Abstract
Mutations of the Plekhm1 gene in humans and rats cause osteopetrosis, an inherited bone disease characterized by diminished bone resorption by osteoclasts. PLEKHM1 binds to RAB7 and is critical for lysosome trafficking. However, the molecular mechanisms by which PLEKHM1 regulates lysosomal pathways remain unknown. Here, we generated germline and conditional Plekhm1-deficient mice. These mice displayed no overt abnormalities in major organs, except for an increase in trabecular bone mass. Furthermore, loss of PLEKHM1 abrogated the peripheral distribution of lysosomes and bone resorption in osteoclasts. Mechanistically, we indicated that DEF8 interacts with PLEKHM1 and promotes its binding to RAB7, whereas the binding of FAM98A and NDEL1 with PLEKHM1 connects lysosomes to microtubules. Importantly, suppression of these proteins results in lysosome positioning and bone resorption defects similar to those of Plekhm1-null osteoclasts. Thus, PLHKEM1, DEF8, FAM98A, and NDEL1 constitute a molecular complex that regulates lysosome positioning and secretion through RAB7.
PLEKHM1 regulates lysosome positioning and osteoclastic bone resorption through a novel complex and is indispensable for bone homeostasis.
Introduction
Osteoclasts are multinucleated cells that are capable of resorbing calcified cartilage and bone matrix during skeletal development, homeostasis, and repair (1). Increased osteoclast number or activity leads to bone loss in metabolic bone diseases, such as postmenopausal osteoporosis (2). Conversely, defects in osteoclast differentiation or function cause accumulation of structurally disorganized bone in osteopetrosis, an inherited marble bone disease (3). Osteoclasts dissolve the bone mineral and digest organic bone matrix by secreting hydrochloric acid and hydrolases through the ruffled border, a highly convoluted plasma membrane structure circumscribed by the sealing zone (4). Genetic and cell biology studies in humans and rodents with osteopetrosis have uncovered key components of the osteoclast acid-secreting machinery, including the a3 subunit of vacuolar proton pumps, the CLC-7 chloride channel, and osteopetrosis-associated transmembrane protein 1 (OSTM1), a β subunit of CLC-7 (3). Notably, these proteins are enriched in the lysosomal membrane, and they translocate to the ruffled border in active osteoclasts (5, 6). Cathepsin K (CTSK), the major lysosomal acidic hydrolase in osteoclasts, is also secreted into the resorption lacuna beneath the ruffled border (7). These findings indicate that lysosome secretion is a major pathway of osteoclast bone resorption (8).
Lysosomes are membrane-bound organelles with versatile functions in cellular clearance, nutrition-sensing/mTOR pathway, and lipid and energy metabolism (9). Traditionally, lysosomes are considered as intracellular degradative compartments and the endpoint of endocytic and autophagic pathways (10). It has become increasingly evident that both conventional lysosomes and various specialized lysosome-related organelles or secretory lysosomes can undergo regulated exocytosis upon stimulation in a number of cell types, especially melanocytes, and cells of hematopoietic origin (11, 12). Lysosome secretion plays an essential role in cellular entry of bacteria and parasites, immune responses, pigmentation, membrane repair, and platelet aggregation. Cellular and molecular studies of Griscelli’s syndrome and its corresponding murine models, Ashen, Dilute, and Leaden mice, have uncovered the key molecular machinery of lysosome secretion (13–15). The small GTPase RAB27 plays a central role, but the precise components of the secretory apparatus vary in a cell type–specific manner. For instance, a RAB27/melanophilin/myosin Va complex is indispensable for melanosome transportation and release in melanocytes (16, 17). RAB27/MUNC13-4, on the other hand, is essential for lysosome secretion in cytotoxic T cells, mast cells, and platelets (18).
Osteoclasts, like most hematopoietic cells, contain secretory lysosomes (19). However, mechanisms underlying lysosome secretion in osteoclasts seem to be unique. A recent study on osteoclasts derived from Ashen mice demonstrates that RAB27A does not directly regulate lysosome secretion during bone resorption (20). Moreover, Chocolate mice harboring a spontaneous mutation in RAB38, which is essential for the biogenesis of lysosome-related organelles, have normal osteoclast differentiation and function (21). RAB7, a protein specifically associated with late endosomes/lysosomes and phagosomes, is localized at the ruffled border of osteoclasts (22). Downregulation of RAB7 expression in osteoclasts impairs ruffled border formation and bone resorption (23). These lines of evidence suggest that lysosome secretion in osteoclasts is regulated by RAB7.
Mutations in the Plekhm1 gene cause osteopetrosis in humans and incisors absent rats (24). Plekhm1-deficient human and rat osteoclasts exhibit diminished bone resorption and an intrinsic defect in ruffled border formation associated with the accumulation of intracellular lysosomes, suggesting that PLEKHM1 is a critical regulator of intracellular lysosomal trafficking and secretion in osteoclasts (24, 25). However, the molecular mechanisms and the step(s) at which PLEKHM1 acts on lysosome secretion in osteoclasts remain unclear. To elucidate the mechanisms by which PLEKHM1 regulates lysosomal secretion, we generated mouse strains with germline and conditional deletion of Plekhm1 using LoxP-Cre technology. We provide herein evidence demonstrating that PLEKHM1 is critical for bone homeostasis. Furthermore, data from a series of cellular and molecular experiments have uncovered that PLEKHM1 is an essential lysosomal adaptor protein regulating lysosome positioning by virtue of its interactions with differentially expressed in FDCP myeloid progenitor cells 8 (DEF8), RAB7, FAM98A, and NDEL1. Nonetheless, the physiological function of DEF8 is unknown (26). It promotes PLEKHM1’s binding to RAB7 through its Rubicon homology (RH) domain (27), FAM98A contains a tubulin-binding calponin homology domain (28), and NDEL1 regulates the activity of microtubule-based motor complexes, cytoplasmic dynein, and kinesins (29, 30). These findings elucidate a molecular complex linking lysosomes to microtubules for lysosome transportation/secretion and bone resorption in osteoclasts.
Results
PLEKHM1 is indispensable for skeletal homeostasis.
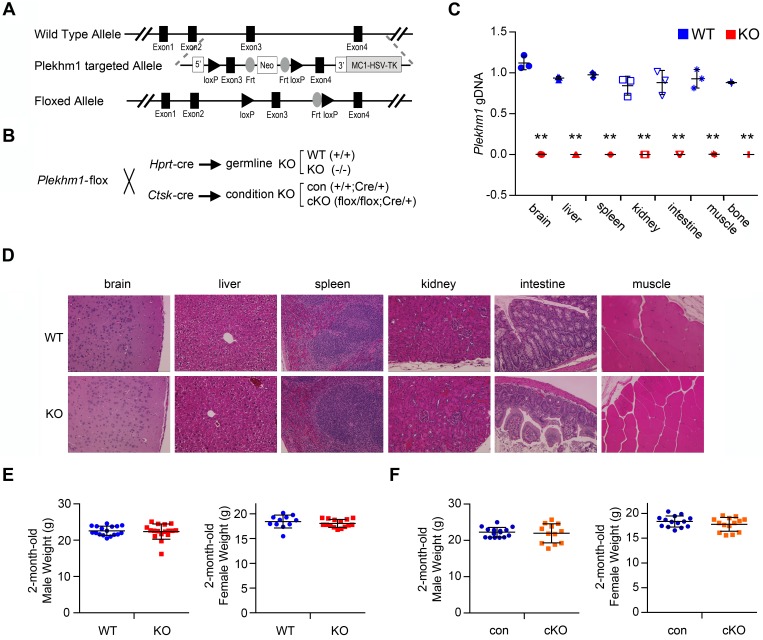
To elucidate the mechanisms by which PLEKHM1 regulates lysosomal pathways, we generated Plekhm1-floxed mice harboring a conditional allele, in which exon 3 of murine Plekhm1 gene was flanked by 2 loxP sites using homologous recombination (Figure 1A). The germline and conditional deletions of Plekhm1 in a C57BL/6 mouse background were generated by crossing Plekhm1-floxed mice with hypoxanthine guanine phosphoribosyl transferase–Cre (Hprt-Cre) and Ctsk-Cre mice, respectively (Figure 1B) (31, 32). Hprt-Cre eliminated Plekhm1 genomic DNA globally (Figure 1C). Ctsk-Cre markedly deleted Plekhm1 in mature osteoclasts, as reported previously (32), but it was less efficient in deleting Plekhm1 from preosteoclasts and osteocytes and had no effect on Plekhm1 in soft tissues and cultured osteoblasts (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.86330DS1). A slight deletion of Plekhm1 by Ctsk-Cre in osteocyte-enriched tibia cortical bone, probably due to the expression of Ctsk in some osteocytes undergoing canaliculae remodeling (33), had no effects on cortical bone thickness (Supplemental Table 2). Both strains of Plekhm1-deficient mice had tooth eruption, developed normally, and were fertile. Histologic analysis of organs that are often affected by lysosomal storage diseases and autophagy defects, such as brain, intestine, kidney, liver, muscle, and spleen, demonstrated normal tissue structures in Plekhm1–/– mice (Figure 1D). Loss of Plekhm1 had no effects on body weight (Figure 1, E and F).
Figure 1. Generation and characterization of Plekhm1 germline and conditional deletion mice.
(A) Strategy to generate a Plekhm1-flox allele. (B) Breeding strategy to generate Plekhm1 wild-type, germline deletion (KO), control (con), and conditional deletion (cKO) mice using Plekhm1-floxed mice and respective Cre-expressing mice. Hprt, hypoxanthine guanine phosphoribosyl transferase; Ctsk, cathepsin K. (C) Quantitative real-time PCR of exon 3 of murine Plekhm1 genomic DNA, normalized to the transferrin receptor 1 locus, using genomic DNA isolated from the indicated soft tissues and tibia (mean ± SD, n = 3). **P < 0.01 versus wild type by Student’s t test. (D) H&E staining of paraffin-embedded tissue sections of wild-type and Plekhm1 germline deletion mice. Representative images of each tissue from 3 different mice/genotype are shown. Original magnification: ×100. (E and F) Scatter dot plot presentations of body weight of 2-month-old male (WT, n = 17; KO, n = 17; con, n = 15; cKO, n = 12) and female mice (WT, n = 11; KO, n = 15; con, n = 14; cKO, n = 14). The mean and SD of each group are overlaid onto each column of dots.
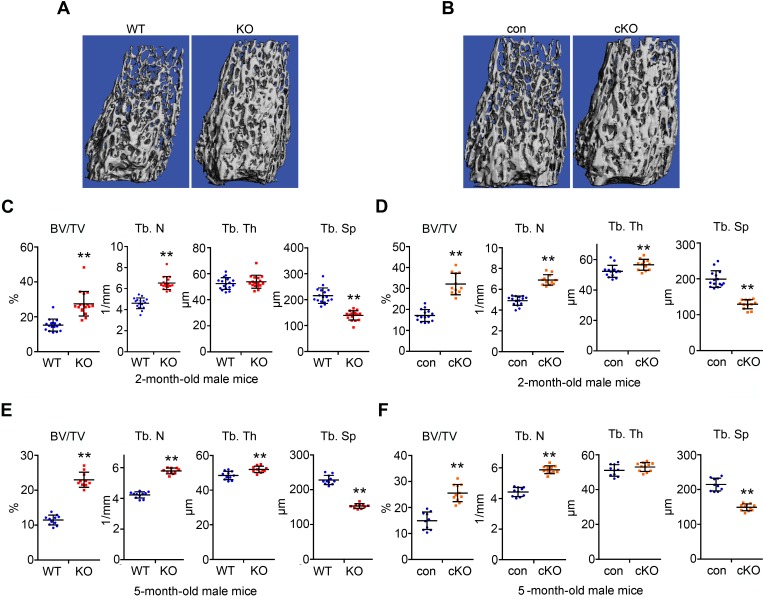
Male and female Plekhm1-deficient mice at 2 months of age exhibited a greater than 60% increase in trabecular bone mass in long bones and vertebrae. This increase was associated with greater trabecular number and decreased trabecular separation, as measured by micro-CT (Figure 2, A–D; Supplemental Figure 2, A–D; and Supplemental Table 1). Loss of Plekhm1 had less effect on cortical bone (Supplemental Table 2). Because the osteopetrotic phenotype in incisors absent rats diminishes as the rats age, we examined the skeletal phenotype in 5-month-old Plekhm1-deficient mice. In contrast to incisors absent rats, increased bone mass persisted in the 5-month-old male and female Plekhm1 germline and conditional deletion mice (Figure 2, E and F, and Supplemental Figure 2, E and F).
Figure 2. Plekhm1 germline and conditional deletion male mice have increased trabecular bone mass at 2- and 5-month-old of age.
(A and C) μ-CT images and analyses of the trabecular bone compartment of the distal femurs of 2-month-old male wild-type and germline deletion (KO) mice (n = 17). (B and D) μ-CT images and analyses of the trabecular bone compartment of the distal femurs of 2-month-old male control (con) and Plekhm1 conditional deletion (cKO) mice (con, n = 15; cKO, n = 12). (E and F) μ-CT analyses of the trabecular bone compartment of the distal femurs of 5-month-old male mice (WT, n = 11; KO, n = 10; con, n = 10; cKO, n = 11). BV/TV, percentage of trabecular bone volume to tissue volume; Tb. N, trabecular number; Tb. Th, trabecular thickness; Tb.Sp, trabecular separation. Data in C–F are presented as scatter dot plots. The mean and SD of each group are overlaid to each column of dots. **P < 0.01 versus WT or con by Student’s t test.
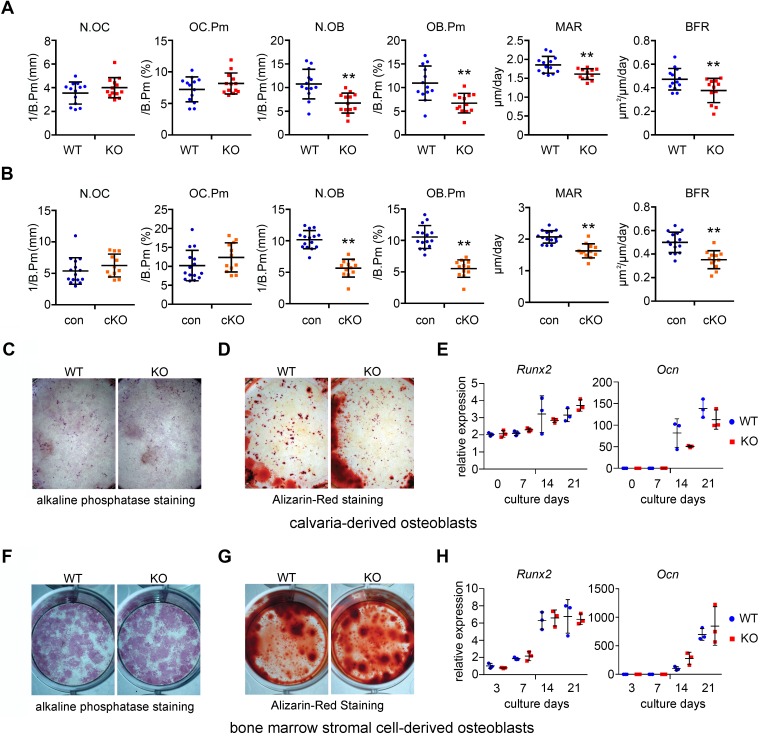
We next performed histomorphometry analysis of femurs of 2-month-old male Plekhm1-deficient mice and their respective controls. Similar to micro-CT data, we observed an increase in trabecular bone mass in both strains of Plekhm1-deficient mice (Supplemental Table 3). Osteoclast number and surface in Plekhm1-deficient mice were similar to those in control mice, suggesting osteoclast differentiation was normal in Plekhm1–/– mice (Figure 3, A and B). Osteoblast number and surface were slightly decreased in Plekhm1–/– mice. Consistent with these findings, the bone formation and mineral apposition rates were low in Plekhm1-deficient mice (Figure 3, A and B). Osteoblast differentiation and bone matrix deposition in vitro were normal, as demonstrated by alkaline phosphatase staining and alizarin red bone nodule staining (Figure 3, C–G). The mRNA expression of osteoblast marker genes, Runx2 and osteocalcin, was indistinguishable between wild-type and Plekhm1-knockout osteoblast cultures (Figure 3, E and H). Therefore, the decreased osteoblast number and bone formation rate in vivo were likely secondary to decreased coupling of bone formation to bone resorption during bone remodeling, rather than cell-autonomous effects of Plekhm1 deficiency on osteoblasts.
Figure 3. Plekhm1 deficiency does not change osteoclast number in vivo but results in decreased bone formation in a non-cell-autonomous manner.
(A and B) Histomorphometry analyses of osteoclast and osteoblast parameters of the femurs of 2-month-old male wild-type, germline knockout (KO), control (con), and conditional knockout (cKO) mice. N.OC, number of osteoclasts; OC.Pm, osteoclast perimeter; N.OB, number of osteoblasts; OB.Pm, osteoblast perimeter; MAR, bone mineral apposition rate; BFR, bone formation rate. The data are presented as scatter dot plots (WT, n = 13; KO, n = 12–13; con, n = 15; cKO, n = 12). The mean and SD of each group are overlaid onto each column of dots. **P < 0.01 versus WT or con by Student’s t test. (C and F) Alkaline phosphatase staining of 14-day cultures of osteoblasts. (D and G) Alizarin red staining of bone nodules of 21-day cultures of osteoblasts. Images are representatives of 3 wells/genotype from 3 independent experiments. Original magnification, ×1. (E and H) qPCR detection of Runx2 and osteocalcin (Ocn) mRNA expression during osteoblast differentiation (n = 3). (C–E) Osteoblasts from calvarias in a 6-well plate. (F–H) Osteoblasts from bone marrow stromal cells in a 12-well plate.
PLEKHM1 plays an intrinsic role in osteoclast function but not differentiation in vitro.
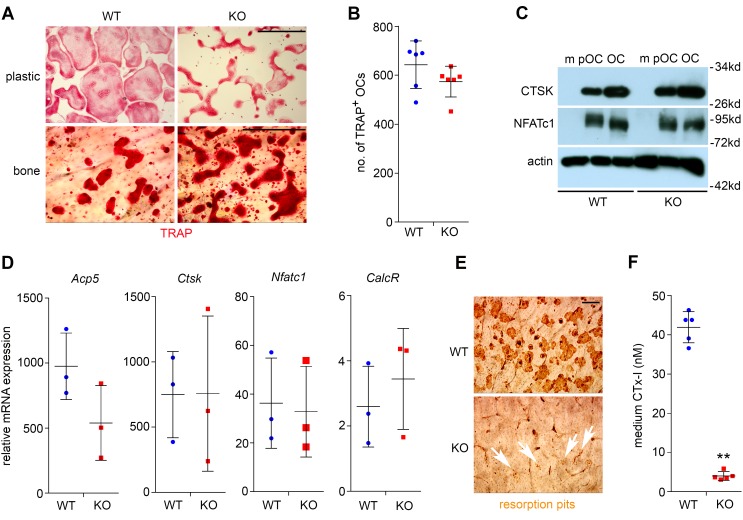
To elucidate the cellular and molecular mechanisms by which PLEKHM1 regulates osteoclasts, we generated osteoclasts in cultures of bone marrow monocytes isolated from mice with germline and conditional deletion of Plekhm1. Loss of Plekhm1 had no effect on the formation of multinucleated mature osteoclasts grown on plastic culture dishes or bovine cortical bone slices (Figure 4A and Supplemental Figure 3A). Nonetheless, the morphology of Plekhm1-deficient osteoclasts was quite different from that of wild-type cells, as was the distribution pattern of tartrate-resistant acid phosphatase (TRAP), an osteoclast-specific acidic phosphatase, which was more aggregated in the perinuclear area of Plekhm1-deficient osteoclasts than wild-type cells (Supplemental Figure 4). The total number of TRAP+ mature osteoclasts with more than 3 nuclei was similar in Plekhm1–/– cultures compared with wild-type cultures (Figure 4B and Supplemental Figure 3B). Furthermore, the protein expression of osteoclast markers, CTSK and nuclear factor of activated T cells, cytoplasmic 1 (NFATc1), and the mRNA levels of osteoclast-specific genes, Acp5 (encoding TRAP), Ctsk, Nfatc1, and CalcR (encoding calcitonin receptor), in Plekhm1-null osteoclasts were comparable to those in wild-type cells (Figure 4, C and D, and Supplemental Figure 3, C and D). See complete unedited blots in the supplemental material. In contrast, the bone resorption capacity of Plekhm1–/– osteoclasts markedly decreased, as demonstrated by resorption pit staining (Figure 4E and Supplemental Figure 3E) and the concentration of CTx-I, a bone resorption marker, in the culture medium (Figure 4F and Supplemental Figure 3F). Taken together, these results indicate that loss of Plekhm1 attenuates osteoclast activity but has no effects on osteoclast differentiation.
Figure 4. Loss of Plekhm1 attenuates bone resorption without affecting osteoclast differentiation.
(A) Tartrate-resistant acid phosphatase (TRAP) staining of wild-type and Plekhm1–/– (KO) osteoclasts cultured on plastic dishes and bovine cortical bone slices. Representative images are from 6 samples/group/culture of 3 independent cultures from different mice. Scale bar: 200 μm. (B) Number of TRAP+ osteoclasts with more than 3 nuclei/well of a 48-well plate (n = 6). (C) Protein expression of osteoclast markers, cathepsin K (CTSK) and nuclear factor of activated T cells, cytoplasmic 1 (NFATc1), as detected by Western blotting during osteoclast differentiation. Actin served as a loading control. m, bone marrow monocytes; pOC, preosteoclasts; OC, mature osteoclasts. The experiment was repeated 3 times. (D) mRNA expression of osteoclast marker genes, Acp5 (also known as TRAP), Ctsk, Nfactc1, calcitonin receptor (CalcR), in mature osteoclasts, as measured by quantitative real-time PCR (n = 3). (E) Resorption pit staining of WT and KO osteoclasts cultured on bone slices. White arrows point out the tiny pits dug by KO osteoclasts. Each image is a representative of 6 bone slices/group/culture of at least 3 independent cultures from different mice. Scale bar: 40 μm. (F) The amount of medium CTx-I, collagen fragments released by osteoclasts during bone resorption, was measured by ELISA (n = 5). **P < 0.01 versus WT by Student t test. Data in B, D, and F are presented as scatter dot plots. The mean and SD of each group are overlaid onto each column of dots.
Mutations in osteopetrotic patients and rats lead to short N-terminal truncated forms of PLEKHM1 (24). An R714C mutation in a human patient increases TRAP secretion (34). We generated recombinant retroviruses expressing HA-tagged full-length murine PLEKHM1 as well as a series of mutants with either mutations in human patients and rats or deletions of individual structural domains (Supplemental Figure 5A). Supplemental Figure 5B shows loss of PLEKHM1 protein in Plekhm1–/– osteoclasts and successful reconstitution of wild-type and different forms of PLEKHM1. Expression of truncated and mutated PLEKHM1 in wild-type osteoclasts had no effect on osteoclast formation and bone resorption (Supplemental Figure 6). This finding is consistent with the recessive effects of these mutants on osteoclasts in humans and rats. Full-length Plekhm1 and the R714C mutant of Plekhm1 rescued the morphology and bone resorbing activity of Plekhm1–/– osteoclasts (Supplemental Figure 7, A–C). However, in contrast to the findings in human osteoclasts (34), R714C mutation in murine osteoclasts failed to exhibit gain of function in vitro. These findings show that PLEKHM1 plays an intrinsic role in regulation of osteoclast function and each structural domain is functionally essential.
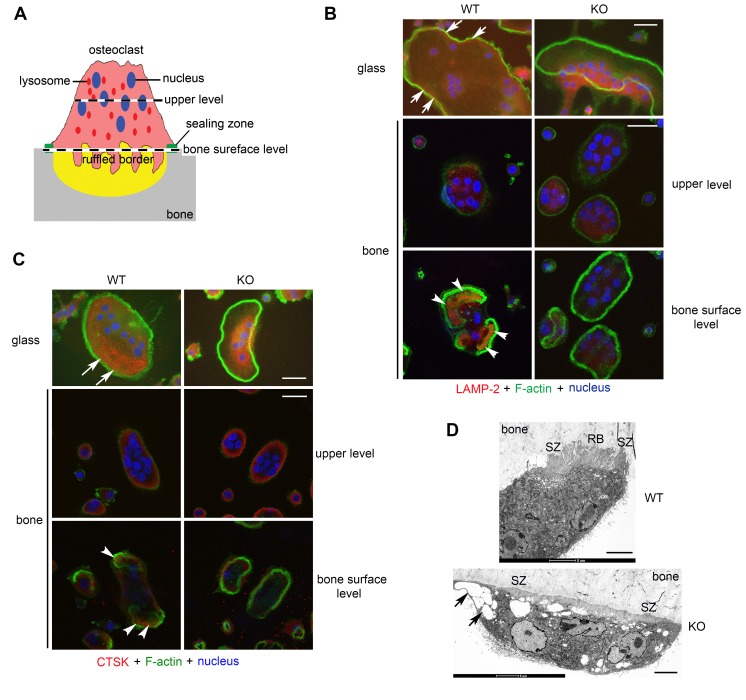
PLEKHM1 regulates lysosome peripheral distribution and ruffled border formation in osteoclasts.
When osteoclasts are cultured on glass coverslips, actin filaments form a peripheral belt of podosomes, which are highly dynamic structures that mediate cell adhesion and migration. Podosomes are clustered into the sealing zone (actin ring) of osteoclasts cultured on bone (35). Microtubules regulate podosome patterning and thus are essential for osteoclast spreading on glass and sealing zone formation on bone (36). Because Plekhm1–/– osteoclasts displayed a dramatic change in morphology and had decreased bone resorption, we investigated whether these phenotypes were caused by defects in cytoskeletal organization. As shown in Supplemental Figure 8A and Supplemental Figure 9A, the organization of actin and microtubules was normal in Plekhm1–/– osteoclasts cultured on glass and bone.
We next examined the distribution of lysosomes by immunofluorescent staining of LAMP-2 and CTSK, two commonly used osteoclast lysosomal markers. While a portion of lysosomes was localized at the cell periphery of wild-type osteoclasts cultured on glass (top left, Figure 5, B and C, and Supplemental Figure 9, B and C), lysosomes in Plekhm1–/– osteoclasts on glass were clustered at the perinuclear area and were absent from the cell periphery (top right, Figure 5, B and C; Supplemental Figure 8, B and C; and top, Supplemental Figure 9, B and C). In wild-type osteoclasts cultured on bone (Figure 5A), LAMP-2 was enriched at the ruffled border inside the actin ring (bottom left, Figure 5B and Supplemental Figure 9B), while CTSK was secreted into the resorption lacuna circumscribed by the actin ring (bottom left, Figure 5C and Supplemental Figure 9C). In contrast, both localization of LAMP-2 at the ruffled border and CTSK secretion were barely seen in Plekhm1–/– osteoclasts (bottom right, Figure 5, B and C, and Supplemental Figure 9, B and C). Consistent with an essential role of lysosome secretion in ruffled border formation, loss of Plekhm1 in osteoclasts resulted in an absence of the ruffled border associated with big vesicles, whereas the sealing zone appeared intact, as observed by electron microscopic images (Figure 5D). These results suggest that impaired bone resorption in Plekhm1–/– osteoclasts is caused by defects in the peripheral positioning and secretion of lysosomes but not organization of cytoskeletons.
Figure 5. PLEKHM1 regulates lysosome peripheral distribution and ruffled border formation in osteoclasts.
(A) An illustration of the polarized structures of an active osteoclast cultured on bone. The levels (upper level and bone surface level) of two representative sections of confocal microscopic images shown in B and C are indicated. (B) Immunofluorescent staining of LAMP-2 and (C) immunofluorescent staining of cathepsin K (CTSK) in wild-type and Plekhm1–/– (KO) osteoclasts cultured on glass coverslips and bone slices. White arrows in top rows point out the distribution of LAMP-2–positive lysosomes and CTSK at the periphery of WT osteoclasts. White arrowheads in the bottom rows point out the localization of LAMP-2 at the ruffled border and secretion of CTSK in the resorption lacuna inside actin rings of WT osteoclasts cultured on bone. Each image is a representative of 6 coverslips or bone slices/group/culture from at least 3 independent cultures from different mice. Scale bar: 20 μm. (D) Electron microscopic images of WT and KO osteoclasts cultured on bone slices in vitro. Images are representatives of 6 osteoclasts/group. SZ, sealing zone; RB, ruffled border. Black arrows in bottom panel point out enlarged vesicles accumulated in KO osteoclasts. Scale bar: 5 μm.
Plekhm1 mRNA was also detected in mouse embryonic fibroblasts (MEFs) but was less abundant than in osteoclasts. Loss of Plekhm1 in MEFs slightly increased the number of lysosomes around the nuclei but significantly inhibited Ca2+-regulated exocytosis of lysosomes, as assayed by release of the lysosomal enzyme β-hexosaminidase in streptolysin-O–permeabilized MEFs (Supplemental Figure 10) (37).
Loss of Plekhm1 has little impact on autophagosome formation or endocytic/phagocytic pathways in osteoclasts.
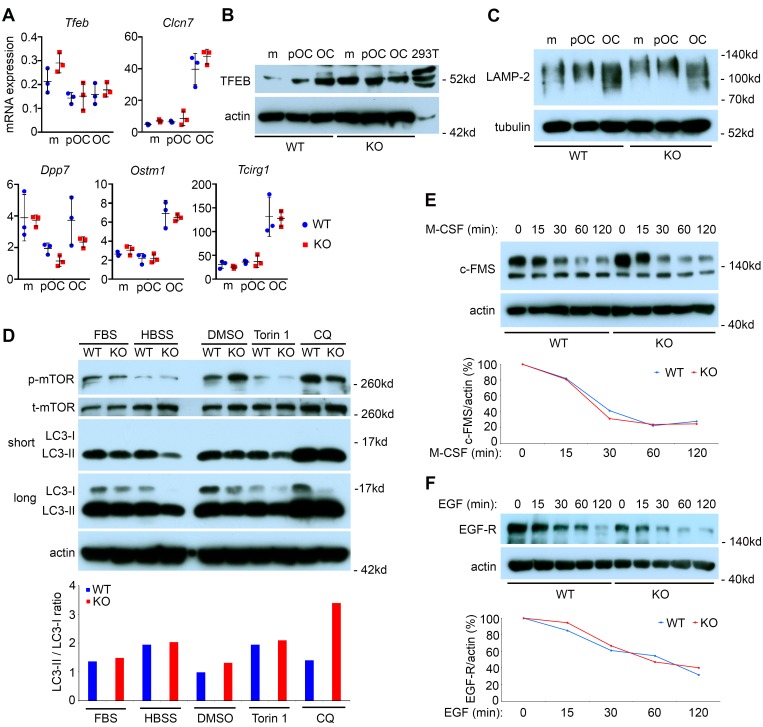
PLEKHM1 is associated with lysosomes in osteoclasts (24, 38), and transcription factor EB (TFEB) regulates osteoclast lysosome biogenesis (39). We, therefore, examined the expression of TFEB in wild-type and Plekhm1–/– osteoclast precursor and mature cells. The mRNA level of Tfeb and its target genes Clcn7 (encoding CLC7), Dpp7 (encoding dipeptidyl peptidase 7), Ostm1, and Tcirg1 (encoding the a3 subunit of vacuolar-ATPase) (39, 40), was indistinguishable between wild-type and Plekhm1–/– osteoclast lineage cells (Figure 6A). The TFEB protein level increased during osteoclast differentiation in wild-type cells. TFEB protein increased in Plekhm1–/– monocytes and preosteoclasts; albeit the level of this transcription factor in mature Plekhm1–/– osteoclasts was similar to that of wild-type cells (Figure 6B). The protein level of LAMP-2 was comparable between wild-type and Plekhm1–/– osteoclast precursors and mature cells (Figure 6C). In contrast to other cell types (40), starvation and mTOR inhibition did not induce nuclear translocation of TFEB in osteoclasts (Supplemental Figure 11). Collectively, these findings indicate that loss of Plekhm1 has little effect on TFEB and lysosome biogenesis.
Figure 6. Loss of Plekhm1 has no effects on TFEB expression, lysosome biogenesis, autophagic, and endocytic degradation pathways in mature osteoclasts.
(A) qPCR detection of mRNA expression of Tfeb and its downstream target genes, Clcn7, Dpp7, Ostm1, and Tcirg1, during osteoclast differentiation in wild-type and Plekhm1–/– (KO) cultures (mean ± SD, n = 3). m, bone marrow monocytes; pOC, preosteoclasts; OC, mature osteoclasts. (B) Western blot detection of protein expression of TFEB during osteoclast differentiation in WT and KO cultures. The lysate of 293-T cells served as a positive control. Actin served as a loading control. The data are representatives of 3 independent experiments. (C) Western blot detection of LAMP-2 expression during osteoclast differentiation. Tubulin served as a loading control. The experiment was repeated 3 times. (D) Mature osteoclasts were either untreated (FBS) or underwent serum and amino acid starvation in HBSS buffer for 2 hours. In another set of experiment, osteoclasts were treated with DMSO, 250 nm of mTOR inhibitor Torin1, and 50 μM of lysosome inhibitor chloroquin (CQ) for 2 hours. The protein levels of phospho-mTOR, LC3-I, and LC3-II were detected by Western blots. The ratio of LC3-II/LC3-I was measured by densitometry of Western blots using NIH ImageJ software. The experiment was repeated twice. (E and F) Receptor degradation assays of c-Fms (E) and EGF receptor (EGF-R) (F) in osteoclasts stimulated with cytokines for the indicated times, as detected by Western blotting and densitometry quantification using NIH ImageJ software. Actin served as loading controls. The experiments were repeated twice.
PLEKHM1 regulates autophagosome-lysosome fusion through interaction with the autophagosome protein LC3 (41). In contrast, depletion of Plekhm1 in A549 lung cells has no effect on autophagy (27), suggesting that the role of PLEKHM1 in autophagy is cell-type specific. Several autophagic proteins participate in lysosomal secretion in osteoclasts (42). Therefore, we tested whether PLEKHM1 regulates autophagy in osteoclasts. Serum and amino acid starvation or treatment of osteoclasts with a potent mTOR inhibitor, Torin1, inhibited the mTOR pathway, as evidenced by decreased levels of phospho-mTOR (Figure 6D). In contrast to the dramatic induction of autophagy in other cell types under the same conditions (41), starvation and Torin1 only slightly increased the LC3-II/LC3-I ratio, a marker of autophagy, in both wild-type and Plekhm1–/– osteoclasts (bottom, Figure 6D). A longer period of starvation or a higher dose of Torin1 caused rapid cell death of osteoclasts (data not shown). Additionally, loss of Plekhm1 had no effect on the morphology and distribution of the Golgi apparatus, early endosomes, or recycling endosomes in osteoclasts cultured on glass or bone (Supplemental Figure 12).
Next, we determined whether PLEKHM1 regulates lysosome degradation activities in osteoclasts. Loss of Plekhm1 in osteoclasts did not alter the expression of the EGF receptor (Supplemental Figure 13A) or the kinetics of degradation of the M-CSF receptor or the EGF receptor stimulated by respective cytokines (Figure 6, E and F). Binding and endocytic trafficking of transferrin, a marker of receptor-mediated endocytosis, to recycling endosomes in Plekhm1–/– osteoclasts was normal (Supplemental Figure 13B). Targeting of transferrin to the ruffled border in wild-type osteoclasts on bone (bottom, Supplemental Figure 12C), which requires normal lysosomal secretion (7), was blocked in Plekhm1–/– osteoclasts. Furthermore, loading of dextran, a liquid-phase endocytic tracer, into LAMP-2–GFP positive lysosomes in Plekhm1–/– osteoclasts was normal (left, Supplemental Figure 13C). While dextran-loaded lysosomes were secreted and disappeared in wild-type osteoclasts after a 2-hour chase, the majority of loaded lysosomes remained in the perinuclear area in Plekhm1–/– osteoclasts (right, Supplemental Figure 13C). Phagolysosome formation is another important cellular process regulated by RAB7 (43). To test whether phagocytosis was affected by Plekhm1 deficiency, we performed bacterial phagocytosis assay in macrophages using the intracellular pathogen Coxiella burnetii (44). Access of C. Burnetii to LAMP-1–positive lysosomes in Plekhm1–/– macrophages was not prevented, indicating that phagolysosome maturation is functional in Plekhm1–/– macrophages (Supplemental Figure 13D).
PLEKHM1 and DEF8 form a complex with RAB7 to regulate lysosome positioning and bone resorption in osteoclasts.
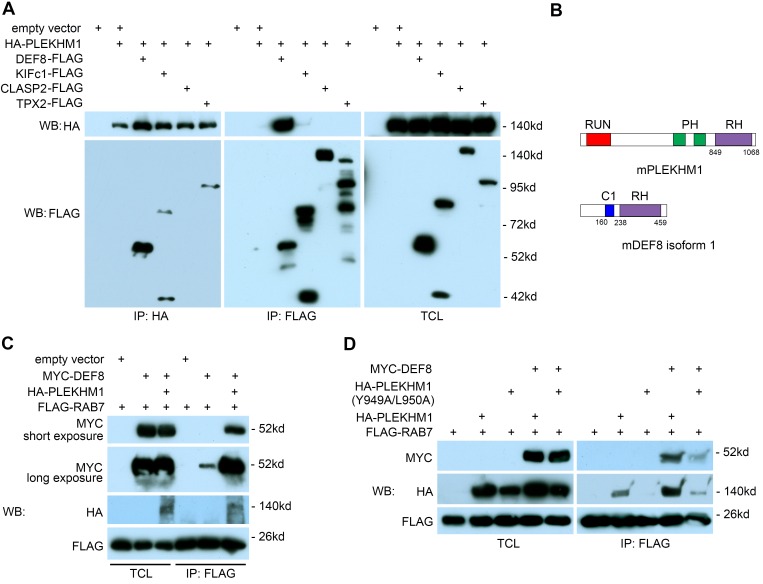
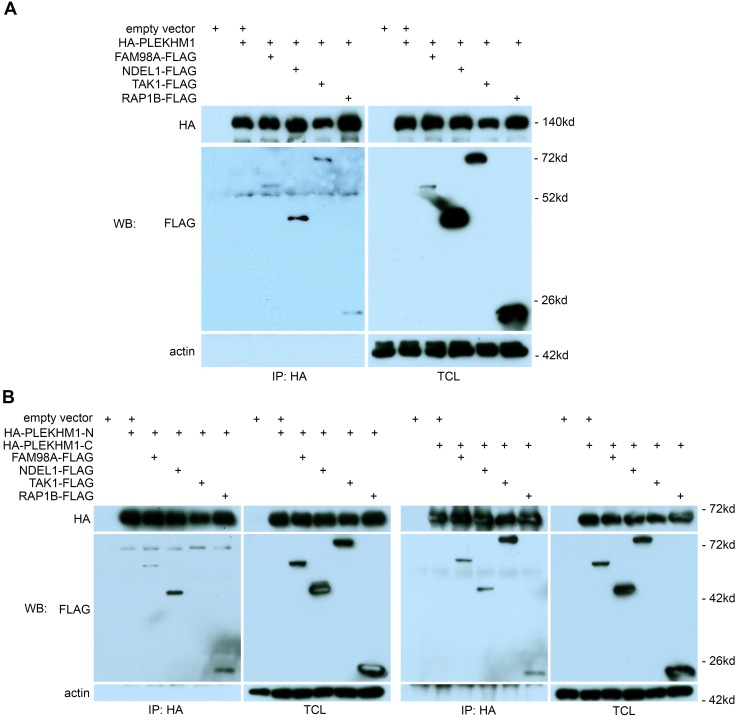
To identify PLEKHM1-interacting proteins, we expressed HA-tagged PLEKHM1 in Plekhm1–/– osteoclasts by retroviral transduction. Reconstituted PLEKHM1, at a similar level to the endogenous one (Supplemental Figure 5B), was IP using anti-HA antibody–coated magnetic beads. Proteomic analysis of HA-PLEKHM1 from osteoclasts identified several PLEKHM1-specific binding proteins (Supplemental Tables 4 and 5). Reciprocal co-IPs of HA-tagged PLEKHM1 and FLAG-tagged proteins in 293-T cells indicated that DEF8 specifically bound to PLEKHM1, whereas KIFc1 and TPX2, two proteins involved in microtubule-based vesicular trafficking, were pulled down only by anti-HA beads and not by anti-FLAG beads. CLASP2, a microtubule plus-end binding protein, did not bind to PLEKHM1 (Figure 7A). The binding region of DEF8 on PLEKHM1 was mapped to the last C-terminal 72 amino acids (Supplemental Figure 14A). Interestingly, both PLEKHM1 and DEF8 contain a C-terminal RH domain, which has been recently identified as a novel RAB7-binding motif (Figure 7B) (27). As shown in Figure 7, C and D, PLEKHM1 and DEF8 individually bound weakly to RAB7 (lane 2 in IP blots). However, when both proteins were coexpressed in 293-T cells, their binding to RAB7 was greatly enhanced (lane 3 in IP blot of Figure 7C and lane 4 in IP blot of Figure 7D), suggesting that PLEKHM11, DEF8, and RAB7 form a ternary complex. Furthermore, an Y949A/L950A double mutation at the N-terminal region of the PLEKHM1 RH domain abolished its binding to RAB7 (lane 3 in IP blot of Figure 7D) but kept the capacity of PLEKHM1 binding to DEF8 (lane 3 in IP blot of Supplemental Figure 14B), indicating that PLEKHM1 interacts with DEF8 and RAB7 through different regions.
Figure 7. DEF8 and PLEKHM1 interact and promote each other’s binding to RAB7.
(A) Co-IP of HA-tagged PLEKHM1 and FLAG-tagged DEF8, KIFc1, CLASP2, and TPX2 in 293-T cells. The lysate of empty vector expressing 293-T cell (lane 1) served as a negative control. TCL, total cell lysate. (B) The structural domains of murine PLEKHM1 and DEF8. The numbers indicate the positions of amino acids. (C and D) Co-IPs of MYC-tagged DEF8, FLAG-tagged RAB7, and HA-tagged PLEKHM1 or PLEKHM1 with Y949A/L950A double mutation in 293-T cells. The experiments were repeated 3 times.
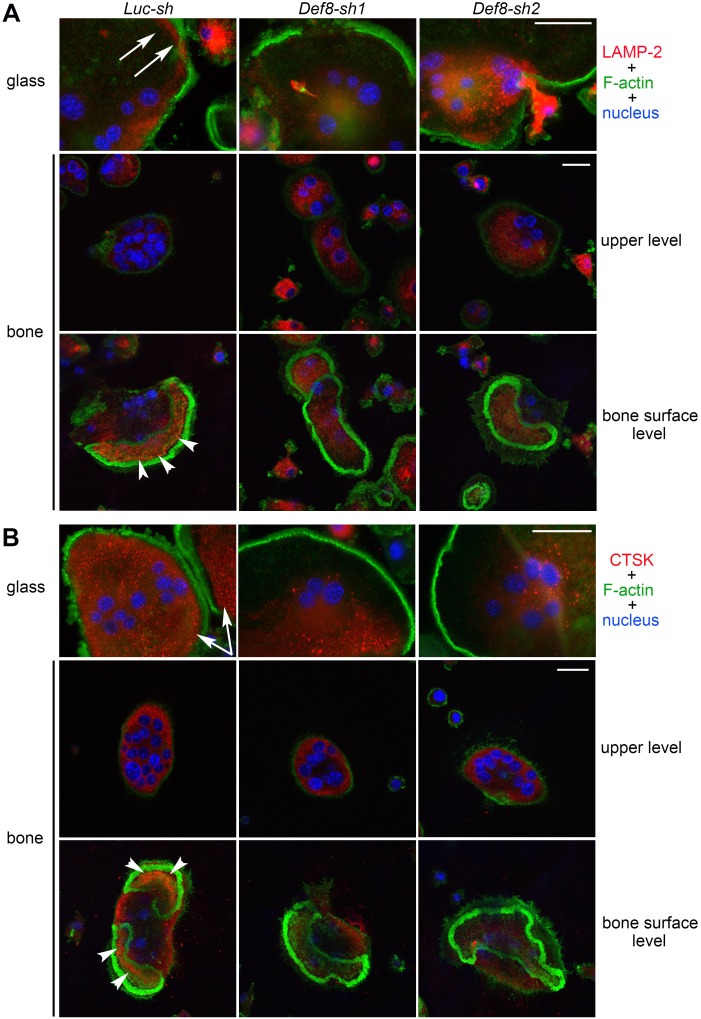
To dissect the role of DEF8 in osteoclast function, we suppressed its expression by lentiviral transduction of two distinct shRNAs specifically targeting murine Def8 mRNA. qPCR showed a dramatic reduction in Def8 mRNA in osteoclast precursor and mature cells, while endogenous Def8 levels slightly decreased during osteoclast differentiation (Supplemental Figure 15A). Depletion of Def8 in bone marrow monocytes inhibited osteoclast formation (Supplemental Figure 15, B–D). To circumvent this, we plated 3 times more control and Def8-depleted monocytes than normal osteoclast cultures. With this maneuver, a similar number of mature osteoclasts was generated from Def8-depleted cultures on both plastic and bone (Supplemental Figure 15E). Nonetheless, the bone resorption capacity of Def8-depleted osteoclasts was significantly decreased (Supplemental Figure 15, F–G). Moreover, loss of Def8 resulted in decreased peripheral distribution of lysosomes in approximately 60% of osteoclasts cultured on glass (Def8-sh1, 63% ± 4.4%; Def8-sh2, 57% ± 4.6%; LUC-sh, 4.0% ± 2.5%; mean ± SD, n = 6) (Figure 8 and Supplemental Figure 15H). Loss of Def8 also diminished localization of LAMP-2 at the ruffled border and secretion of CTSK into the resorption lacuna in osteoclasts cultured on bone (Figure 8). Collectively, these results indicate that DEF8, in concert with PLEKHM1, regulates lysosome positioning, secretion, and bone resorption in osteoclasts.
Figure 8. Loss of DEF8 attenuates the peripheral distribution of lysosomes and their targeting to the ruffled border in osteoclasts.
(A and B) Immunofluorescent staining of LAMP-2 (A) and cathepsin K (CTSK) (B) in control osteoclasts (Luc-sh) and osteoclasts in which Def8 has been knocked down (Def8-sh1 and Def8-sh2) osteoclasts cultured on glass coverslips and bovine cortical bone slices. White arrows in the top rows point out the peripheral distribution of LAMP-2 and CTSK in control cells cultured on glass coverslips. White arrowheads in the bottom rows point out the ruffled border localization of LAMP-2 and the secretion of CTSK into the resorption lacuna in control cells cultured on bone slices. Each image is a representative of 6 bone slices/group/culture from at least 3 independent cultures from different mice. Scale bar: 20 μm.
FAM98A and NDEL1 interact with PLEKHM1 and function in lysosome positioning in osteoclasts.
To determine how PLEKHM11 regulates lysosome positioning in osteoclasts, we focused on 4 potential PLEKHM1-binding proteins identified by proteomics of PLEKHM1 immunoprecipitates from osteoclasts (Supplemental Table 4): FAM98A, a microtubule-associated protein with unknown function (45); NDEL1, a regulatory protein of microtubules and their motor complexes, cytoplasmic dynein and kinesins (29, 30); TAK1, an important downstream signaling molecule of RANKL (46); and RAP1B, a small GTPase essential for integrin signaling and osteoclast function (47).
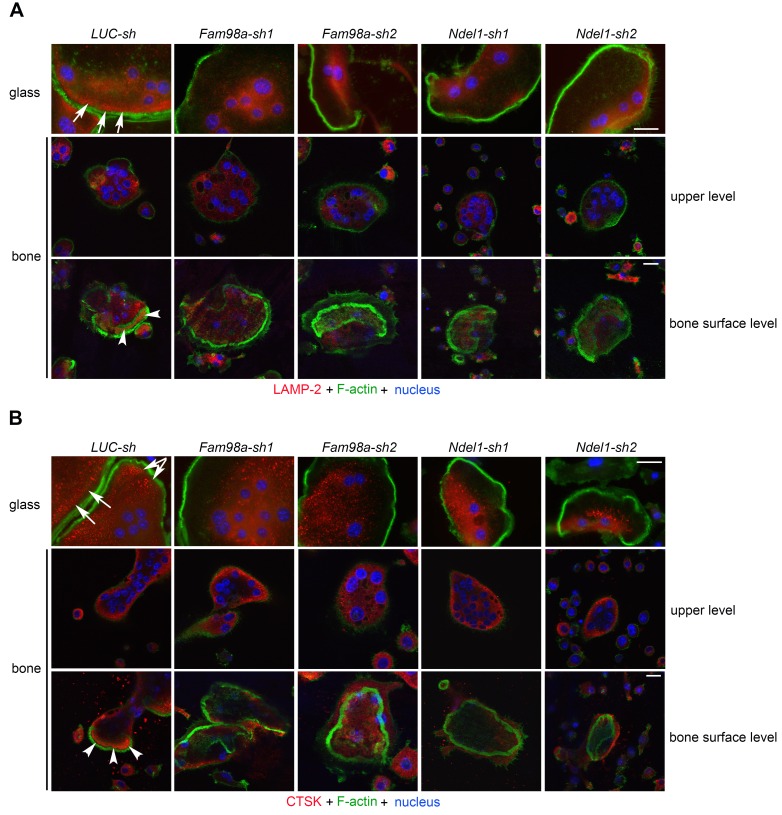
IP studies in 293-T cells showed that the full-length PLEKHM1 bound to all 4 proteins (Figure 9A). FAM98A, NDEL1, and RAP1B bound to both N- and C-terminal fragments of PLEKHM1, while TAK1 interacted with only the C-terminal portion of PLEKHM1 (Figure 9B). Furthermore, deletion of RUN and PH2 domains reduced the binding of FAM98A to PLEKHM1 (Supplemental Figure 16A). Similarly, deletion of the RUN domain dramatically decreased the binding of NDEL1 to PLEKHM1 (Supplemental Figure 16B). To elucidate the functional role of FAM98A and NDEL1 in osteoclasts, we knocked down their expression by lentivirus-mediated shRNA transduction. Depletion of Fam98a or Ndel1 in osteoclast precursors inhibited osteoclast formation (Supplemental Figure 17, A and B). Some mature osteoclasts did form in Fam98a- and Ndel1-silenced cultures. Immunofluorescent staining of mature osteoclasts on glass showed that loss of Fam98a or Ndel1 had no effect on podosome-belt formation or organization of the microtubule network (Supplemental Figure 17C). Similar to Plekhm1–/– osteoclasts, lysosomes were clustered at the perinuclear region, and the number of peripherally distributed lysosomes in Fam98a- or Ndel1-depleted osteoclasts cultured on glass coverslips was markedly reduced (Figure 10 and Supplemental Figure 17, D and E). Loss of Fam98a or Ndel1 attenuated the targeting of lysosomes to the ruffled border in osteoclasts cultured on bone (Figure 10). Therefore, PLEKHM1 likely interacts with FAM98A and NDEL1 to couple lysosomes to microtubules for their peripheral positioning and secretion.
Figure 9. PLEKHM1 interacts with FAM98A, NDEL1, TAK1, and RAP1B.
Co-IP of HA-tagged full-length PLEKHM1 (A) or the N-terminal and the C-terminal portions of PLEKHM1 (B) with FLAG-tagged FAM98A, NDEL1, TAK1, and RAP1B in 293-T cells. The lysate of empty vector expressing 293-T cells (lane 1) served as a negative control. TCL, total cell lysate. Actin served as loading controls. The experiments were repeated 4 times.
Figure 10. Decreased expression of FAM98A and NDEL1 inhibits the peripheral distribution of lysosomes and their targeting to the ruffled border in osteoclasts.
Immunofluorescent staining of LAMP-2 (A) and cathepsin K (CTSK) (B) in control (Luc-sh), Fam98a- (Fam98a-sh1 and Fam98a-sh2), and Ndel1- (Ndel1-sh1 and Ndel1-sh2) knocked down osteoclasts cultured on glass coverslips and cortical bovine bone slices. White arrows in the top rows point out the peripheral distribution of LAMP-2 and CTSK in control cells cultured on glass coverslips. White arrowheads in the bottom rows point out the ruffled border localization of LAMP-2 and the secretion of CTSK into the resorption lacuna in control cells cultured on bone slices. Each image is a representative of 6 bone slices/group/culture from at least 3 independent cultures from different mice. Scale bar: 20 μm.
Discussion
Bone regenerates throughout life by remodeling — a process during which resorption mediated by osteoclasts is coupled with formation mediated by osteoblasts in time and space. Under normal conditions, the amount of bone resorbed is refilled by an equal amount of new bone. This so-called “balance” between resorption and formation is critical for skeletal homeostasis (48). Mutations of PLEKHM1 in humans and rats cause either an intermediate form (24) or a malignant form (49) of osteopetrosis, suggesting that PLEKHM1 is essential for osteoclast function. In this study, we have generated Plekhm1 germline and conditional deletion mouse models. Detailed characterization of these mouse lines in vivo and in vitro revealed that PLEKHM1 is a critical regulator of osteoclast lysosome section and bone resorption. Loss of Plekhm1 increased cancellous bone mass due to decreased bone resorption without overt defects in other tissues and organs. In vitro mechanistic studies showed that PLEKHM1 regulates lysosome peripheral distribution (positioning) and secretion through a protein complex comprising PLEKHM1, DEF8, RAB7, FAM98A, and NDEL1 at a minimum. These proteins interact with each other and link lysosomes to microtubules for their transportation to the ruffled border during bone resorption (Supplemental Figure 17F). Elucidation of PLEKHM1 and its interacting proteins provides insights not only into osteoclast biology, but in lysosome biology as well, especially in the quickly expending field of lysosome positioning (50–58). Moreover, the bone-specific phenotype of mice with germline and conditional Plekhm1 deficiency suggests that PLEKHM1-mediated lysosome secretion may be a new therapeutic target for bone diseases with increased resorption.
Growth factors released from the bone matrix during osteoclastic bone resorption have been long implicated in the coupling of resorption to formation, as evidenced by the ability of such factors to stimulate osteoblast generation (59, 60). Loss of function of CLCN7 and TCIRG1 — genes that encode key components of the osteoclast acid-secreting machinery — leads to complete abrogation of bone resorption in both humans and mice, in spite of increased osteoclast number; this condition, therefore, is termed osteoclast-rich osteopetrosis (61). Nevertheless, both humans and mice with osteoclast-rich osteopetrosis have increased bone formation (62–64). In contrast, loss-of-function mutations of RANK lead to complete absence of osteoclasts and “osteoclast-poor” osteopetrosis. However, in the latter form of osteopetrosis, osteoblast number and bone formation are decreased (65). Based on these lines of evidence, it is possible that the nonfunctional osteoclasts in the osteoclast-rich osteopetrosis are still capable of secreting factors that stimulate osteoblasts (66, 67). In the studies reported herein, osteoblasts derived from either calvaria or bone marrow stromal cells from Plekhm1–/– mice were able to differentiate and form bone nodules normally in vitro. Nonetheless, osteoblast number and bone formation were attenuated, albeit slightly, in vivo in both germline and conditional Plekhm1-deficient mice (Figure 3). Whether the attenuated bone formation in Plekhm1–/– mice is secondary to a decrease in coupling factors released from Plekhm1–/– osteoclasts or independent of them will await further studies.
The involvement of lysosomes in osteoclastic bone resorption was suggested almost 30 years ago by the demonstration of a lysosomal membrane protein at the ruffled border (68) and the distribution of mannose-6-phosphate receptors, which are essential for the delivery of newly synthesized lysosomal enzymes to endosomes (69), from the trans-Golgi network to the ruffled border. These findings led to a model in which the ruffled border and resorption lacuna underneath it form an extracellular lysosome-like compartment (70). In this case, mutations in genes controlling biogenesis of secretory lysosomes, as occurs in patients with Hermansky-Pudlak syndrome and corresponding animal models (71), should impair ruffled border formation and bone resorption. However, there is no evidence of defective bone resorption in such humans and mice. Similarly, mutations of RAB27 and its effectors, which control exocytosis of secretory lysosomes in Griscelli’s syndrome and respective mouse models, have no effect on osteoclast function (20). In contrast, inhibition of intracellular trafficking of lysosomes and their fusion with the plasma membrane in osteoclasts attenuate the ruffled border formation and bone resorption (Figures 4 and 5 and previous reports, refs. 7, 23, 24, 72). Taken together, these lines of evidence indicate that lysosome secretion plays a central role in osteoclast function, but the molecular machinery governing this process in osteoclasts is different from other cell types, such as melanocytes and other hematopoietic cells.
Lysosome positioning is important for lysosomal function and cell migration (53, 73). RAB7 and its effector RILP propel lysosomes towards the nucleus by recruiting the microtubule motor dynein to lysosomes (74), whereas the BORC-ARL8/SKIP/kinesin-1 protein module moves lysosomes to the cell periphery (57, 58). Interestingly, SKIP, also known as PLEKHM2, has similar structural domains to those of PLEKHM1 but lacks the RAB7-binding RH domain. Instead, PLEKHM2 interacts with another small GTPase ARL8, and together they regulate lysosome positioning. Loss-of-function mutations of PLEKHM2 cause lysosome/autophagy defects and human cardiomyopathy (56). In the present studies (Figures 7–10, Supplemental Figure 14–17, and Supplemental Tables 4 and 5), we have uncovered a RAB7-associated molecular machinery, comprising PLEKHM1, DEF8, FAM98A, and NDEL1, that mediates lysosome peripheral positioning in osteoclasts. The functional relationship of PLEKHM2/BORC/ARL8 (57, 58) and PLEKHM1/DEF8/RAB7 (this study) in osteoclasts and other cell types in vitro and in vivo warrants detailed characterization in the future.
DEF8, a protein of unknown function in hematopoietic cells, contains a RAB7-binding RH domain and forms a ternary complex with PLEKHM1 and RAB7. FAM98A has an N-terminal calponin homology domain, a motif mediating microtubule binding (28, 75). At present, the physiological functions of FAM98A and its interaction with PLEKHM1 are unknown. Notably, both FAM98A and PLEKHM1 have been implicated in ovarian cancer progression (45, 76), suggesting a possible functional interplay of these two proteins. NDEL1, in coordination with NDE1 and LIS1, regulates the distribution and activity of cytoplasmic dynein (29, 77, 78). NDEL1 also links to kinesin-1 through DISC1 (30, 79). Interestingly, both NDE1 and DISC1 were immunoprecipitated with PLEKHM1 from osteoclast lysates in our studies (Supplemental Table 4). We have previously reported that LIS1 binds to the RUN and PH1 domains of PLEKHM1 in osteoclasts and knocking down LIS1 expression in vitro abrogates lysosome distribution and bone resorption (38). We also found herein that, in both osteoclasts and 293-T cells, PLEKHM1 binds to TAK1 and RAP1B, two signaling molecules downstream of the RANKL/RANK and integrin signaling pathways, which are essential for osteoclast activation and function. Therefore, through its interaction with the proteins identified in our work, PLEKHM1 may link lysosomes to microtubules and microtubule motor proteins upon osteoclast activation.
Depletion of PLEKHM1-interacting proteins in osteoclast precursor cells inhibited osteoclast formation in vitro (Supplemental Figures 15 and 17), whereas loss of Plekhm1 had no effect on osteoclast differentiation in vitro or in vivo (Figures 3 and 4). It is, therefore, likely that these proteins may have additional functions independent of PLEKHM1 and lysosome trafficking. Consistent with this possibility, we have previously found that LIS1 regulates the survival of osteoclast precursors by modulating JNK activation (38). Similarly, disturbance of dynein function impairs osteoclast differentiation and function (80). Loss of Plekhm1 caused dramatic morphological changes in osteoclasts but did not affect the number of mature osteoclasts cultured on plastic (Figure 4 and Supplemental Figure 3). The exact mechanisms underlying these phenomena are unknown. It remains possible, however, that the defective lysosome positioning and secretion in osteoclasts affects osteoclast motility and/or the adhesion and spreading of osteoclasts (73).
Finally, PLEKHM1 has been recently implicated in autophagy (41) and EGF receptor degradation in other cell types (27). In osteoclasts, however, loss of Plekhm1 did not affect autophagy or the endocytic and lytic functions of lysosomes (Figure 6 and Supplemental Figures 12 and 13), suggesting that the regulation of PLEKHM1 in these cellular processes is cell-type and tissue-type specific. Given that Plekhm–/– mice are viable, fertile, and have no overt abnormalities in other tissues and organs, except high trabecular bone mass, the function of PLEKHM1 in other cells and tissues may be compensated by other proteins. The role of PLEKHM1 in other cells and tissues needs more detailed characterization under physiological and pathological conditions.
In closing, the findings in this study indicate that PLEKHM1 is indispensable for bone homeostasis in mice, as is the case in rats and humans. Furthermore, our work has uncovered a molecular apparatus linking lysosomes to microtubules through PLEKHM1/DEF8/RAB7 for lysosome transportation/secretion and bone resorption in osteoclasts.
Methods
Mice.
The Plekhm1-floxed mice were generated at the Gene Targeting and Transgenic Facility of University of Connecticut Health Center at Farmington. We crossed Plekhm1-floxed mice with C57BL/6 mice for 10 generations before crossing them with Cre-expressing mice.
Osteoclast and osteoblast cultures.
Whole bone marrow was extracted from tibias and femurs of one or two 8- to 10-week-old mice (81). Primary osteoblasts were harvested from calvaria or cultured from bone marrow stromal cells, as described in the Supplemental Methods.
Plasmid transfection and retroviral/lentiviral transduction.
The HA-tagged, full-length murine Plekhm1 and FLAG-tagged Rab7 were amplified by PCR with Pfx DNA polymerase (Life Technologies) from a mature osteoclast cDNA library in our laboratory. The LKO.1 lentiviral vectors expressing shRNA sequences targeting specific mRNAs were purchased from Sigma-Aldrich. Retroviral and lentiviral transduction was performed as described previously (82) and in the Supplemental Methods.
Western blotting and IP.
For Western blotting, cultured cells were washed with ice-cold PBS twice and lysed in 1× RIPA buffer (R-0278, Sigma-Aldrich) containing 1 mM DTT and Complete Mini EDTA-free protease inhibitor cocktail (04693159001, Roche). The HA- or FLAG-tagged proteins were IP with protein G magnetic beads (S1430, New England BioLabs) precoated with mouse monoclonal antibodies. For detailed protocols, see the Supplemental Methods.
Immunofluorescence and laser confocal scanning microscopy.
For staining of LAMP-2, cells were fixed with 4% paraformaldehyde in PBS for 20 minutes, followed by incubation with PBS/0.2% BSA/0.1% Saponin for 30 minutes. For EEA1 labeling, cells were fixed and penetrated at −20°C with methanol for 5 minutes and acetone for 30 seconds, followed by blocking with PBS/0.2% BSA for 30 minutes. For rest of the immunofluorescent staining, cells were fixed with 4% paraformaldehyde in PBS for 20 minutes and permeabilized with 0.1% Triton X-100/PBS for 10 minutes. For details, see the Supplemental Methods and ref. 83.
Micro-CT.
The left femurs and L4 vertebrae were cleaned of soft tissues and fixed in 10% Millonig’s formalin with 0.5% sucrose for 24 hours. The bone samples were gradually dehydrated into 100% ethanol. The bones were loaded into a 12.3-mm diameter scanning tube and were imaged in a μCT (model μCT40, Scanco Medical). For details, see the Supplemental Methods.
Histology and bone histomorphometry.
30 μg of tetracycline per gram of mouse weight was injected into mice at day 4 and day 8 prior to sacrifice. The femurs were fixed in 10% Millonig’s formalin with 0.5% sucrose for 24 hours and were gradually dehydrated into 100% ethanol. The histological staining and bone histomorphometry were performed as described in the Supplemental Methods.
Statistics.
For all graphs, data are represented as the mean ± SD. For comparison of 2 groups, data were analyzed using a 2-tailed Student’s t test. For comparison of more than 2 groups, data were analyzed using 1-way ANOVA, and the Bonferroni procedure was used for Tukey comparison. A P value of less than 0.05 was considered significant.
Study approval.
All animal protocols and procedures used in this study were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences. The protocols for generation and use of recombinant DNAs, retroviruses, and lentiviruses were approved by Institutional Biosafety Committee of the University of Arkansas for Medical Sciences.
Author contributions
TF, SY, and HZ designed and conducted experiments in mice and osteoclasts and analyzed data. TCG and NWA performed lysosome exocytosis assays in MEFs. CGW and DEV performed bacterial phagocytosis in macrophages. KIV helped with analysis of Plekhm1 structure and construction of mutants. SGM helped with mass spectrometry and proteomics analysis. YF helped with shRNAs and lentiviral transduction. NP helped with confocal and time-lapse microscopy. TN helped with mouse models. NWA, DEV, SCM, and HZ supervised the experiments and analyzed data. SCM and HZ conceived the project. HZ wrote the manuscript with the help of all coauthors.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR062012 and AR068509) and the National Institute of Aging (P01 AG13918). We acknowledge Jeff Kamykowski at the University of Arkansas for Medical Sciences Digital Microscopy Lab for help with image analysis. The University of Arkansas for Medical Sciences Digital Microscopy Lab is supported by NIH grants 1S10OD018065 and P20 GM103429. The University of Arkansas for Medical Sciences Proteomics Core is supported by NIH grants P20GM103429 and P20GM103625.
Footnotes
DEV and CGW were supported by funding from the Arkansas Biosciences Institute and a University of Arkansas for Medical Sciences Department of Microbiology and Immunology Microgrant for Collaborative Exploratory Research. The authors would like to thank Charles A. Obrien and GibAnn Berryhill for critical reading of the manuscript prior submission. We thank Erin Hogan for her help with microscopes. We acknowledge Siu-Pok Yee for help in generating Plekhm1-flox mice.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(17):e86330. doi:10.1172/jci.insight.86330.
Contributor Information
Toshifumi Fujiwara, Email: TFujiwara@uams.edu.
Shiqiao Ye, Email: sye@uams.edu.
Thiago Castro-Gomes, Email: tcgbio@yahoo.com.br.
Yunfeng Feng, Email: fengyunfeng3@gmail.com.
Nathan Pavlos, Email: nathan.pavlos@uwa.edu.au.
Takashi Nakamura, Email: tnk@keio.jp.
Haibo Zhao, Email: hzhao@uams.edu.
References
- 1.Nakashima T, Hayashi M, Takayanagi H. New insights into osteoclastogenic signaling mechanisms. Trends Endocrinol Metab. 2012;23(11):582–590. doi: 10.1016/j.tem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377(9773):1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobacchi C, Schulz A, Coxon FP, Villa A, Helfrich MH. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol. 2013;9(9):522–536. doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- 4.Väänänen HK, Zhao H, Mulari M, Halleen JM. The cell biology of osteoclast function. J Cell Sci. 2000;113(Pt 3):377–381. doi: 10.1242/jcs.113.3.377. [DOI] [PubMed] [Google Scholar]
- 5.Lange PF, Wartosch L, Jentsch TJ, Fuhrmann JC. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature. 2006;440(7081):220–223. doi: 10.1038/nature04535. [DOI] [PubMed] [Google Scholar]
- 6.Toyomura T, et al. From lysosomes to the plasma membrane: localization of vacuolar-type H+ -ATPase with the a3 isoform during osteoclast differentiation. J Biol Chem. 2003;278(24):22023–22030. doi: 10.1074/jbc.M302436200. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Väänänen HK. Pharmacological sequestration of intracellular cholesterol in late endosomes disrupts ruffled border formation in osteoclasts. J Bone Miner Res. 2006;21(3):456–465. doi: 10.1359/JBMR.051204. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H. Membrane trafficking in osteoblasts and osteoclasts: new avenues for understanding and treating skeletal diseases. Traffic. 2012;13(10):1307–1314. doi: 10.1111/j.1600-0854.2012.01395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 11.Andrews NW. Regulated secretion of conventional lysosomes. Trends Cell Biol. 2000;10(8):316–321. doi: 10.1016/S0962-8924(00)01794-3. [DOI] [PubMed] [Google Scholar]
- 12.Dell’Angelica EC, Mullins C, Caplan S, Bonifacino JS. Lysosome-related organelles. FASEB J. 2000;14(10):1265–1278. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- 13.Ménasché G, et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet. 2000;25(2):173–176. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- 14.Stinchcombe J, Bossi G, Griffiths GM. Linking albinism and immunity: the secrets of secretory lysosomes. Science. 2004;305(5680):55–59. doi: 10.1126/science.1095291. [DOI] [PubMed] [Google Scholar]
- 15.Wilson SM, et al. A mutation in Rab27a causes the vesicle transport defects observed in ashen mice. Proc Natl Acad Sci U S A. 2000;97(14):7933–7938. doi: 10.1073/pnas.140212797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277(14):12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- 17.Wu XS, et al. Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4(4):271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 18.Ménager MM, et al. Secretory cytotoxic granule maturation and exocytosis require the effector protein hMunc13-4. Nat Immunol. 2007;8(3):257–267. doi: 10.1038/ni1431. [DOI] [PubMed] [Google Scholar]
- 19.van Meel E, et al. Disruption of the Man-6-P targeting pathway in mice impairs osteoclast secretory lysosome biogenesis. Traffic. 2011;12(7):912–924. doi: 10.1111/j.1600-0854.2011.01203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimada-Sugawara M, et al. Rab27A regulates transport of cell surface receptors modulating multinucleation and lysosome-related organelles in osteoclasts. Sci Rep. 2015;5:e86330. doi: 10.1038/srep09620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charles JF, et al. The collection of NFATc1-dependent transcripts in the osteoclast includes numerous genes non-essential to physiologic bone resorption. Bone. 2012;51(5):902–912. doi: 10.1016/j.bone.2012.08.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palokangas H, Mulari M, Väänänen HK. Endocytic pathway from the basal plasma membrane to the ruffled border membrane in bone-resorbing osteoclasts. J Cell Sci. 1997;110(Pt 15):1767–1780. doi: 10.1242/jcs.110.15.1767. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Laitala-Leinonen T, Parikka V, Väänänen HK. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J Biol Chem. 2001;276(42):39295–39302. doi: 10.1074/jbc.M010999200. [DOI] [PubMed] [Google Scholar]
- 24.Van Wesenbeeck L, et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest. 2007;117(4):919–930. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinholt FP, Hultenby K, Heinegård D, Marks SC, Norgård M, Anderson G. Extensive clear zone and defective ruffled border formation in osteoclasts of osteopetrotic (ia/ia) rats: implications for secretory function. Exp Cell Res. 1999;251(2):477–491. doi: 10.1006/excr.1999.4585. [DOI] [PubMed] [Google Scholar]
- 26.Hotfilder M, Baxendale S, Cross MA, Sablitzky F. Def-2, -3, -6 and -8, novel mouse genes differentially expressed in the haemopoietic system. Br J Haematol. 1999;106(2):335–344. doi: 10.1046/j.1365-2141.1999.01551.x. [DOI] [PubMed] [Google Scholar]
- 27.Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, Yoshimori T. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell. 2010;21(23):4162–4172. doi: 10.1091/mbc.E10-06-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schou KB, Andersen JS, Pedersen LB. A divergent calponin homology (NN-CH) domain defines a novel family: implications for evolution of ciliary IFT complex B proteins. Bioinformatics. 2014;30(7):899–902. doi: 10.1093/bioinformatics/btt661. [DOI] [PubMed] [Google Scholar]
- 29.Niethammer M, et al. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28(3):697–711. doi: 10.1016/S0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 30.Taya S, et al. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3epsilon complex through kinesin-1. J Neurosci. 2007;27(1):15–26. doi: 10.1523/JNEUROSCI.3826-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32(3):199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, et al. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 2007;130(5):811–823. doi: 10.1016/j.cell.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 33.Qing H, et al. Demonstration of osteocytic perilacunar/canalicular remodeling in mice during lactation. J Bone Miner Res. 2012;27(5):1018–1029. doi: 10.1002/jbmr.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Fattore A, et al. A new heterozygous mutation (R714C) of the osteopetrosis gene, pleckstrin homolog domain containing family M (with run domain) member 1 (PLEKHM1), impairs vesicular acidification and increases TRACP secretion in osteoclasts. J Bone Miner Res. 2008;23(3):380–391. doi: 10.1359/jbmr.071107. [DOI] [PubMed] [Google Scholar]
- 35.Destaing O, Saltel F, Géminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14(2):407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Destaing O, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118(Pt 13):2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez A, Webster P, Ortego J, Andrews NW. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J Cell Biol. 1997;137(1):93–104. doi: 10.1083/jcb.137.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye S, et al. LIS1 regulates osteoclast formation and function through its interactions with dynein/dynactin and Plekhm1. PLoS ONE. 2011;6(11):e86330. doi: 10.1371/journal.pone.0027285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferron M, et al. A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27(8):955–969. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McEwan DG, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57(1):39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 42.DeSelm CJ, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21(5):966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem. 1997;272(20):13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- 44.Hussain SK, Voth DE. Coxiella subversion of intracellular host signaling. Adv Exp Med Biol. 2012;984:131–140. doi: 10.1007/978-94-007-4315-1_7. [DOI] [PubMed] [Google Scholar]
- 45.Akter KA, Mansour MA, Hyodo T, Ito S, Hamaguchi M, Senga T. FAM98A is a novel substrate of PRMT1 required for tumor cell migration, invasion, and colony formation. Tumour Biol. 2016;37(4):4531–4539. doi: 10.1007/s13277-015-4310-5. [DOI] [PubMed] [Google Scholar]
- 46.Mizukami J, et al. Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol Cell Biol. 2002;22(4):992–1000. doi: 10.1128/MCB.22.4.992-1000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou W, et al. Talin1 and Rap1 are critical for osteoclast function. Mol Cell Biol. 2013;33(4):830–844. doi: 10.1128/MCB.00790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21(2):115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 49.Bo T, et al. 2016. Characterization of a Relatively Malignant form of Osteopetrosis Caused by a Novel Mutation in the PLEKHM1 Gene. J Bone Miner Res. doi: 10.1002/jbmr.2885. [published online ahead of print June 13, 2016]. doi: 10.1002/jbmr.2885. [DOI] [PubMed] [Google Scholar]
- 50.Caviston JP, Zajac AL, Tokito M, Holzbaur EL. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol Biol Cell. 2011;22(4):478–492. doi: 10.1091/mbc.E10-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Encarnação M, et al. A Rab3a-dependent complex essential for lysosome positioning and plasma membrane repair. J Cell Biol. 2016;213(6):631–640. doi: 10.1083/jcb.201511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khatter D, Sindhwani A, Sharma M. Arf-like GTPase Arl8: Moving from the periphery to the center of lysosomal biology. Cell Logist. 2015;5(3):e86330. doi: 10.1080/21592799.2015.1086501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13(4):453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016;18(4):404–417. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGourty K, Thurston TL, Matthews SA, Pinaud L, Mota LJ, Holden DW. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science. 2012;338(6109):963–967. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muhammad E, et al. PLEKHM2 mutation leads to abnormal localization of lysosomes, impaired autophagy flux and associates with recessive dilated cardiomyopathy and left ventricular noncompaction. Hum Mol Genet. 2015;24(25):7227–7240. doi: 10.1093/hmg/ddv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33(2):176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell. 2011;21(6):1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oreffo RO, Mundy GR, Seyedin SM, Bonewald LF. Activation of the bone-derived latent TGF beta complex by isolated osteoclasts. Biochem Biophys Res Commun. 1989;158(3):817–823. doi: 10.1016/0006-291X(89)92795-2. [DOI] [PubMed] [Google Scholar]
- 60.Tang Y, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15(7):757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villa A, Guerrini MM, Cassani B, Pangrazio A, Sobacchi C. Infantile malignant, autosomal recessive osteopetrosis: the rich and the poor. Calcif Tissue Int. 2009;84(1):1–12. doi: 10.1007/s00223-008-9196-4. [DOI] [PubMed] [Google Scholar]
- 62.Kornak U, et al. Mutations in the a3 subunit of the vacuolar H(+)-ATPase cause infantile malignant osteopetrosis. Hum Mol Genet. 2000;9(13):2059–2063. doi: 10.1093/hmg/9.13.2059. [DOI] [PubMed] [Google Scholar]
- 63.Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23(4):447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- 64.Neutzsky-Wulff AV, et al. Severe developmental bone phenotype in ClC-7 deficient mice. Dev Biol. 2010;344(2):1001–1010. doi: 10.1016/j.ydbio.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 65.Guerrini MM, et al. Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am J Hum Genet. 2008;83(1):64–76. doi: 10.1016/j.ajhg.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henriksen K, Karsdal MA, Martin TJ. Osteoclast-derived coupling factors in bone remodeling. Calcif Tissue Int. 2014;94(1):88–97. doi: 10.1007/s00223-013-9741-7. [DOI] [PubMed] [Google Scholar]
- 67.Thudium CS, et al. A comparison of osteoclast-rich and osteoclast-poor osteopetrosis in adult mice sheds light on the role of the osteoclast in coupling bone resorption and bone formation. Calcif Tissue Int. 2014;95(1):83–93. doi: 10.1007/s00223-014-9865-4. [DOI] [PubMed] [Google Scholar]
- 68.Baron R, Neff L, Louvard D, Courtoy PJ. Cell-mediated extracellular acidification and bone resorption: evidence for a low pH in resorbing lacunae and localization of a 100-kD lysosomal membrane protein at the osteoclast ruffled border. J Cell Biol. 1985;101(6):2210–2222. doi: 10.1083/jcb.101.6.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4(3):202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 70.Baron R. Molecular mechanisms of bone resorption by the osteoclast. Anat Rec. 1989;224(2):317–324. doi: 10.1002/ar.1092240220. [DOI] [PubMed] [Google Scholar]
- 71.Dell’Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21(4):552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 72.Zhao H, Ito Y, Chappel J, Andrews NW, Teitelbaum SL, Ross FP. Synaptotagmin VII regulates bone remodeling by modulating osteoclast and osteoblast secretion. Dev Cell. 2008;14(6):914–925. doi: 10.1016/j.devcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schiefermeier N, et al. The late endosomal p14-MP1 (LAMTOR2/3) complex regulates focal adhesion dynamics during cell migration. J Cell Biol. 2014;205(4):525–540. doi: 10.1083/jcb.201310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jordens I, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11(21):1680–1685. doi: 10.1016/S0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 75.Bhogaraju S, et al. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341(6149):1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Permuth-Wey J, et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun. 2013;4:e86330. doi: 10.1038/ncomms2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J Cell Sci. 2010;123(Pt 2):202–212. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- 78.Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J Neurosci. 2011;31(47):17207–17219. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamiya A, et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet. 2006;15(22):3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- 80.Ng PY, et al. Disruption of the dynein-dynactin complex unveils motor-specific functions in osteoclast formation and bone resorption. J Bone Miner Res. 2013;28(1):119–134. doi: 10.1002/jbmr.1725. [DOI] [PubMed] [Google Scholar]
- 81.Zhou J, Ye S, Fujiwara T, Manolagas SC, Zhao H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. J Biol Chem. 2013;288(42):30064–30074. doi: 10.1074/jbc.M113.478750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujiwara T, Zhou J, Ye S, Zhao H. RNA-binding protein Musashi2 induced by RANKL is critical for osteoclast survival. Cell Death Dis. 2016;7:e86330. doi: 10.1038/cddis.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao H, Ettala O, Väänänen HK. Intracellular membrane trafficking pathways in bone-resorbing osteoclasts revealed by cloning and subcellular localization studies of small GTP-binding rab proteins. Biochem Biophys Res Commun. 2002;293(3):1060–1065. doi: 10.1016/S0006-291X(02)00326-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.