of TGF-β in myeloid cells or blockade of TGF-β signaling in AECs through deletion of the cognate receptor TGFBR2 protected HPS mice from AEC apoptosis and bleomycin-induced fibrosis. Together, these data reveal a feedback loop in which increased MCP-1 production by dysfunctional AECs results in recruitment and activation of the production of TGF-β in lung macrophages, which thereby amplifies the fibrotic cascade through AEC apoptosis and stimulation of fibrotic remodeling (Figure 8).
Figure 8. Epithelial-macrophage interactions in HPS pulmonary fibrosis.
Schematic depiction of the mechanisms by which macrophages transduce alveolar epithelial cell (AEC) dysfunction, thereby promoting pulmonary fibrosis in HPS through a MCP-1/CCR2/TGF-β–mediated feedback cycle.
There is strong evidence that macrophage-mediated inflammation precedes pulmonary fibrosis in HPS patients, though the mechanisms by which activated macrophages contribute to lung fibrosis in HPS are unknown. A key study from the National Institutes of Health Clinical Center demonstrated that BAL fluid from HPS1 patients with relatively preserved lung function contained increased numbers of activated macrophages (9). Macrophage-mediated inflammation is also present in HPS1 and HPS2 mouse models, which have been shown to reliably model the subtype-specific fibrotic susceptibility observed in HPS patients (7, 8, 20). Therefore, we utilized HPS mouse models as a platform to define the role of activated macrophages in fibrosis in a human disease model. We utilized both HPS1 and HPS2 models in these studies; inclusion of both models provided additional confidence regarding mechanisms important for HPS pulmonary fibrosis. We identified increased numbers of macrophages with cell surface markers consistent with the interstitial macrophage phenotype in the lungs of unchallenged HPS mice. In addition, we found increased recruitment of interstitial macrophages into the airways and a propensity for expression of M2 markers in HPS macrophages after bleomycin treatment, including increased TGF-β production. Utilizing genetic approaches and bone marrow transplantation experiments, we directly tested the contribution of macrophage TGF-β to fibrosis in HPS mice. Our studies demonstrate that macrophages, through generation of TGF-β, substantially contribute to AEC apoptosis and lung fibrosis in HPS mice. Together, these studies provide definitive evidence that macrophages play an important role in HPS pulmonary fibrosis and mechanistically link excess local production of TGF-β by macrophages in the alveolar space with AEC apoptosis and fibrotic susceptibility in HPS mice.
We have previously established that HPS fibrotic susceptibility and macrophage activation are direct consequences of type II AEC dysfunction and not due to intrinsic defects in macrophage function (7). Data that support this conclusion include that the constitutive activation phenotype of HPS macrophages was attenuated by targeted transgenic correction of the HPS2 defect in the alveolar epithelium, whereas the abnormalities in BAL macrophages in both HPS1 and HPS2 mice were not corrected by transplantation of WT bone marrow and were not conferred by transfer of HPS marrow into WT mice (7). Further, BAL fluid from HPS1 and HPS2 mice induced activation of WT macrophages (8). Thus, having established in the current study that HPS macrophages substantially contribute to pulmonary fibrosis, we sought to define the mechanisms of paracrine activation of macrophages by HPS AECs. We identify excess MCP-1 production by HPS AECs as a mechanism for both macrophage activation and recruitment in both HPS1 and HPS2 mice. We found that primary type II AECs from unchallenged HPS1 and HPS2, but not HPS3, mice secrete excess MCP-1 and that the accumulation of excess macrophages in the lungs of HPS mice tracks with AEC MCP-1 production and HPS subtype-specific fibrotic susceptibility. The mechanisms underlying how HPS mutations result in AEC dysfunction, including MCP-1 production, remain incompletely understood; however, recent publications have implicated lysosomal stress (21); mistrafficking of specific cargo, such as Prdx6 (22) or IL-13Rα2 (23); and autophagy (24). In our current studies, eliminating MCP-1/CCR2 signaling, macrophage TGF-β production, or TGF-β signaling in AECs reduced apoptosis of AECs in HPS mice. Our data also indicate that the TGF-β signaling axis contributes to excess MCP-1 production by HPS AECs through direct interactions that are amplified with subsequent injury through a feedback loop. Taken together, these data suggest that, while intrinsic trafficking abnormalities are responsible for HPS AEC dysfunction, cell-cell interactions also play a role and further investigation is required to define the mechanisms of AEC dysfunction that lead to paracrine activation of macrophages in HPS pulmonary fibrosis.
In addition to macrophage recruitment, MCP-1 contributes to angiogenesis and fibroblast recruitment, survival, and differentiation. CCR2 deficiency has been shown to attenuate fibrosis in WT mice in a variety of fibrosis models, including bleomycin, FITC, IL-10 overexpression, and diphtheria toxin–targeted epithelial injury (13, 14, 25–29). Our results in HPS are largely consistent with these prior studies, but there are several key differences and advances, starting with the elucidation of cell-specific mechanisms in unchallenged HPS mice. The immune consequences of CCR2 deficiency in mice are complex, and MCP-1 signaling regulates both stromal cells and bone marrow–derived populations in the lung. Therefore, we utilized complementary approaches with MCP-1 and CCR2 as well as bone marrow transplantation experiments using CCR2-deficient mice as bone marrow donors. This approach enabled us to dissect central cell-specific relationships in HPS pulmonary fibrosis: MCP-1 specifically from the lung epithelium is sufficient for fibrotic susceptibility in HPS mice, and CCR2 signaling in myeloid cells directly contributes to fibrosis in HPS mice. Further, our data reveal that epithelial-macrophage interactions are coordinated through the MCP-1/CCR2 pathway in regulating macrophage TGF-β production. The extent of correction achieved with interruption of MCP-1/CCR2 signaling was similar to that observed with the HPS2 epithelial-specific transgenic correction in HPS mice (7), suggesting that these mechanisms are the predominant explanation for how AEC dysfunction leads to fibrotic susceptibility in HPS mice.
Elevated levels of MCP-1 have been found in the blood, BAL, and lungs of patients with HPS and idiopathic pulmonary fibrosis (IPF), and concentrations correlate with severity of pulmonary function impairment (9, 30–34). Substantial preclinical data exist regarding targeting of the MCP-1/CCR2 axis in animal models of diabetes, inflammatory disorders, and malignancies (35–38). Notably, a phase II trial in IPF (NCT00786201) using a human monoclonal antibody to CCL2 (MCP-1) showed no effect on disease progression, pulmonary function parameters, or mortality. However, free CCL2 levels remained elevated above baseline in subjects in this trial, suggesting pharmacologic failure of this particular therapeutic agent (39). Thus, further studies will be needed to determine whether the CCL2/CCR2 axis can be effectively targeted in the lung. In this regard, the recognizable clinical features of HPS (i.e., oculocutaneous albinism) provide a uniquely promising opportunity to identify patients at early stages of disease and implement targeted therapeutic and/or preventative strategies before significant pulmonary fibrosis occurs. Thus, we propose that strategies to target macrophage activation in HPS pulmonary fibrosis may be more promising than those in advanced stages of IPF.
While we identified MCP-1 as a key driver of macrophage recruitment and activation in HPS mice, BAL fluid from HPS patients also contains elevated levels of both M-CSF and GM-CSF (9). Although HPS AECs produced only modestly elevated levels of GM-CSF, it is possible that these and other factors in addition to MCP-1 also play a role in macrophage recruitment and activation in HPS. For example, levels of chitinase 3-like 1 (CHI3L1) are increased in the plasma of HPS patients and may also contribute to M2 macrophage polarization and AEC apoptosis (23). In addition to excessive TGF-β production, we recognize that there are other potential mechanisms by which HPS macrophages could contribute to lung fibrosis. For example, BAL macrophages from HPS patients produce excess levels of several cytokines and chemokines ex vivo (9), and our previously published data demonstrate that murine HPS macrophages are primed, exhibiting hyperresponsiveness to LPS, TLR2, and TLR3 agonists (8). Our findings are consistent with a recent publication demonstrating that macrophage-derived TGF-β contributes to fibrosis in WT mice and that macrophages from IPF patients exhibit increased TGF-β expression (40). While our data indicate that macrophage TGF-β promotes fibrosis through a major effect on the lung epithelium in HPS, additional roles for macrophage-derived TGF-β are likely, including activation and differentiation of fibroblasts (40).
In summary, our studies have dissected the mechanism by which macrophages contribute to fibrosis in HPS, revealing that MCP-1 production by dysfunctional AECs results in recruitment and activation of lung macrophages that produce TGF-β, thus amplifying the fibrotic cascade through AEC apoptosis and stimulation of fibrotic remodeling. This feedback loop identifies promising targets for therapeutic intervention and prevention of pulmonary fibrosis in HPS and may inform approaches to more common fibrotic disorders.
Methods
Mice.
HPS mice with homozygous mutations on a congenic C57BL/6J background were used in these studies (7, 41–44), with C57BL/6J WT mice from Jackson Laboratories or littermate controls as indicated. The HPS2 model was predominantly utilized for studies relating to our previously published epithelial-specific correction model, with mice on the HPS2 background (7), while we also performed key confirmatory studies in the HPS1 model, as HPS1 is the most common subtype associated with fibrosis in humans. Other murine models utilized were bred onto the HPS backgrounds as summarized in Supplemental Table 1, including the following models from Jackson Laboratories: global CCR2-deficient (CCR2–/–) mice (stock 004999), mice with loxP sites flanking exon 6 of the Tgfb1 gene (TGF-β1f/f; stock 010721), and mice with loxP sites flanking exons 2–3 of the Ccl2 gene (encoding MCP-1; MCP1f/f; stock 016849). LysM.Cre mice were as previously described (45), and mice with Cre recombinase under the control of the surfactant protein C promoter (SPC.Cre) were a gift from Brigid Hogan, Duke University (Durham, North Carolina, USA) (19, 46). Mice were housed in the animal facility at Vanderbilt University, and studies were performed in both male and female mice.
Antibodies and reagents.
Recombinant mouse MCP-1 and recombinant mouse TGF-β1 were from R&D Systems. Rabbit polyclonal psmad2 was from Cell Signaling, Cy3-conjugated secondary antibody was from Jackson Immunoresearch Laboratories, and DAPI with Vectashield mounting medium was from Vector Laboratories.
BAL.
BAL with total and differential cell counting was performed as previously described (6, 8).
Isolation of murine macrophages and type II AECs.
BAL macrophages were isolated and cultured on plastic as previously described (8). Type II AECs were isolated from 5- to 6-week-old mice according to published methods using dispase and negative selection on antibody-coated plates to separate leukocyte and monocyte populations (7, 47). Type II AECs were plated at a density of 1 × 106 in 12-well plates coated with 500 μl of Cultrex basement membrane extract (Trevigen) and cultured in Bronchial Epithelial Cell Growth Media (Lonza Walkersville Inc.) with 5% fetal bovine serum and 10 ng/ml of recombinant KGF (Peprotech).
Quantitative real-time PCR analysis.
Total RNA for quantitative real-time PCR was isolated and reverse transcribed by standard methods using the Qiagen RNEasy kit (Qiagen) and the Bio-Rad iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was performed using Sybr Green PCR Master Mix (Applied Biosystems) with quantification on an Applied Biosystems Step One Plus Cycler. PCR primers used were as follows: Tnfa (F: 5′-AAGCCTGTAGCCCACGTCGTA-3′, R: 5′-GGCACCACTAGTTGGTTGTCTTTG-3′), CCL3 (F: 5′-TGCCCTTGCTGTTCTTCTCT-3′, R: 5′-GATGAATTGGCGTGGAATCT-3′), CXCL10 (F: 5′-TGAATCCGGAATCTAAGACCATCAA-3′, R: 5′-AGGACTAGCCATCCACTGGGTAAAG-3′), iNOS (F: 5-CACCTTGGAGTTCACCCAGT-3′; R: 5′-ACCACTCGTACTTGGGATGC-3′), Il10 (F: 5′-ACCTGCTCCACTGCCTTGCT-3′; R: 5′-GGTTGCCAAGCCTTATCGGA-3′), Fizz1 (F: 5′-TCCCAGTGAATACTGATGAGA-3′; R: 5′-CCACTCTGGATCTCCCAAGA-3′), Ym1 (F: 5′-GGGCATACCTTTATCCTGAG-3′; R: 5′-CCACTGAAGTCATCCATGTC-3′), CCL17 (F: 5′-TGCTTCTGGGGACTTTTCTG-3′, R: 5′-CATCCCTGGAACACTCCACT-3′), Arg1 (F: 5′-ATGGAAGAGACCTTCAGCTAC-3′, R: 5′-GCTGTCTTCCCAAGAGTTGGG-3′), TGF-β1 (F: 5′-CAATACGTCAGACATTCGGGAAGC-3′, R: 5′-CTGGTAGAGTTCTACGTGTTGCTC-3′), and GAPDH (F: 5′-TGAGGACCAGGTTGTCTCCT-3′; R: 5′-CCCTGTTGCTGTAGCCGTAT-3′). The PCR reactions and relative quantifications were performed in triplicate using 5 ng of cDNA per reaction. For each sample, expression was normalized to Gapdh or Actin.
Measurement of protein concentrations.
Cytokine levels were measured in cell culture media supernatant or BAL fluid by ELISA (R&D Systems : MCP-1, #MJE00; MCP-5, #MCC120; MIP-1α, # MMA00; GM-CSF, #MGM00; RANTES, #MMR00; TGFβ1, MB100B; and Abcam: MCP-3, #ab205571; and M-CSF, #ab155457).
Flow cytometry.
Lungs were perfused with chilled PBS, harvested, and digested in DNAse type I and collagenase type XI (Sigma-Aldrich) for 40 minutes at 37°C, followed by passage through a 70-μM filter to generate a single-cell suspension as previously described (6, 48). Cells were treated with ACK red blood cell lysis buffer (Lonza Group) for 5 minutes, washed with FACS buffer (PBS, 0.5% BSA, and 2mM EDTA), and incubated with anti-CD16/CD32 (BD Biosciences) prior to antibody staining. In additional studies, BAL was performed to collect cells for flow cytometry. Cell surface antibodies used for staining with FACS buffer included CD45-APC, F4/80-PE, CD11b-AlexaFluor 700, CD11c-PerCP-Cy5.5, Gr-1-APC/Cy7, CD14-PE/Cy7, CD4-FITC, and CD8-APC and DAPI as a viability dye (eBioscience Inc.). Samples were analyzed using a LSR II flow cytometer (BD Biosciences) and Flow Jo v.10.1 software (Tree Star).
Bleomycin challenge.
Pharmaceutical-grade bleomycin sulfate (0.03 units, unless otherwise noted) was administered by IT instillation to 8- to 12-week-old male and female mice (6).
Lung histology, immunohistochemistry, and evaluation of apoptosis.
Lung tissues were prepared and studied as previously described (7). A modified Ashcroft fibrosis score was determined as previously described, based on scoring of the entire right lung by two observers (49). For evaluation of apoptosis, lungs were harvested 24 hours after IT bleomycin challenge. TUNEL staining was performed using the In Situ Cell Death Detection Kit (Roche Diagnostics). Quantitation of TUNEL+ AECs was determined by scoring of a minimum of 10 images of ×20 magnification fields by two independent observers as previously described (6, 7).
Quantitation of lung collagen content.
Whole left lung total soluble collagen was measured using the Sircol assay (Biocolor, Accurate Chemical And Scientific Corporation) according to manufacturer’s instructions and as previously described (7).
Bone marrow transplantation.
Bone marrow transplantation was performed as previously described (7) with minor modifications, such that recipient mice were exposed to 11 Gy of whole body irradiation, followed by tail vein injection of whole marrow from donors. Transplanted mice were studied a minimum of 60 days after transplantation.
Statistics.
Numeric data are presented as mean ± SEM or box-and-whisker Tukey plots. Statistical analyses were performed using GraphPad Prism. Parametric data were evaluated by ANOVA with Tukey’s post-hoc analysis, and nonparametric data were evaluated by the Kruskal-Wallis test with Dunn’s multiple comparisons post-test or by the Mann-Whitney test. A 2-sided P value of less than 0.05 was regarded as significant.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee at Vanderbilt University.
Author contributions
LRY was responsible for the overall project and wrote the manuscript. PMG and CWS performed the majority of the animal and cell culture experiments. HT, AQ, TS, APM, RZ, JTB, SVN, and WEL contributed to specific subsets of experiments, including bone marrow transplantation, macrophage phenotyping, and studies of AECs. TSB contributed to the experimental design, data analysis, and manuscript preparations. All authors edited the final manuscript.
Supplementary Material
Acknowledgments
Funding was provided by the NIH/National Heart, Lung, and Blood Institute (R01 HL-119503 to LRY; R01 HL 085317 and P01 HL 092870, to TSB; RO1 HL 105479, to WEL) and NIH/ National Cancer Institute (R01 CA 200681, to SVN); the American Thoracic Society/Hermansky-Pudlak Syndrome Network (to HT); the K12 Research Scholar Award (NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development 6K12HD087023-02, to JTB); and the Department of Veterans Affairs (to TSB and WEL).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(17):e88947. doi:10.1172/jci.insight.88947.
Contributor Information
Harikrishna Tanjore, Email: harikrishna.tanjore@vanderbilt.edu.
Taylor Sherrill, Email: taylor.sherrill@vanderbilt.edu.
Aidong Qi, Email: aidong.qi@vanderbilt.edu.
Rinat Zaynagetdinov, Email: rinat.zaynagetdinov@hotmail.com.
John T. Benjamin, Email: john.benjamin@vanderbilt.edu.
References
- 1.Gahl WA, et al. Genetic defects and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky-Pudlak syndrome) N Engl J Med. 1998;338(18):1258–1264. doi: 10.1056/NEJM199804303381803. [DOI] [PubMed] [Google Scholar]
- 2.Cullinane AR, et al. A BLOC-1 mutation screen reveals that PLDN is mutated in Hermansky-Pudlak Syndrome type 9. Am J Hum Genet. 2011;88(6):778–787. doi: 10.1016/j.ajhg.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Gochuico BR, et al. Interstitial lung disease and pulmonary fibrosis in Hermansky-Pudlak syndrome type 2, an adaptor protein-3 complex disease. Mol Med. 2012;18:56–64. doi: 10.2119/molmed.2011.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantly M, Avila NA, Shotelersuk V, Lucero C, Huizing M, Gahl WA. Pulmonary function and high-resolution CT findings in patients with an inherited form of pulmonary fibrosis, Hermansky-Pudlak syndrome, due to mutations in HPS-1. Chest. 2000;117(1):129–136. doi: 10.1378/chest.117.1.129. [DOI] [PubMed] [Google Scholar]
- 5.Shotelersuk V, Dell’Angelica EC, Hartnell L, Bonifacino JS, Gahl WA. A new variant of Hermansky-Pudlak syndrome due to mutations in a gene responsible for vesicle formation. Am J Med. 2000;108(5):423–427. doi: 10.1016/S0002-9343(99)00436-2. [DOI] [PubMed] [Google Scholar]
- 6.Young LR, Pasula R, Gulleman PM, Deutsch GH, McCormack FX. Susceptibility of Hermansky-Pudlak mice to bleomycin-induced type II cell apoptosis and fibrosis. Am J Respir Cell Mol Biol. 2007;37(1):67–74. doi: 10.1165/rcmb.2006-0469OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young LR, et al. The alveolar epithelium determines susceptibility to lung fibrosis in Hermansky-Pudlak syndrome. Am J Respir Crit Care Med. 2012;186(10):1014–1024. doi: 10.1164/rccm.201207-1206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young LR, Borchers MT, Allen HL, Gibbons RS, McCormack FX. Lung-restricted macrophage activation in the pearl mouse model of Hermansky-Pudlak syndrome. J Immunol. 2006;176(7):4361–4368. doi: 10.4049/jimmunol.176.7.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouhani FN, et al. Alveolar macrophage dysregulation in Hermansky-Pudlak syndrome type 1. Am J Respir Crit Care Med. 2009;180(11):1114–1121. doi: 10.1164/rccm.200901-0023OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasse A, et al. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am J Respir Crit Care Med. 2006;173(7):781–792. doi: 10.1164/rccm.200509-1518OC. [DOI] [PubMed] [Google Scholar]
- 11.Gangadharan B, et al. Murine gammaherpesvirus-induced fibrosis is associated with the development of alternatively activated macrophages. J Leukoc Biol. 2008;84(1):50–58. doi: 10.1189/jlb.0507270. [DOI] [PubMed] [Google Scholar]
- 12.Murray LA, et al. Serum amyloid P therapeutically attenuates murine bleomycin-induced pulmonary fibrosis via its effects on macrophages. PLoS One. 2010;5(3):e88947. doi: 10.1371/journal.pone.0009683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tighe RM, et al. Recruited exudative macrophages selectively produce CXCL10 after noninfectious lung injury. Am J Respir Cell Mol Biol. 2011;45(4):781–788. doi: 10.1165/rcmb.2010-0471OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L, et al. New concepts of IL-10-induced lung fibrosis: fibrocyte recruitment and M2 activation in a CCL2/CCR2 axis. Am J Physiol Lung Cell Mol Physiol. 2011;300(3):L341–L353. doi: 10.1152/ajplung.00122.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons MA, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med. 2011;184(5):569–581. doi: 10.1164/rccm.201010-1719OC. [DOI] [PubMed] [Google Scholar]
- 16.Morales-Nebreda L, Misharin AV, Perlman H, Budinger GR. The heterogeneity of lung macrophages in the susceptibility to disease. Eur Respir Rev. 2015;24(137):505–509. doi: 10.1183/16000617.0031-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrne AJ, Maher TM, Lloyd CM. Pulmonary Macrophages: A New Therapeutic Pathway in Fibrosing Lung Disease? Trends Mol Med. 2016;22(4):303–316. doi: 10.1016/j.molmed.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Murray LA, et al. TGF-beta driven lung fibrosis is macrophage dependent and blocked by Serum amyloid P. Int J Biochem Cell Biol. 2011;43(1):154–162. doi: 10.1016/j.biocel.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Degryse AL, et al. TGFβ signaling in lung epithelium regulates bleomycin-induced alveolar injury and fibroblast recruitment. Am J Physiol Lung Cell Mol Physiol. 2011;300(6):L887–L897. doi: 10.1152/ajplung.00397.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atochina-Vasserman EN, et al. Early alveolar epithelial dysfunction promotes lung inflammation in a mouse model of Hermansky-Pudlak syndrome. Am J Respir Crit Care Med. 2011;184(4):449–458. doi: 10.1164/rccm.201011-1882OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahavadi P, et al. Epithelial stress and apoptosis underlie Hermansky-Pudlak syndrome-associated interstitial pneumonia. Am J Respir Crit Care Med. 2010;182(2):207–219. doi: 10.1164/rccm.200909-1414OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kook S, et al. Impaired lysosomal integral membrane protein 2-dependent peroxiredoxin 6 delivery to lamellar bodies accounts for altered alveolar phospholipid content in adaptor protein-3-deficient pearl mice. J Biol Chem. 2016;291(16):8414–8427. doi: 10.1074/jbc.M116.720201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, et al. Chitinase 3-like-1 and its receptors in Hermansky-Pudlak syndrome-associated lung disease. J Clin Invest. 2015;125(8):3178–3192. doi: 10.1172/JCI79792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahuja S, et al. MAP1LC3B overexpression protects against Hermansky-Pudlak syndrome type-1-induced defective autophagy in vitro. Am J Physiol Lung Cell Mol Physiol. 2016;310(6):L519–L531. doi: 10.1152/ajplung.00213.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gharaee-Kermani M, McCullumsmith RE, Charo IF, Kunkel SL, Phan SH. CC-chemokine receptor 2 required for bleomycin-induced pulmonary fibrosis. Cytokine. 2003;24(6):266–276. doi: 10.1016/j.cyto.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Moore BB, et al. Protection from pulmonary fibrosis in the absence of CCR2 signaling. J Immunol. 2001;167(8):4368–4377. doi: 10.4049/jimmunol.167.8.4368. [DOI] [PubMed] [Google Scholar]
- 27.Osterholzer JJ, et al. Implicating exudate macrophages and Ly-6C(high) monocytes in CCR2-dependent lung fibrosis following gene-targeted alveolar injury. J Immunol. 2013;190(7):3447–3457. doi: 10.4049/jimmunol.1200604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore BB, et al. CCR2-mediated recruitment of fibrocytes to the alveolar space after fibrotic injury. Am J Pathol. 2005;166(3):675–684. doi: 10.1016/S0002-9440(10)62289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuma T, et al. C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases. J Pathol. 2004;204(5):594–604. doi: 10.1002/path.1667. [DOI] [PubMed] [Google Scholar]
- 30.Car BD, Meloni F, Luisetti M, Semenzato G, Gialdroni-Grassi G, Walz A. Elevated IL-8 and MCP-1 in the bronchoalveolar lavage fluid of patients with idiopathic pulmonary fibrosis and pulmonary sarcoidosis. Am J Respir Crit Care Med. 1994;149(3 Pt 1):655–659. doi: 10.1164/ajrccm.149.3.8118632. [DOI] [PubMed] [Google Scholar]
- 31.Suga M, Iyonaga K, Ichiyasu H, Saita N, Yamasaki H, Ando M. Clinical significance of MCP-1 levels in BALF and serum in patients with interstitial lung diseases. Eur Respir J. 1999;14(2):376–382. doi: 10.1034/j.1399-3003.1999.14b23.x. [DOI] [PubMed] [Google Scholar]
- 32.Capelli A, Di Stefano A, Gnemmi I, Donner CF. CCR5 expression and CC chemokine levels in idiopathic pulmonary fibrosis. Eur Respir J. 2005;25(4):701–707. doi: 10.1183/09031936.05.00082604. [DOI] [PubMed] [Google Scholar]
- 33.Shinoda H, et al. Elevated CC chemokine level in bronchoalveolar lavage fluid is predictive of a poor outcome of idiopathic pulmonary fibrosis. Respiration. 2009;78(3):285–292. doi: 10.1159/000207617. [DOI] [PubMed] [Google Scholar]
- 34.Mercer PF, et al. Pulmonary epithelium is a prominent source of proteinase-activated receptor-1-inducible CCL2 in pulmonary fibrosis. Am J Respir Crit Care Med. 2009;179(5):414–425. doi: 10.1164/rccm.200712-1827OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Göser S, et al. Critical role for monocyte chemoattractant protein-1 and macrophage inflammatory protein-1alpha in induction of experimental autoimmune myocarditis and effective anti-monocyte chemoattractant protein-1 gene therapy. Circulation. 2005;112(22):3400–3407. doi: 10.1161/CIRCULATIONAHA.105.572396. [DOI] [PubMed] [Google Scholar]
- 36.Weisberg SP, et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest. 2006;116(1):115–124. doi: 10.1172/JCI24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia M, Sui Z. Recent developments in CCR2 antagonists. Expert Opin Ther Pat. 2009;19(3):295–303. doi: 10.1517/13543770902755129. [DOI] [PubMed] [Google Scholar]
- 38.Xia Y, Entman ML, Wang Y. CCR2 regulates the uptake of bone marrow-derived fibroblasts in renal fibrosis. PLoS One. 2013;8(10):e88947. doi: 10.1371/journal.pone.0077493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghu G, et al. CC-chemokine ligand 2 inhibition in idiopathic pulmonary fibrosis: a phase 2 trial of carlumab. Eur Respir J. 2015;46(6):1740–1750. doi: 10.1183/13993003.01558-2014. [DOI] [PubMed] [Google Scholar]
- 40.Larson-Casey JL, Deshane JS, Ryan AJ, Thannickal VJ, Carter AB. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity. 2016;44(3):582–596. doi: 10.1016/j.immuni.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGarry MP, Reddington M, Novak EK, Swank RT. Survival and lung pathology of mouse models of Hermansky-Pudlak syndrome and Chediak-Higashi syndrome. Proc Soc Exp Biol Med. 1999;220(3):162–168. doi: 10.1046/j.1525-1373.1999.d01-24.x. [DOI] [PubMed] [Google Scholar]
- 42.Feng L, et al. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum Mol Genet. 1999;8(2):323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- 43.Feng L, Rigatti BW, Novak EK, Gorin MB, Swank RT. Genomic structure of the mouse Ap3b1 gene in normal and pearl mice. Genomics. 2000;69(3):370–379. doi: 10.1006/geno.2000.6350. [DOI] [PubMed] [Google Scholar]
- 44.Yang W, Li C, Ward DM, Kaplan J, Mansour SL. Defective organellar membrane protein trafficking in Ap3b1-deficient cells. J Cell Sci. 2000;113(Pt 22):4077–4086. doi: 10.1242/jcs.113.22.4077. [DOI] [PubMed] [Google Scholar]
- 45.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 46.Tanjore H, et al. Contribution of epithelial-derived fibroblasts to bleomycin-induced lung fibrosis. Am J Respir Crit Care Med. 2009;180(7):657–665. doi: 10.1164/rccm.200903-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice WR, Conkright JJ, Na CL, Ikegami M, Shannon JM, Weaver TE. Maintenance of the mouse type II cell phenotype in vitro. Am J Physiol Lung Cell Mol Physiol. 2002;283(2):L256–L264. doi: 10.1152/ajplung.00302.2001. [DOI] [PubMed] [Google Scholar]
- 48.Zaynagetdinov R, et al. Identification of myeloid cell subsets in murine lungs using flow cytometry. Am J Respir Cell Mol Biol. 2013;49(2):180–189. doi: 10.1165/rcmb.2012-0366MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Degryse AL, et al. Repetitive intratracheal bleomycin models several features of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2010;299(4):L442–L452. doi: 10.1152/ajplung.00026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.