Abstract
A unique feature of rheumatoid arthritis (RA) is the presence of anti-citrullinated protein antibodies (ACPA). Several risk factors for RA are known to increase the expression or activity of peptidyl arginine deiminases (PADs), which catalyze citrullination and, when dysregulated, can result in hypercitrullination. However, the consequence of hypercitrullination is unknown and the function of each PAD has yet to be defined. Th cells of RA patients are hypoglycolytic and hyperproliferative due to impaired expression of PFKFB3 and ATM, respectively. Here, we report that these features are also observed in peripheral blood mononuclear cells (PBMCs) from healthy at-risk individuals (ARIs). PBMCs of ARIs are also hypercitrullinated and produce more IL-2 and Th17 cytokines but fewer Th2 cytokines. These abnormal features are due to impaired induction of PTPN22, a phosphatase that also suppresses citrullination independently of its phosphatase activity. Attenuated phosphatase activity of PTPN22 results in aberrant expression of IL-2, ATM, and PFKFB3, whereas diminished nonphosphatase activity of PTPN22 leads to hypercitrullination mediated by PADs. PAD2- or PAD4-mediated hypercitrullination reduces the expression of Th2 cytokines. By contrast, only PAD2-mediated hypercitrullination can increase the expression of Th17 cytokines. Taken together, our data depict a molecular signature of preclinical RA that is triggered by impaired induction of PTPN22.
PTPN22 regulates the molecular features of preclinical rheumatoid arthritis through its phosphatase and non-phosphatase activities.
Introduction
The presence of anti-citrullinated protein antibodies (ACPA) is a unique feature of rheumatoid arthritis (RA) (1, 2). Citrullinated proteins are generated by deimination catalyzed by peptidyl arginine deiminases (PADs), including PAD1–4 and PAD6 (3). Although the etiology of RA is still not fully understood, alterations in protein citrullination probably contribute to certain risk factors in RA. For example, smoking, a major risk factor of RA, can increase the level of extracellular PAD2 and intracellular citrullinated proteins in lung lavage (4, 5). Porphyromonas gingivalis, the most common pathogen of periodontitis, expresses a PAD-like enzyme that is capable of citrullinating host proteins (6, 7). Several epidemiological studies have suggested an association between periodontitis and risk of developing ACPA or RA (8–10). In addition, a functional haplotype stabilizing the mRNA from padi4, the gene encoding PAD4, is associated with a higher risk of RA and the presence of ACPA in RA patients (11–14). Hematopoietic cells express mainly PAD2 and PAD4. These two PADs have overlapping but not identical substrate spectra (15, 16). However, it is still unknown if either PAD has a unique contribution to the pathogenesis of RA.
It was recently demonstrated that Th cells of ACPA+ RA patients expressed less PFKFB3, a rate-limiting enzyme of glycolysis, and ATM, a cell cycle checkpoint kinase, but a higher level of G6PD, which catalyzes the pentose phosphate pathway and promotes the generation of NADPH and glutathione (17, 18). Therefore, RA Th cells are hypoglycolytic, hyperproliferative, and under reductive stress, but the cause of these features is still unknown. In addition, it is unclear whether these features appear before or after the development of ACPA or prior to the onset of clinical symptoms. In this regard, it is notable that ACPA are found in the serum of asymptomatic individuals for an average of 3–5 years prior to the onset of clinically apparent arthritis and the classification of subjects as having RA (19, 20).
Citrullination converts positively charged arginines to neutral citrullines and is expected to alter protein folding and function. Indeed, citrullination critically regulates embryogenesis (21, 22), epithelial-to-mesenchymal transformation (23), and pluripotency of embryonic stem cells (24). In addition, PAD4-mediated citrullination of histones is essential for the formation of neutrophil extracellular traps (NETs) (25–27), which can induce the expression of inflammatory cytokines, such as IL-6 and IL-18, as well as CCL20 and ICAM-1 from fibroblast-like synoviocytes (28). NETs are also a rich source of citrullinated antigens. Thus, PADs can contribute to RA pathogenesis by promoting the generation of citrullinated antigens and aggravating inflammation through the formation of NETs. However, it is still unclear if citrullination and PADs have other regulatory roles in immune cells in addition to promoting NETosis.
We recently discovered that PTPN22, a nonreceptor protein tyrosine phosphatase, can interact with and inhibit the activity of PAD4 (29). This function of PTPN22 is independent of its phosphatase activity. A C-to-T SNP, located at position 1,858 of human PTPN22 cDNA and replacing an arginine (R620) with a tryptophan (W620), carries the highest risk among all non-HLA genetic variations that are associated with RA (30–33). This R-to-W conversion renders PTPN22 unable to interact with PAD4 and suppress citrullination. Accordingly, the C1858T SNP is associated with hypercitrullination in peripheral blood mononuclear cells (PBMCs) and heightened propensity for forming NETs even in healthy donors. Whether PTPN22 also inhibits the activity of PAD2 remains to be determined.
The frequency of the C1858T SNP in individuals of European descent is approximately 10%, highest among of all ethnicities. It is unclear if hypercitrullination is also present in PBMCs of other at-risk individuals (ARIs) who do not carry the C1858T SNP. If it is, what is the cause and functional consequence of hypercitrullination? To answer these questions, we studied PBMCs obtained from ARIs, including unaffected RA first-degree relatives (FDRs) and ACPA+ individuals without RA. Positive family history and positive ACPA status each carries an odds ratio of approximately 4 to 5 or higher (34–36), which is higher than that of the C1858T SNP. Thus, these individuals are at high risk of developing RA. We found in two independent cohorts of ARIs that hypercitrullination in PBMCs is common. Their PBMCs, regardless of ACPA status, have a defect in the induction of PTPN22, display an aberrant Th cytokine profile, and exhibit a phenotype very similar but not identical to that of RA Th cells. We establish the chronological and causal relationships of these abnormal features and demonstrate that attenuation of both phosphatase and nonphosphatase activities of PTPN22 caused by impaired induction of this protein is likely to be at the root of these abnormal features.
Results
Hypercitrullination in PBMCs of healthy ARIs.
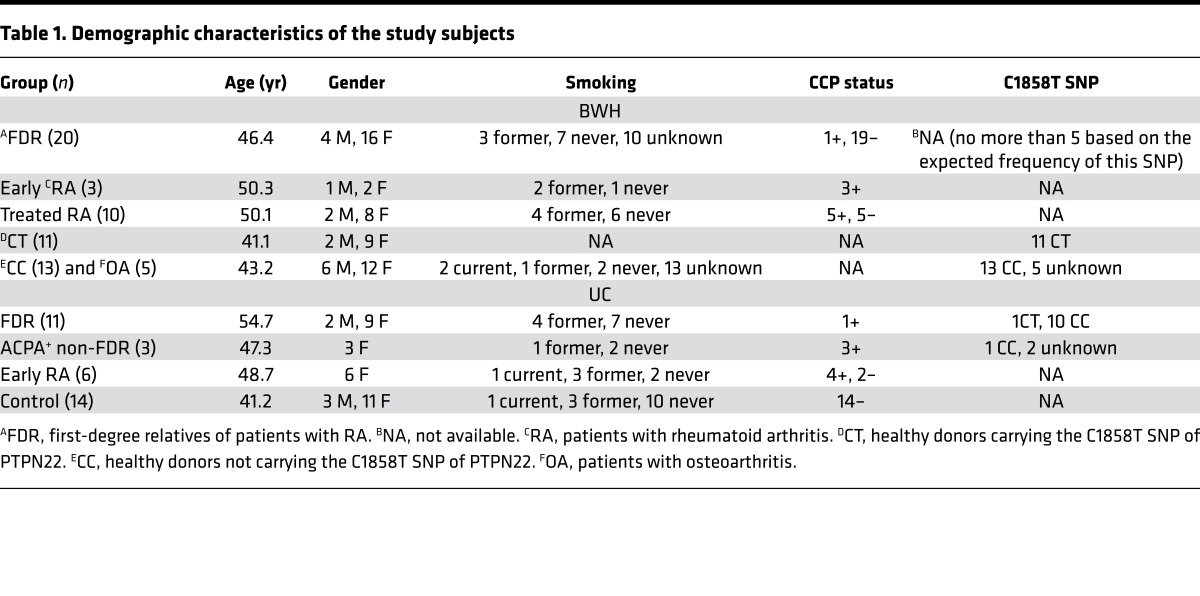
The observation that PBMCs from healthy donors carrying the C1858T SNP contain a high level of citrullinated proteins prompted us to postulate that hypercitrullination is a precondition of RA. We therefore set to determine if hypercitrullination was also detected in PBMCs from healthy FDRs of RA patients recruited through the Personalized Risk Estimator for Rheumatoid Arthritis Family Study conducted at Brigham and Women’s Hospital (BWH) (37). The demographic characteristics of FDRs are shown in Table 1 (BWH cohort). The level of citrullinated histone H3 (cit-H3) in PBMCs was examined with anti-cit-H3 antibody in Western blotting (Figure 1A). We chose cit-H3 as a readout of citrullinated proteins because we have found that the commercial anti-cit-H3 is sensitive and reliable. We also included PBMCs from healthy control donors carrying (CT donors) or not carrying (CC donors) the C1858T SNP (Figure 1A), osteoarthritis patients (Figure 1B), treated RA patients (Figure 1B), and early/untreated RA patients (Figure 1C). These donors were recruited through the PhenoGenetic Project and Partners HealthCare Biobank (38). The level of cit-H3 was quantified with densitometry and then normalized against that of total H3 (Figure 1D). We included in each Western blot 1–2 samples from other blots. By doing so, we were able to compare the level of normalized cit-H3 among experiments. We also included in the analysis 3 CC and 4 CT samples that were published previously (29). Consistent with our hypothesis, we found that PBMCs from nearly all FDRs had a high level of cit-H3, even higher than that of CT donors. By contrast, PBMCs from osteoarthritis patients had a low level of cit-H3 comparable to that of CC donors (Figure 1D). Ten of the FDRs were from ACPA+ RA probands and the other ten were from ACPA– RA probands. There was no difference in the level of cit-H3 between these two groups (Figure 1E). Only one FDR (#4) was positive for ACPA. Therefore, the hypercitrullination is unlikely to be related to ACPA status.
Table 1. Demographic characteristics of the study subjects.
Figure 1. Hypercitrullination in PBMCs of healthy first-degree relatives of rheumatoid arthritis patients recruited at Brigham and Women’s Hospital.
PBMCs obtained from healthy first-degree relatives (FDRs) (A), healthy donors carrying (CT) or not carrying (CC) the C1858T SNP (A and B), patients with osteoarthritis (OA) (B), treated rheumatoid arthritis (tRA) patients (B), and early/untreated rheumatoid arthritis (eRA) patients (C) were directly analyzed by Western blots for the level of citrullinated histone H3 (cit-H3) or total histone H3 (H3). The identity of each donor within each group is denoted with Arabic numerals. The level of cit-H3 was quantified with densitometry and normalized against that of H3. The normalized density of CT #4 in A was arbitrarily set as 1. The normalized density of cit-H3 from all donors is shown in D. The normalized cit-H3 levels of FDR of anti-citrullinated protein antibody (ACPA+) or ACPA– probands are shown in E. Statistical analysis was performed with 1-way ANOVA followed by multiple comparison tests (D) or with 2-tailed Student’s t test (E). The CC group was used as the control group. ***P < 0.001; ****P < 0.0001. The bars shown in D and E represent mean ± SD.
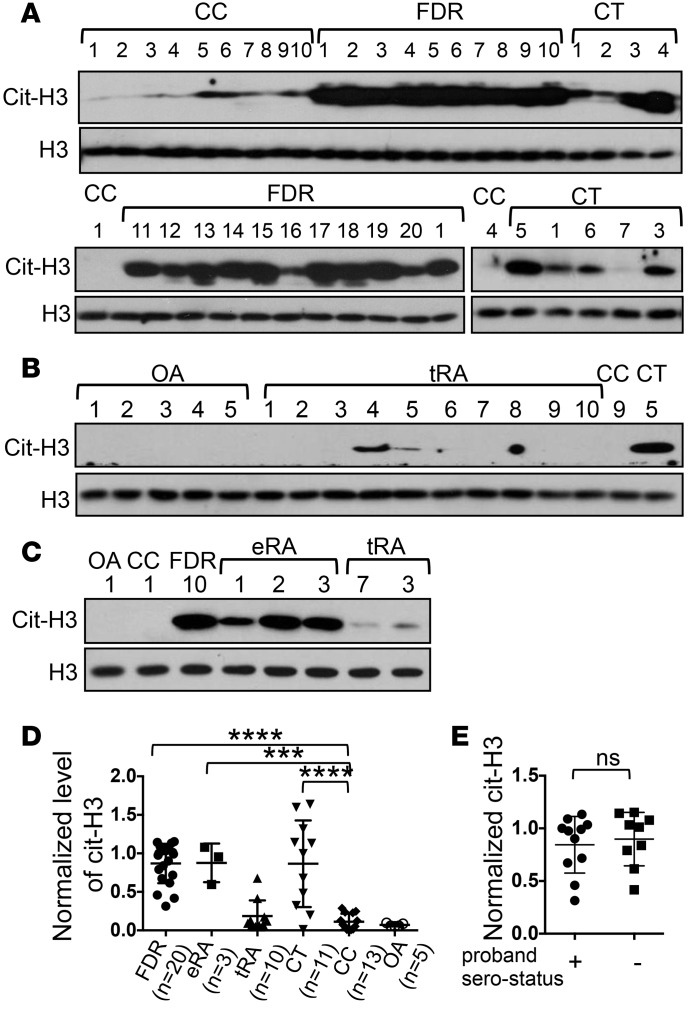
To further confirm our observation, we examined PBMCs from a different cohort of ARIs recruited through the Studies of the Etiology of Rheumatoid Arthritis at University of Colorado (UC cohort, Table 1). This cohort includes 11 healthy FDRs and 3 healthy ACPA+/non-FDR individuals. It also includes 14 healthy ACPA–/non-FDR donors (controls) and 6 early/untreated RA patients. One of the FDRs is also positive for ACPA (FDR #11). Again, we detected more cit-H3 in PBMCs from this cohort of ARIs compared with the 14 healthy controls (Figure 2, A and B). There was no difference in the level of cit-H3 between ACPA+ and ACPA– ARIs (Figure 2C). There was no known current smoker among ARIs in either the BWH or UC cohort. The genotype of the C1858T SNP was determined in 12 of the 14 ARIs of the UC cohort; only one ARI carried this SNP. While the genotype of the C1858T SNP was not determined in the FDRs of the BWH cohort, its frequency is approximately 20% in RA patients in North America, so there should be no more than 5 CT FDRs in the BWH cohort. All ARIs had no systemic inflammation at the time of blood draw. Therefore, the elevated level of cit-H3 in ARIs is unlikely to be due to smoking, the C1858T SNP, or systemic inflammation.
Figure 2. Hypercitrullination in PBMCs from healthy at-risk individuals recruited at the University of Colorado.
PBMCs from first-degree relatives (FDR), individuals positive for anti-citrullinated protein antibodies (ACPA+)/non-FDR (ACPA+), individuals with early rheumatoid arthritis (eRA), and controls were analyzed by Western blots for the level of citrullinated histone H3 (cit-H3) and total histone H3 (H3) (A). The density of cit-H3 was measured and normalized against that of H3. The normalized density of FDR donor #8 was arbitrarily set as 1. The normalized density of cit-H3 of all donors is shown in B. The normalized cit-H3 density from ACPA+ and ACPA– at-risk individuals (ARIs) is shown in C. Statistical analysis was performed with 1-way ANOVA followed by multiple comparison tests (B) and with 2-tailed Student’s t test (C). *P < 0.05. The bars shown in B and C represent mean ± SD.
If hypercitrullination in PBMCs is a precondition of RA, then we should expect to see hypercitrullination in early RA patients, particularly before any treatment. Consistent with our hypothesis, PBMCs from early RA patients also displayed hypercitrullination compared with controls (Figure 1, C and D, and Figure 2, A and B). Interestingly, no hypercitrullination was detected in 10 treated RA patients (Figure 1, B and D). These treated RA patients had a mean CDAI of 2.6. This observation strongly suggests that effective treatment reduces the level of cit-H3. In the following experiments, ARI samples from the BWH and UC cohorts were used indistinguishably. Early RA patients were not analyzed because they were treated soon after diagnosis.
Cellular sources of cit-H3 in PBMCs.
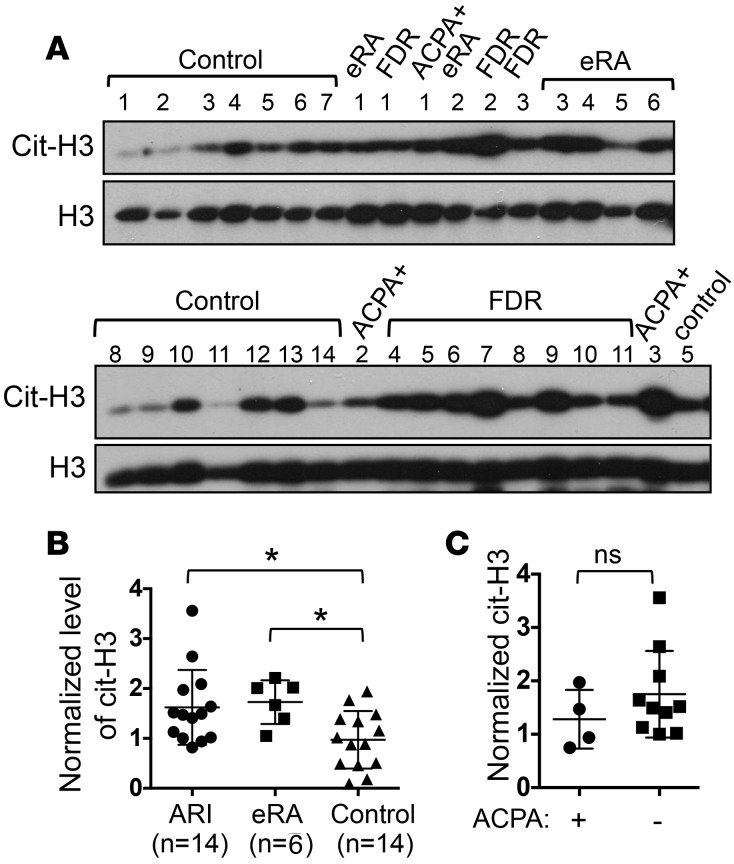
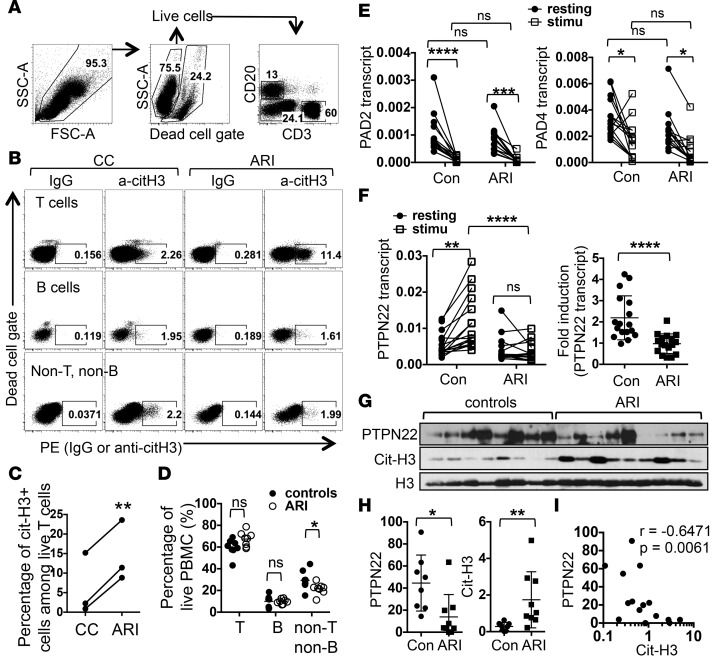
The hypercitrullination seen in ARI PBMCs can come from a single population or multiple populations of blood cells. To identify the source of cit-H3 in PBMCs, we decided to use intracellular staining to quantify the level of cit-H3 on a single-cell basis. We stimulated splenocytes collected from WT mice or mice deficient in PAD4 (PAD4KO), the only PAD bearing a nuclear localization signal, with PMA and then subjected the cells to intracellular staining of cit-H3. PMA increased the staining of cit-H3 in T, B, and non-T/non-B cells (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.90045DS1). This increase was almost completed abrogated in the absence of PAD4. There was still trace cit-H3 staining in PAD4KO splenic T cells. This residual staining is most likely caused by PAD2, which is also expressed in mouse T cells and can also be found in the nucleus (39). We then stained PBMCs from ARIs and control donors with anti-cit-H3 or control IgG. We excluded dead cells and used CD3 and CD20 to identify T, B, and non-T/non-B cells (Figure 3A). Despite interexperimental variations in the level of cit-H3 staining, we reproducibly detected more cit-H3 in T cells from ARIs compared with those from control donors (Figure 3, B and C). No difference in the level of cit-H3 between ARIs and control donors was observed in B and non-T/non-B cells, indicating that T cells are the major contributors of cit-H3 in PBMCs of ARIs.
Figure 3. Sources and causes of citrullinated histone H3 in PBMCs of at-risk individuals.
(A–D) PBMCs were subjected to intracellular staining with control IgG or anti–citrullinated histone H3 (anti-cit-H3). The gating strategy is shown in A. The cit-H3 staining of live T, B, and non-T/non-B cells is shown in B. The percentages of cit-H3+ T cells from 3 paired experiments are shown in C. The percentages of T, B, and non-T/non-B cells in PBMCs from 8 paired experiments are shown in D. (E and F) PBMCs from 14 at-risk individuals (ARIs) and 14 control donors (con) were left unstimulated or stimulated with anti-CD3 for 24 hours. Transcript levels of PAD2 and PAD4 (E) and PTPN22 (F) in PBMCs were measured with real-time PCR. Normalized values from the same donors are connected with lines. The fold induction of PTPN22 is shown in the right panel of F. (G–I) Anti-CD3 stimulated PBMCs from 9 ARIs and 8 control donors were analyzed with Western blotting for PTPN22, cit-H3, and total histone H3 (H3). The density of PTPN22 and cit-H3 was normalized against that of H3 and shown in H. The normalized PTPN22 values were plotted against normalized cit-H3 values and shown in I. Statistical analysis was performed with Student’s 2-tailed t test in C, D, the right panel of F, and H; 1-way ANOVA in E and the left panel of F; and Spearman’s test in I. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. The bars shown in H represent mean ± SD.
Impaired induction of PTPN22 in ARI PBMCs.
As T cells are the major source of cit-H3, it is possible that ARI PBMCs have a higher percentage of T cells, which causes the higher level of cit-H3. However, in the 3 pairs of donors analyzed in Figure 3A and 5 additional pairs of donors, we did not see any major difference in the distribution of T or B cells, even though the percentage of non-T/non-B cells was slightly lower in ARIs (Figure 3D). We then examined the expression of PAD2 and PAD4, two dominant PADs in hematopoietic cells (40). Interestingly, anti-CD3 stimulation led to a reduction in the transcript level of PAD2 and PAD4 (Figure 3E). However, the levels of PAD2 and PAD4 in either resting or stimulated PBMCs were very comparable between controls and ARIs.
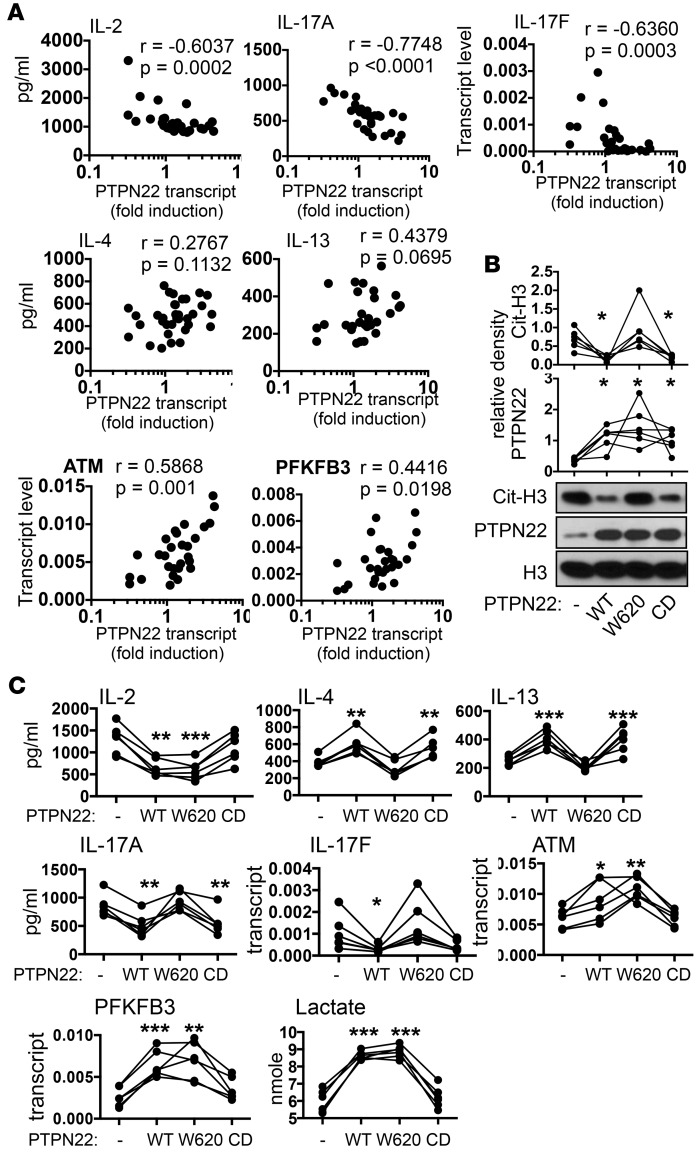
Figure 5. Phosphatase and nonphosphatase activities of PTPN22.
(A) The levels of cytokines and genes in PBMCs shown in Figure 4 were plotted against the fold induction of PTPN22 shown in the right panel of Figure 3F. Statistical analysis was performed with Spearman’s correlation test. (B and C) PBMCs from at-risk individuals were transfected with plasmid vector expressing PTPN22, W620-PTPN22, CD-PTPN22, or the empty vector (–), and then stimulated with anti-CD3 for 24 hours. The levels of PTPN22, citrullinated histone H3 (cit-H3), and total histone H3 (H3) in transfected/stimulated cells were quantified by Western blotting (B). Representative blots of 6 independent experiments are shown. The density of PTPN22 and cit-H3 was quantified with densitometry and normalized against that of H3. The normalized levels of PTPN22 and cit-H3 are shown. The expression of indicated cytokines and genes was measured with ELISA or real-time PCR (C). The concentration of lactate in supernatant was quantified with a colorimetric assay (C). The data values from the same donors were connected with lines. Statistical analysis for B and C was performed with 1-way ANOVA followed by multiple comparison tests using the empty vector–transfected groups as controls. *P < 0.05; **P < 0.01; ***P < 0.001.
An alternative explanation for the hypercitrullination seen in ARI PBMCs is impaired expression of PTPN22. We found that the level of PTPN22 transcript in resting PBMCs was comparable between controls and ARIs (Figure 3F). Anti-CD3 stimulation expectedly increased the level of PTPN22 transcript by almost 2-fold in control PBMCs. Surprisingly, no such induction was detected in ARIs PBMCs (Figure 3F). The level of PTPN22 transcript in half of the ARIs was actually reduced by anti-CD3 stimulation. There was no difference in the induction of PTPN22 between ACPA+ and ACPA– ARIs (Supplemental Figure 2). We were able to examine the level of PTPN22 protein after anti-CD3 stimulation in 9 ARIs and 8 controls (Figure 3G). Indeed, the level of PTPN22 protein was significantly lower in stimulated ARI PBMCs (Figure 3H). Contrarily, the level of cit-H3 in stimulated ARI PBMCs was higher than that in stimulated control PBMCs (Figure 3H). There was a reverse correlation between the level of PTPN22 and cit-H3 when ARIs and controls were analyzed together (Figure 3I). Those with a higher level of PTPN22 tends to have a lower level of cit-H3, suggesting that the impaired induction of PTPN22 contributes to the hypercitrullination.
Abnormal phenotype of ARI PBMCs.
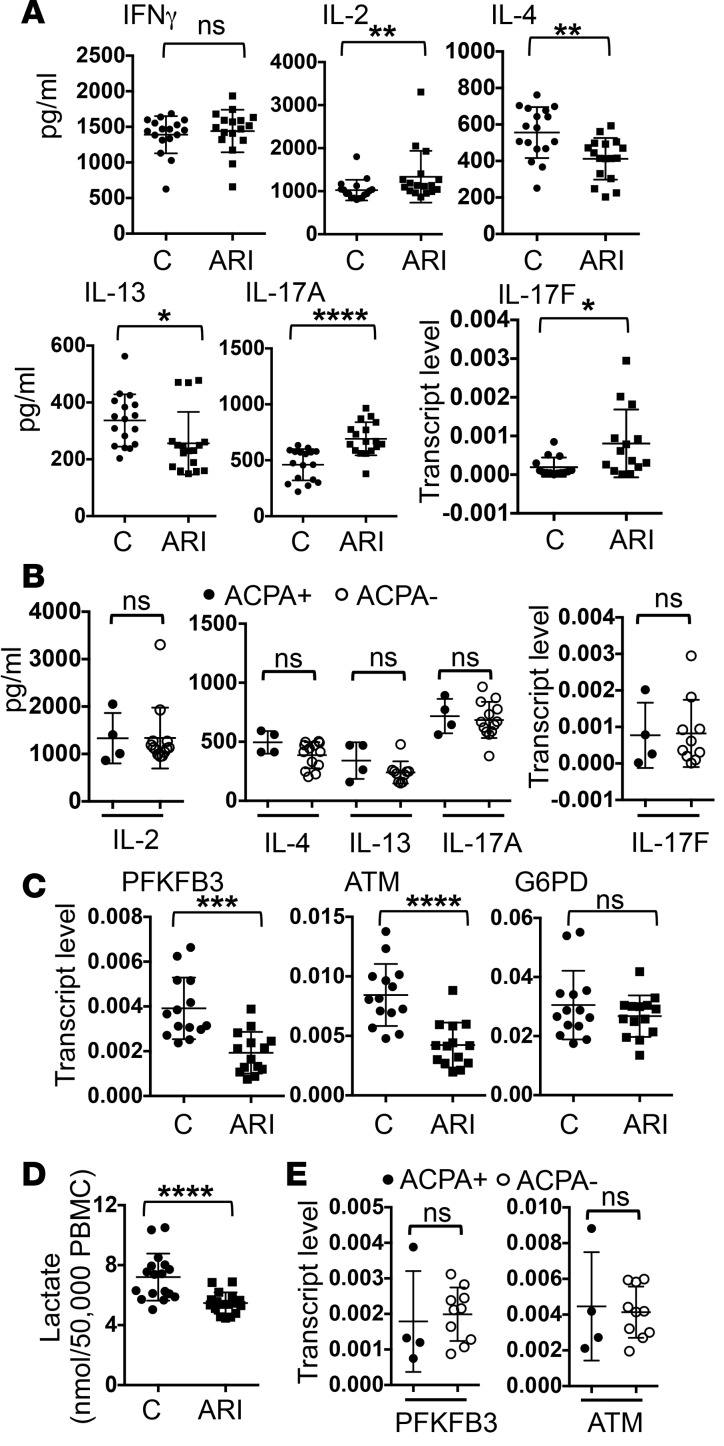
Th cytokines, such as IL-17, IL-4, and IFN-γ, play a critical role in the pathogenesis of RA and many other autoimmune diseases. To determine if ARI PBMCs have any defect in the expression of Th cytokines, the PBMCs were stimulated with anti-CD3 for 24 hours and the production of Th cytokines was measured with ELISA or real-time PCR. We found no difference in the level of IFN-γ between ARIs and controls (Figure 4A). Interestingly, ARI PBMCs produced more IL-2 and Th17 cytokines, including IL-17A and IL-17F, but almost 50% less Th2 cytokines, such as IL-4 and IL-13. There was no difference in cytokine production between ACPA+ and ACPA– ARIs (Figure 4B).
Figure 4. Abnormal phenotype of at-risk individual PBMCs.
PBMCs from control donors (C) and at-risk individuals (ARIs) (14–17 per group) were stimulated with anti-CD3 for 24 hours. The expression of indicated cytokines was quantified with ELISA or real-time PCR (A). The levels of cytokines from ARIs who are ACPA+ and ARIs who are ACPA– are compared in B. In addition, the transcript levels of PFKFB3, ATM, and G6PD measured with real-time PCR are shown in C, and the concentration of lactate in supernatant is shown in D. The transcript levels of PFKFB3 and ATM in ACPA+ and ACPA– ARIs are compared in E. Statistical analyses were performed with Student’s 2-tailed t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. The bars shown represent mean ± SD.
A recent study indicates that naive Th cells from ACPA+ RA patients express less PFKFB3 and ATM but more G6PD in response to stimulation (18). We found that PBMCs of ARIs also had a reduced level of PFKFB3 and ATM after stimulation (Figure 4C). Accordingly, PBMCs of ARIs generated less lactic acid upon stimulation (Figure 4D). The levels of PFKFB3, ATM, and lactate were comparable between ACPA+ and ACPA– ARIs (Figure 4E and data not shown), suggesting that these changes do not require the development of ACPA. Interestingly, the expression of G6PD was normal in ARIs (Figure 4C). This observation indicates that the aberrant expression of G6PD is not coupled to the attenuated expression of PFKFB3 and ATM and occurs independently of ACPA.
Distinct effects of phosphatase and nonphosphatase activities of PTPN22.
When we pooled ARI and control samples together, we found that the levels of IL-2, IL-17A, and IL-17F reversely correlated with the induction of PTPN22 (Figure 5A). There appeared to be a threshold effect for IL-2 and IL-17F. When the induction of PTPN22 dropped to below 1, the level of IL-2 and IL-17F started to rise. Contrarily, there was a positive correlation between the induction of PTPN22 and the level of ATM or PFKFB3. There was also a trend of positive correlation between Th2 cytokines and the induction of PTPN22, but this trend did not reach statistical significance.
These observations prompted us to investigate if impaired induction of PTPN22 was responsible for the phenotype of ARI PBMCs. We transfected PBMCs from 6 randomly selected ARIs with PTPN22 expression vector (Figure 5, B and C). The transfected cells were then stimulated with anti-CD3. In agreement with our hypothesis, forced expression of PTPN22 reduced the level of cit-H3 (Figure 5B); increased the level of Th2 cytokines, PFKFB3, ATM, and lactate; and reduced the level of IL-2 and Th17 cytokines (Figure 5C).
PTPN22 is a phosphatase and is known to attenuate activation signals in lymphocytes. Thus, impaired induction of PTPN22 is expected to augment activation signals in T cells. However, the reciprocal changes in Th2 and Th17 cytokines observed in ARI PBMCs cannot be explained by stronger activation signals in T cells. In addition to acting as a phosphatase, PTPN22 also has nonphosphatase activities, including suppressing citrullination and promoting TLR-induced expression of type I interferon (29, 41). To determine if PTPN22 shaped the phenotype of ARI PBMCs through its phosphatase or nonphosphatase activity, we also expressed W620-PTPN22 or a catalytic dead PTPN22 (CD-PTPN22) in ARI PBMCs. CD-PTPN22 carries two point mutations at the protein tyrosine phosphatase domain of PTPN22 and has little phosphatase activity (42). However, it is still fully capable of suppressing citrullination (29), whereas W620-PTPN22 retains the phosphatase activity but no longer has the nonphosphatase activities (29, 41). In agreement with our published data (29), CD-PTPN22 but not W620-PTPN22 reduced the level of cit-H3 in ARI PBMCs as efficiently as WT-PTPN22 (Figure 5B). In addition, CD-PTPN22 was able to normalize the level of Th2 and Th17 cytokines but not IL-2, ATM, PFKFB3, or lactate (Figure 5C). Although the effect of CD-PTPN22 on IL-17F did not reach statistical significance in 1-way ANOVA analysis, it carried a P value of 0.0487 when directly compared with empty vector control in Student’s t test analysis. By contrast, W620-PTPN22 was able to normalize the level of IL-2, ATM, PFKFB3, and lactate but not Th2 or Th17 cytokines (Figure 5C). These results indicate that the aberrant expression of IL-2, ATM, and PFKFB3 is caused by attenuated phosphatase activity of PTPN22, whereas the abnormal Th2 and Th17 cytokine profile is due to attenuated nonphosphatase activity of PTPN22.
PTPN22 regulates the Th2/Th17 cytokine profile by suppressing citrullination.
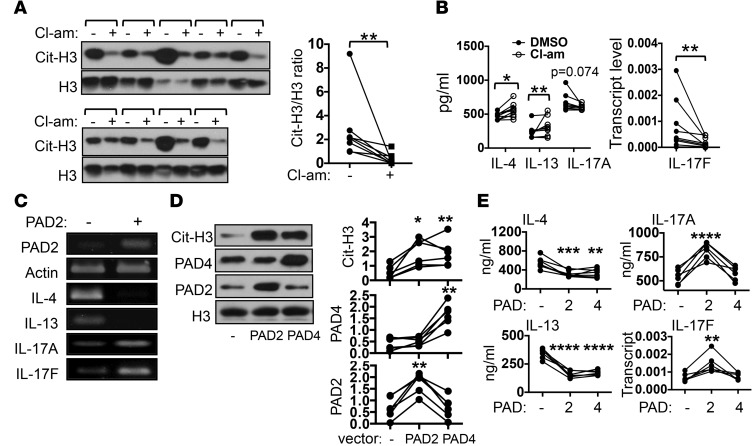
Attenuation of nonphosphatase activities of PTPN22 is expected to lead to hypercitrullination and impaired TLR-induced expression of type I interferon. The observation that normalization of Th2 and Th17 cytokines correlated with a reduction in the level of cit-H3 prompted us to postulate that hypercitrullination, but not dysregulated expression of type I interferon, is the cause of the abnormal Th2/Th17 cytokine profile of ARI PBMCs. To test this hypothesis, we randomly selected 9 ARIs and examined the Th2 and Th17 cytokine profile of their PBMCs after stimulation with anti-CD3 in the absence or presence of Cl-amidine, a pan-PAD inhibitor. Expectedly, Cl-amidine reduced the level of cit-H3 (Figure 6A). It also increased the level of IL-4 and IL-13 and decreased the level of IL-17F (Figure 6B). There was also a trend toward reducing the level of IL-17A. However, the effect of Cl-amidine treatment is rather modest. This modest effect may be expected considering that hypercitrullination already existed in ARI PBMCs before stimulation.
Figure 6. PTPN22 regulates the Th2/Th17 cytokine profile by suppressing citrullination.
(A and B) PBMCs from 9 at-risk individuals were stimulated with anti-CD3 in the presence or absence of Cl-amidine (Cl-am, 50 μM). The level of citrullinated histone H3 (cit-H3) and total histone H3 (H3) was analyzed by Western blotting (A). Each bracket represents one donor. The cit-H3/H3 density ratios from all donors are shown to the right. The levels of indicated cytokines are shown in B. (C) Jurkat cells expressing (+) or not expressing (–) a doxycycline-inducible PAD2 were treated with doxycycline (50 μM) for 12 hours. The transcript levels of indicated genes from 1 of 3 independent experiments are shown. (D and E) Control PBMCs were transfected with an empty vector (–) or a vector expressing PAD2 or PAD4 and then stimulated with anti-CD3. The levels of PAD2, PAD4, cit-H3, and H3 were measured by Western blotting (D). Representative blots from 6 independent experiments and normalized density of PAD2, PAD4, and cit-H3 are shown. The levels of indicated cytokines in the PBMCs are shown in E. Statistical analysis was performed with paired Student’s 2-tailed t test (A and B) or 1-way ANOVA using empty vector–transfected groups as controls (D and E). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We therefore took two additional approaches to further examine the impact of hypercitrullination on the expression Th cytokines. We generated a line of Jurkat cells that express an exogenous PAD2 in an inducible manner (43). We found that inducing the expression of PAD2 increased the expression of Th17 cytokines but inhibited the expression of Th2 cytokines (Figure 6C). In addition to PAD2, human PBMCs also express PAD4. Attenuated nonphosphatase activity of PTPN22 may lead to hypercitrullination through PAD2 and/or PAD4. To recapitulate the cytokine phenotype of ARI PBMCs and to distinguish between the effects of PAD2-mediated and PAD4-mediated hypercitrullination, we overexpressed PAD2 or PAD4 in control PBMCs, which were then stimulated with anti-CD3. Exogenous PAD2 and PAD4 equally increased the level of cit-H3 (Figure 6D) and comparably suppressed the expression of IL-4 and IL-13 (Figure 6E). However, only PAD2, but not PAD4, was able to enhance the expression of Th17 cytokines.
Discussion
A molecular signature of preclinical RA has emerged from our data (Supplemental Figure 3). Impaired induction of PTPN22 leads to attenuated phosphatase and nonphosphatase activities of PTPN22. Attenuated phosphatase activity of PTPN22 results in augmented expression of IL-2 but diminished expression of ATM and PFKFB3, which subsequently leads to hypoglycolysis. By contrast, attenuated nonphosphatase activity of PTPN22 causes hypercitrullination, which is responsible for aberrant production of Th2 and Th17 cytokines. PAD2- or PAD4-mediated hypercitrullination suppresses the expression of Th2 cytokines, whereas only PAD2-mediated hypercitrullination is able to augment the production of Th17 cytokines. All these molecular events very likely take place before the heightened expression of G6PD and independently of ACPA.
Unfortunately, there are only 4 ACPA+ ARIs in our study, a number that is probably too small to show any difference between ACPA+ and ACPA– ARIs. In addition, we were unable to examine the molecular signature in purified ARI Th cells, given the limited amount of blood that we were allowed to collect from each donor. It is possible that the molecular signature is influenced by non-T cells in PBMCs. This possible scenario may explain the normal expression of G6PD in ARIs. Recruiting more ACPA+ ARIs and collecting more blood from each ARI will be needed to address these issues.
The observation that PBMCs of ARIs contain more citrullinated proteins further strengthens the notion that hypercitrullination is a precondition of RA, regardless of the status of ACPA. Our data indicate that impaired induction of PTPN22, but not smoking, systemic inflammation, abnormal expression of PADs, or the C1858T SNP, is the cause of hypercitrullination in the ARIs of this study. Then, what is the cause of the impaired induction of PTPN22? PTPN22 is expressed mainly in hematopoietic cells, and its expression in T cells is induced after anti-CD3 stimulation in vitro. There is no other SNP, in addition to the C1858T within the PTPN22 gene, that carries a significant risk of RA. Thus, it is unlikely that ARIs share a SNP in the PTPN22 gene that prevents its induction by anti-CD3. We propose that yet-to-be identified environmental factors play a key role in regulating the expression of PTPN22. Recently, it was reported that gut and oral microbiomes are different between RA patients and healthy controls (44, 45). Specifically, Prevotella copri and Lactobacillus salivarius were overrepresented in RA patients, whereas Haemophilus spp. were depleted. These observations not only highlight the critical role of the environment in the pathogenesis of RA, but also raise the possibility that the microbiome may influence the expression of PTPN22. Thorough investigation into the molecular mechanism regulating the expression of PTPN22 will be the first step to test this hypothesis.
Our data indicate that phosphatase and nonphosphatase activities of PTPN22 are equally important in shaping the phenotype of PBMCs. However, the detailed mechanism of action of PTPN22 is still poorly understood. Attenuated phosphatase activity of PTPN22 is expected to strengthen the activation signals in lymphocytes (42, 46–48). This scenario can explain the higher level of IL-2. However, the impaired expression of ATM and PFKFB3, which are also induced by anti-CD3 stimulation, in ARI PBMCs cannot be explained by stronger activation signals and suggests a novel mechanism. The phosphatase activity of PTPN22 also inhibits the signals induced by type I interferon (49), modulates macrophage polarization (50), and activates the inflammasome by dephosphorylating NLRP3 (51). A role of NLRP3 in Th cells was recently discovered (52). It is possible that PTPN22 promotes the expression of PFKFB3 through the inflammasome in T cells. This hypothesis remains to be tested. Interestingly, several SNPs located between pfkfb3 and prkcq are associated with a higher risk of RA in genome-wide association studies (53, 54). Our finding that PTPN22 directly or indirectly regulates the expression of PFKFB3 further provides a molecular link between these two RA-associated genes.
Thus far, two nonphosphatase activities of PTPN22 have been identified: suppressing citrullination and promoting LPS-induced production of type I interferon by myeloid cells (29, 41). The observation that the abnormal Th2/Th17 cytokine profile of ARI PBMCs was partly normalized by a pan-PAD inhibitor and was recapitulated by overexpression of PADs strongly suggests that failure to suppress citrullination, but not failure to produce type I interferon, is the cause of the abnormal cytokine profile. In addition, the Th2/Th17 cytokine profile of ARI PBMCs is almost a mirror image of the ex vivo cytokine profile of mice treated with another pan-PAD inhibitor, BB-Cl-amidine (55). While it is still unclear how hypercitrullination causes the aberrant expression of Th2 and Th17 cytokines, our data have expanded the role of hypercitrullination in RA pathogenesis. Hypercitrullination not only enlarges the pool of citrullinated antigens, but also actively modulates the expression of Th cytokines.
We demonstrate again that PTPN22 is an inhibitor of citrullination. The C1858T SNP ablates this function of PTPN22, resulting in hypercitrullination and excessive production of citrullinated antigens. This mechanism can explain the synergy between the C1858T SNP and HLA shared epitopes in ACPA+ RA (56). Through hypercitrullination, this SNP also increases the propensity of the formation of NETs, which have been shown to play a pathogenic role in SLE and other autoimmune diseases (57, 58). This latter mechanism satisfactorily explains the association between the C1858T SNP and a higher risk of several other autoimmune diseases (59, 60), which do not have ACPA. It will be very interesting to examine the phenotype of PBMCs from healthy donors carrying this SNP. As the conversion of R620 to W620 alters mainly nonphosphatase activities of PTPN22, one would expect that those PBMCs should have abnormal expression of Th2 and Th17 cytokines but normal levels of IL-2, ATM, and PFKFB3.
Impaired induction of PTPN22 leads to augmented expression of Th17 cytokines, and only PAD2-mediated, but not PAD4-mediated, hypercitrullination recapitulates this feature. These data strongly suggest that PTPN22 also inhibits the activity of PAD2. This scenario remains to be confirmed. The latter observation also demonstrates that PAD2 and PAD4 have different roles in regulating the differentiation and function of Th cells. It is also consistent with previous reports showing that these two PAD enzymes have overlapping but not identical substrate spectra (15, 16). Accordingly, PAD2 deficiency should preferentially attenuate Th17 response. However, PAD2-deficient mice are still sensitive to experimental autoimmune encephalomyelitis (61), a model of multiple sclerosis that is heavily dependent on Th17 cells. This discrepancy may originate from the fundamental difference between gain-of-function and loss-of-function approaches or from the intrinsic difference between mice and humans. Identifying the substrates of PAD2 and PAD4 in mouse and human Th cells will clarify this issue.
Approximately 50% of the ARIs in our study had impaired induction of PTPN22. This finding is reminiscent of our recent observation showing that nearly 40% of ACPA– FDRs already had detectable levels of APCA in their sputum (62). It will be of great interest to examine if the presence of APCA in sputum correlates with the impaired induction of PTPN22 in PBMCs of ARIs. While the Th cytokine profile of ARIs is different from that of controls, the difference is modest overall. This is not surprising, given that all ARIs are free of systemic inflammation. However, the functional consequence of this modest difference is still unclear. The odds ratio for developing RA in our ARIs is 4 to 5. Accordingly, only approximately 5%–10% of ARIs are expected to develop RA. This discrepancy strongly suggests that the molecular signature we discovered in this study is necessary but not sufficient for the development of RA and that additional “hits” are needed. One potential candidate is heightened expression of G6PD. It is a feature of Th cells from ACPA+ RA patients and positively correlated with disease activity. However, the level of G6PD was normal in the ARIs of our study, suggesting that the heightened expression of G6PD is a late event that appears after the development of the molecular signature of preclinical RA. It remains to be determined if this feature occurs before or after the onset of clinical symptoms. Longitudinally studies following ARIs will be needed to test this hypothesis.
Methods
Human subjects.
PBMCs were obtained from the following sources: (a) BWH PhenoGenetic Project (38); (b) Partners HealthCare Biobank, an enterprise biobank of consented patient samples at Partners HealthCare (Massachusetts General Hospital and BWH), according to IRB-approved protocols; (c) Personalized Risk Estimator for Rheumatoid Arthritis Family Study, an NIH-funded prospective, randomized, controlled trial designed to evaluate whether personalized RA risk education affects willingness to change RA-related behaviors among unaffected FDRs of RA patients (37); (d) Profiling of Cell Subsets in Human Diseases, a research initiative of BWH comparing immune cells in blood from patients with or without inflammatory diseases; and (e) Studies of the Etiology of Rheumatoid Arthritis, a multicenter study designed to examine the role of environmental and genetic factors in the development and progression of RA-related autoimmunity (63).
ACPA status was determined with anti-CCP2 and anti-CCP3.1 ELISA assays (Diastat, Axis-Shield Diagnostics Ltd.).
Purification, stimulation, and transfection of PBMCs.
PBMCs were isolated from whole human peripheral blood by Ficoll-Paque PLUS (17-1440-03, GE Healthcare) density gradient centrifugation. Transfection was performed with Amaxa nucleofection (Amaxa Biosystems) according to the manufacturer’s instructions. Briefly, 2–2.5 million PBMCs were suspended in 100 μl of Human T cell Nucleofector solution (VPA-1002, Amaxa Biosystems) and transfected with 5 μg of plasmid DNA. PBMCs were plated in 24-well plates (2–2.5 million/1 ml/well) precoated with anti-CD3 (2.5 μg/ml, HIT3a clone, Biolegend) for 24 hours before harvesting.
Plasmid.
The expression vectors of WT and W620 PTPN22 were described previously (29). CD-PTPN22 (D195A/C227S) was generated through site-specific mutagenesis. The expression vectors for PAD2 and PAD4 were provided by Hyejeong Lee at Vanderbilt University (Nashville, Tennessee, USA) and Anthony Rosen at The Johns Hopkins School of Medicine (Baltimore, Maryland, USA) (64, 65), respectively.
Intracellular cit-H3 staining.
PBMCs were washed twice with 1% BSA in PBS, resuspended in 100 μl of 1% BSA in PBS, and incubated with anti-CD3 and anti-CD20 at 4°C for 30 minutes. The cells were washed twice with cold PBS, fixed with 100 μl of fixation buffer (eBioscience IC Fixation Buffer) at room temperature for 20 minutes in the dark, mixed with 2 ml permeabilization buffer (0.5% Triton X-100 in 1% BSA/PBS), centrifuged at 300 to 500 g and washed again with 2 ml permeabilization buffer. The washed cells were resuspended in 100 μl permeabilization buffer containing 1:300 rabbit IgG or anti-cit-H3 (ab5103, Abcam) at 4°C in the dark for 30 to 60 minutes, washed twice with 2 ml permeabilization buffer, and resuspended in 100 μl permeabilization buffer containing 1:400 goat anti-rabbit IgG-PE (sc-3739, Santa Cruz Biotechnology) at 4°C in the dark for 30 to 60 minutes. The stained cells were washed twice with 2 ml permeabilization buffer at 4°C and resuspended in PBS for flow cytometry.
Western blotting.
Whole cell extract was obtained by lysing cells with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% TritonX-100, 0.5% DOC, 0.1% SDS, and 1 mM EDTA) containing 0.5 mM PMSF and complete protease inhibitor cocktail (Roche). The following antibodies were used: human PTPN22 antibody (AF3428, R&D Systems); cit-H3 antibody (ab5103, Abcam); histone H3 antibody (601902, Biolegend); PAD4 antibody (ab50247, Abcam); and PAD2 antibody (ab50257, Abcam). Densitometry readings of Western blots were obtained and analyzed with UN-SCAN-IT 6.0 software (Silk Scientific Inc.) and normalized against those of loading controls (total histone H3).
Quantitative RNA analysis.
RNA isolation, reverse transcription, and real-time PCR were performed as described previously (66). The transcript levels detected were normalized against that of actin from the same sample. The sequences of primers used in quantitative PCR are listed in Supplemental Tables 1 and 2.
ELISA.
Sandwich ELISA was performed with the following antibodies: anti-human IL-2 (catalog 555051)/biotin-anti-human IL-2 (catalog 555040), and anti-human IFN-γ (catalog 551221)/biotin-anti-human IFN-γ (catalog 554550) from BD Pharmingen and anti-human IL-4 (catalog 14-7049)/biotin-anti-human IL-4 (catalog 13-7048), anti-human IL-13 (catalog 14-7139)/biotin-anti-human IL-13 (catalog 13-7138), and anti-human IL-17A (catalog 14-7178)/biotin-anti-human IL-17A (catalog 13-7179) from eBioscience.
Measurement of lactate concentration.
Lactate concentration was measured with the Lactate Assay Kit (K607-100, BioVision) according to the manufacturer’s instructions. Briefly, 50 μl of supernatant was mixed with 50 μl of the Reaction Mix (BioVision) for 30 minutes, and OD 570 nm was measured for colorimetric assay.
Statistics.
Statistical analyses were performed with 1-way ANOVA followed by multiple comparison tests, 2-tailed Student’s t test, or Spearman’s correlation test, as indicated in the figure legends. A P value less than 0.05 is considered significant.
Study approval.
This study has been approved by the Partners Human Research Committee, Boston, Massachusetts, USA, and the Colorado Multiple Institution Review Board, UC Denver, Anschutz Medical Campus, Aurora, Colorado, USA. Informed consent was obtained from participants prior to inclusion in the studies.
Author contributions
HHC, GYL, HCH, and ICH designed the experiments. HHC, ND, BS, GYL, and HCH conducted the experiments. PRT generated Cl-amidine. HHC, ND, BS, GYL, HCH, and ICH analyzed the data. YO, JDK, KDD, MKD, JMN, JAS, DAR, EWK, and VHM recruited donors and isolated PBMCs. HHC, GYL, PRT, VMH, and ICH wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank Miriam Shelef from the University of Wisconsin for providing us with lymphoid organs from PAD4-deficient mice. This work was supported by a bridge grant from BWH (to ICH), grants from the NIH (AR049880, AR052403, AR070253 , and AR047782 to EWK; AR069688 and AR066953 to JAS; AR051394 to VMH and JMN; AI101981 to VMH; AR07534 to JDK), the Rheumatology Research Foundation Scientist Development Award (to JAS), and The Ministry of Science and Technology, Taiwan (MOST 101-2311-B-005-005-MY3 to HCH and 102-2320-B-040-018-MY3 to GYL).
Footnotes
Jennifer D. Kinslow’s present address is: Department of Immunology/Microbiology, Rush University Medical Center, Chicago, Illinois, USA.
Conflict of interest: P. Thompson is a consultant for Bristol-Myers Squibb.
Reference information:JCI Insight. 2016;1(17):e90045. doi:10.1172/jci.insight.90045.
Contributor Information
Hui-Hsin Chang, Email: HCHANG0@PARTNERS.ORG.
Guang-Yaw Liu, Email: liugy@csmu.edu.tw.
Nishant Dwivedi, Email: NDWIVEDI@PARTNERS.ORG.
Bo Sun, Email: BSUN4@PARTNERS.ORG.
Yuko Okamoto, Email: YUKO.OKAMOTO@UCDENVER.EDU.
Jennifer D. Kinslow, Email: jenniferd.larson@gmail.com.
Kevin D. Deane, Email: kevin.deane@ucdenver.edu.
Jill M. Norris, Email: jill.norris@ucdenver.edu.
Jeffrey A. Sparks, Email: JASPARKS@PARTNERS.ORG.
Deepak A. Rao, Email: DARAO@PARTNERS.ORG.
Elizabeth W. Karlson, Email: EKARLSON@PARTNERS.ORG.
Hui-Chih Hung, Email: hchung@dragon.nchu.edu.tw.
V. Michael Holers, Email: Michael.Holers@ucdenver.edu.
I-Cheng Ho, Email: iho@partners.org.
References
- 1.Demoruelle MK, Deane K. Antibodies to citrullinated protein antigens (ACPAs): clinical and pathophysiologic significance. Curr Rheumatol Rep. 2011;13(5):421–430. doi: 10.1007/s11926-011-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uysal H, et al. Antibodies to citrullinated proteins: molecular interactions and arthritogenicity. Immunol Rev. 2010;233(1):9–33. doi: 10.1111/j.0105-2896.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- 3.Bicker KL, Thompson PR. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers. 2013;99(2):155–163. doi: 10.1002/bip.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makrygiannakis D, et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann Rheum Dis. 2008;67(10):1488–1492. doi: 10.1136/ard.2007.075192. [DOI] [PubMed] [Google Scholar]
- 5.Damgaard D, Friberg Bruun Nielsen M, Quisgaard Gaunsbaek M, Palarasah Y, Svane-Knudsen V, Nielsen CH. Smoking is associated with increased levels of extracellular peptidylarginine deiminase 2 (PAD2) in the lungs. Clin Exp Rheumatol. 2015;33(3):405–408. [PubMed] [Google Scholar]
- 6.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun. 1999;67(7):3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenstein ED, Greenwald RA, Kushner LJ, Weissmann G. Hypothesis: the humoral immune response to oral bacteria provides a stimulus for the development of rheumatoid arthritis. Inflammation. 2004;28(6):311–318. doi: 10.1007/s10753-004-6641-z. [DOI] [PubMed] [Google Scholar]
- 8.Mikuls TR, et al. Porphyromonas gingivalis and disease-related autoantibodies in individuals at increased risk of rheumatoid arthritis. Arthritis Rheum. 2012;64(11):3522–3530. doi: 10.1002/art.34595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Smit M, Westra J, Vissink A, Doornbos-van der Meer B, Brouwer E, van Winkelhoff AJ. Periodontitis in established rheumatoid arthritis patients: a cross-sectional clinical, microbiological and serological study. Arthritis Res Ther. 2012;14(5):e90045. doi: 10.1186/ar4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutger Persson G, Rheumatoid arthritis and periodontitis — inflammatory and infectious connections. Review of the literature. J Oral Microbiol. 2012;4:e90045. doi: 10.3402/jom.v4i0.11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki A, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 12.Kang CP, Lee HS, Ju H, Cho H, Kang C, Bae SC. A functional haplotype of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Koreans. Arthritis Rheum. 2006;54(1):90–96. doi: 10.1002/art.21536. [DOI] [PubMed] [Google Scholar]
- 13.Hou S, et al. PADI4 polymorphisms and susceptibility to rheumatoid arthritis: a meta-analysis. Mod Rheumatol. 2013;23(1):50–60. doi: 10.3109/s10165-012-0639-4. [DOI] [PubMed] [Google Scholar]
- 14.Hoppe B, et al. Influence of peptidylarginine deiminase type 4 genotype and shared epitope on clinical characteristics and autoantibody profile of rheumatoid arthritis. Ann Rheum Dis. 2009;68(6):898–903. doi: 10.1136/ard.2008.091983. [DOI] [PubMed] [Google Scholar]
- 15.Darrah E, Rosen A, Giles JT, Andrade F. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann Rheum Dis. 2012;71(1):92–98. doi: 10.1136/ard.2011.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensen SM, Pruijn GJ. Methods for the detection of peptidylarginine deiminase (PAD) activity and protein citrullination. Mol Cell Proteomics. 2014;13(2):388–396. doi: 10.1074/mcp.R113.033746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210(10):2119–2134. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci Transl Med. 2016;8(331):e90045. doi: 10.1126/scitranslmed.aad7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rantapää-Dahlqvist S, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003;48(10):2741–2749. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 20.Nielen MM, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004;50(2):380–386. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 21.Esposito G, et al. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273(1–2):25–31. doi: 10.1016/j.mce.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Yurttas P, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135(15):2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadler SC, et al. Dysregulation of PAD4-mediated citrullination of nuclear GSK3β activates TGF-β signaling and induces epithelial-to-mesenchymal transition in breast cancer cells. Proc Natl Acad Sci U S A. 2013;110(29):11851–11856. doi: 10.1073/pnas.1308362110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christophorou MA, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507(7490):104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leshner M, et al. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front Immunol. 2012;3:e90045. doi: 10.3389/fimmu.2012.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184(2):205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207(9):1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khandpur R, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5(178):e90045. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HH, Dwivedi N, Nicholas AP, Ho IC. The W620 polymorphism in PTPN22 disrupts its interaction with peptidylarginine deiminase type 4 and enhances citrullination and NETosis. Arthritis Rheumatol. 2015;67(9):2323–2334. doi: 10.1002/art.39215. [DOI] [PubMed] [Google Scholar]
- 30.Kunz M, Ibrahim SM. Non-major histocompatibility complex rheumatoid arthritis susceptibility genes. Crit Rev Immunol. 2011;31(2):99–114. doi: 10.1615/CritRevImmunol.v31.i2.20. [DOI] [PubMed] [Google Scholar]
- 31.Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orozco G, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52(1):219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 33.Lee AT, et al. The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun. 2005;6(2):129–133. doi: 10.1038/sj.gene.6364159. [DOI] [PubMed] [Google Scholar]
- 34.Koumantaki Y, et al. Family history as a risk factor for rheumatoid arthritis: a case-control study. J Rheumatol. 1997;24(8):1522–1526. [PubMed] [Google Scholar]
- 35.Hemminki K, Li X, Sundquist J, Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009;60(3):661–668. doi: 10.1002/art.24328. [DOI] [PubMed] [Google Scholar]
- 36.Arkema EV, et al. Anti-citrullinated peptide autoantibodies, human leukocyte antigen shared epitope and risk of future rheumatoid arthritis: a nested case-control study. Arthritis Res Ther. 2013;15(5):e90045. doi: 10.1186/ar4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sparks JA, et al. Personalized Risk Estimator for Rheumatoid Arthritis (PRE-RA) Family Study: rationale and design for a randomized controlled trial evaluating rheumatoid arthritis risk education to first-degree relatives. Contemp Clin Trials. 2014;39(1):145–157. doi: 10.1016/j.cct.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, et al. A 17q12 allele is associated with altered NK cell subsets and function. J Immunol. 2012;188(7):3315–3322. doi: 10.4049/jimmunol.1102775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cherrington BD, Morency E, Struble AM, Coonrod SA, Wakshlag JJ. Potential role for peptidylarginine deiminase 2 (PAD2) in citrullination of canine mammary epithelial cell histones. PLoS One. 2010;5(7):e90045. doi: 10.1371/journal.pone.0011768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Wang Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim Biophys Acta. 2013;1829(10):1126–1135. doi: 10.1016/j.bbagrm.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity. 2013;39(1):111–122. doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, Jallal B. The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol. 2002;30(3):237–244. doi: 10.1016/S0301-472X(01)00794-9. [DOI] [PubMed] [Google Scholar]
- 43.Hsu PC, Liao YF, Lin CL, Lin WH, Liu GY, Hung HC. Vimentin is involved in peptidylarginine deiminase 2-induced apoptosis of activated Jurkat cells. Mol Cells. 2014;37(5):426–434. doi: 10.14348/molcells.2014.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 45.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e90045. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gjörloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol. 1999;29(12):3845–3854. doi: 10.1002/(SICI)1521-4141(199912)29:12<3845::AID-IMMU3845>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat Struct Biol. 2001;8(11):998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, et al. Identification of substrates of human protein-tyrosine phosphatase PTPN22. J Biol Chem. 2006;281(16):11002–11010. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 49.Holmes DA, et al. Autoimmunity-associated protein tyrosine phosphatase PEP negatively regulates IFN-α receptor signaling. J Exp Med. 2015;212(7):1081–1093. doi: 10.1084/jem.20142130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang HH, et al. PTPN22 modulates macrophage polarization and susceptibility to dextran sulfate sodium-induced colitis. J Immunol. 2013;191(5):2134–2143. doi: 10.4049/jimmunol.1203363. [DOI] [PubMed] [Google Scholar]
- 51.Spalinger MR, et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest. 2016;126(5):1783–1800. doi: 10.1172/JCI83669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arbore G, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science. 2016;352(6292):e90045. doi: 10.1126/science.aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coenen MJ, et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum Mol Genet. 2009;18(21):4195–4203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 54.Raychaudhuri S, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawalkowska J, et al. Abrogation of collagen-induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci Rep. 2016;6:e90045. doi: 10.1038/srep26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kallberg H, et al. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet. 2007;80(5):867–875. doi: 10.1086/516736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lood C, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22(2):146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grayson PC, Kaplan MJ. At the Bench: Neutrophil extracellular traps (NETs) highlight novel aspects of innate immune system involvement in autoimmune diseases. J Leukoc Biol. 2016;99(2):253–264. doi: 10.1189/jlb.5BT0615-247R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kyogoku C, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75(3):504–507. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Criswell LA, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76(4):561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raijmakers R, et al. Experimental autoimmune encephalomyelitis induction in peptidylarginine deiminase 2 knockout mice. J Comp Neurol. 2006;498(2):217–226. doi: 10.1002/cne.21055. [DOI] [PubMed] [Google Scholar]
- 62.Willis VC, et al. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum. 2013;65(10):2545–2554. doi: 10.1002/art.38066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hughes-Austin JM, et al. Multiple cytokines and chemokines are associated with rheumatoid arthritis-related autoimmunity in first-degree relatives without rheumatoid arthritis: Studies of the Aetiology of Rheumatoid Arthritis (SERA) Ann Rheum Dis. 2013;72(6):901–907. doi: 10.1136/annrheumdis-2012-201505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee HJ, et al. Peptidylarginine deiminase 2 suppresses inhibitory {kappa}B kinase activity in lipopolysaccharide-stimulated RAW 264.7 macrophages. J Biol Chem. 2010;285(51):39655–39662. doi: 10.1074/jbc.M110.170290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris ML, et al. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum. 2008;58(7):1958–1967. doi: 10.1002/art.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang HH, et al. PTPN22.6, a dominant negative isoform of PTPN22 and potential biomarker of rheumatoid arthritis. PLoS One. 2012;7(3):e90045. doi: 10.1371/journal.pone.0033067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.