Abstract
Cancer registries are an important source of population-level information on brain tumor incidence and survival. Surveillance, Epidemiology, and End Results (SEER) registries currently collect data on specific required factors related to brain tumors as defined by the American Joint Commission on Cancer, including World Health Organization (WHO) grade, MGMT methylation and 1p/19q codeletion status. We assessed ‘completeness’, defined as having valid values over the time periods that they have been collected, overall, by year, histology, and registry. Data were obtained through a SEER custom data request for four factors related to brain tumors for the years 2004–2012 (3/4 factors were collected only from 2010 to 2012). SEER*Stat was used to generate frequencies of ‘completeness’ for each factor overall, and by year, histology and registry. The four factors varied in completeness, but increased over time. WHO grade has been collected the longest, and showed significant increases in completeness. Completeness of MGMT and 1p/19q codeletion was highest for glioma subtypes for which testing is recommended by clinical practice guidelines. Completeness of all factors varied by histology and cancer registry. Overall, several of the factors had high completeness, and all increased in completeness over time. With increasing focus on ‘precision medicine’ and the incorporation of molecular parameters into the 2016 WHO CNS tumor Classification, it is critical that the data are complete, and factors collected at the population level are fully integrated into cancer reporting. It is critical that cancer registries continue to collect established and emerging prognostic and predictive factors.
Keywords: WHO grade, Brain tumors, Cancer registration, MGMT methylation, 1p/19q codeletion
Introduction
Cancer registries are a critical resource for monitoring cancer incidence and estimating cancer survival on a population level. The minimum level of data required for these registries to fulfill this function is the collection of basic demographic information, tumor site, and assigned histology for every new cancer case diagnosed within their geographic region. As cancer research progresses, investigators have uncovered many other factors of significance to the natural history of a disease and in estimating prognosis. Many of these—including basic treatment information—are currently required data elements collected for all cases by registries within the National Cancer Institute's (NCI) Surveillance, Epidemiology and End Results (SEER) registry program. SEER registries also collect additional cancer-specific variables, such as tumor markers, not required by other surveillance organizations.
Cancer staging is a critical component of determining cancer prognosis and proper course of treatment and provides a rubric for evaluating how much cancer is in a person's body and where the cancer is located. The American Joint Committee on Cancer (AJCC) has developed a classification and staging system used for all cancers except those within the central nervous system (CNS). For these tumors, AJCC has developed the Collaborative Staging (CS) schema, an important standard for cancer registration. SEER registries regularly collect information according to site-specific CS schema as defined by the Seventh Edition of AJCC, though the time when data collection began for each factor and whether or not these are required data varies significantly [1]. Brain and other CNS tumors have been classified according to the 2000 [2] and 2007 [3] editions of the World Health Organization (WHO). The 2007 Classification of Tumours of the CNS [3] assigns a WHO grade (I through IV) based on predicted clinical behavior. Grading assignments are recorded by cancer registrars as Collaborative Stage Site-Specific Factor 1—WHO Grade Classification according to the AJCC CS schema [4] At the time of this study, the Eighth Edition of AJCC is under revision with publication due by the end of 2016. 2016 WHO Classification of Tumours of the CNS was revised and published in May 2016 [5]. Both these revisions will impact future cancer registration practices.
For brain and CNS tumors, SEER registries currently collect information on eight site-specific factors (SSF) defined by the CS schema, including: WHO grade (SSF 1), Ki-67/IB-1 Labeling Index (SSF 2), Karnofsky performance status (KPS) (SSF 3), promoter methylation status of the gene O-6-methylguanine-DNA methyltransferase (MGMT, SSF 4), deletion of the short (p) arm of chromosome 1 (SSF 5), deletion [also called loss of heterozygosity (LOH)] of the long (q) arm of chromosome 19 (SSF 6), extent of surgical resection (SSF 7), and whether a tumor was unifocal or multifocal (SSF 8) (please see supplementary Table 1 for more information on these factors). However, only 4 of these 8 factors have mandatory collection: WHO grade, methylation of the MGMT gene promoter, 1p deletion and 19q deletion. These factors are important to track on a population level, as they are associated with clinical outcomes for specific brain and CNS histologies.
Cancer registrars obtain patient information through review of patients' pathology reports and other clinical records. Though some of these factors are relevant to brain tumor prognosis, they may not be recorded in the medical records of all patients. This analysis aimed to determine the availability and completeness of CS SSF data collection for brain and CNS cases in the SEER database.
Methods
Data were obtained from the SEER program for all 18 registries from the years 2004–2012 [6]. Registries included within this program represent approximately 28 % of the US as of the 2010 census [7]. They are funded by the NCI to conduct active follow-up for clinical outcomes on all cases, as well as to collect additional pieces of data that may not be collected by other central cancer registries. Data for the SSF 1 variable are currently available in the public access SEER data release. Data for the other SSFs were requested from SEER and approved by their custom data review group. This current study was conducted under exempt approval from the University Hospitals of Cleveland Institutional Review Board (IRB).
All cases of malignant and non-malignant brain and CNS tumors diagnosed in the SEER 18 registries between the years of 2004 and 2012 were included in this analysis. Brain and CNS tumors were defined as tumors occurring in the following categories as listed by ICD-O-3 (International Classification of Diseases for Oncology, 3rd edition) [8] three site codes and CS schema: C70.0, C71.0–C71.9 (Brain [9]), C70.1, C70.9, C72.0–C72.5, C72.8–C72.9 (CNS Other [10]), C75.1, C75.2, C75.3 (Intracranial Glands [11]). SEER*Stat 8.2.1 [12] was used to generate a list of cases. SSFs 4–6 were collected for brain and CNS tumors only after 2010, and, as a result, only cases diagnosed from 2010 to 2012 were used for these analyses. SSF 4–6 were examined for brain and CNS tumors in the broad category of gliomas (ICD-O-3 histology codes: 9380–9384, 9391–9460, 9480 [13]) only. Completeness of each site specific factor (SSF) was assessed for brain tumor cases overall, as well as by histologic types defined by the CBTRUS histologic grouping scheme [13], and year of diagnosis. Applicable years, sites, and histologies are listed in supplementary Table 1. Definitions of complete and incomplete varied by SSF and are also listed in supplementary Table 1.
Results
Frequencies, percent complete and percent incomplete for each SSF by histology are presented in Table 1. Frequencies and percent completeness by each of the SEER 18 registries are presented in Table 2. The percentage of records with complete WHO grade for all gliomas, embryonal tumors, meningioma, and nerve sheath tumors by year are presented in Fig. 1.
Table 1. Frequency, percent complete and percent incomplete by required site specific factors (SSF) and histology (SEER 2004-2012, varying).
| Variable | World Health Organization (WHO) grade classification |
Methylation of 06-methylguanine- methyltransferase (MGMT) |
Chromosome lp: loss of heterozygosity (LOH) |
Chromosome 19q: loss of heterozygosity (LOH) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSF | 1 | 4 | 5 | 6 | ||||||||
|
|
|
|
|
|||||||||
| Histology | Total | Complete (%) |
Incomplete (%) |
Total | Complete (%) |
Incomplete (%) |
Total | Complete (%) |
Incomplete (%) |
Total | Complete (%) |
Incom- plete (%) |
| Pilocytic astrocytoma | 2291 | 64.1 | 35.9 | 796 | 1.1 | 98.9 | 796 | 1.5 | 98.5 | 796 | 1.4 | 98.6 |
| Diffuse astrocytoma | 3760 | 76.9 | 23.1 | 1141 | 5.7 | 94.3 | 1141 | 8.8 | 91.2 | 1141 | 8.9 | 91.1 |
| Anaplastic astrocytoma | 2724 | 86.5 | 13.5 | 1004 | 8.4 | 91.6 | 1004 | 15.3 | 84.7 | 1004 | 15.4 | 84.6 |
| Unique astrocytoma variants | 416 | 59.6 | 40.4 | 144 | 1.4 | 98.6 | 144 | 2.1 | 97.9 | 144 | 2.1 | 97.9 |
| Glioblastoma | 22,528 | 69.3 | 30.7 | 8083 | 12.2 | 87.8 | 8083 | 4.7 | 95.3 | 8083 | 4.7 | 95.3 |
| Oligodendroglioma | 1972 | 84.1 | 15.9 | 589 | 6.1 | 93.9 | 589 | 49.4 | 50.6 | 589 | 49.7 | 50.3 |
| Anaplastic oligodendroglioma | 883 | 87.3 | 12.7 | 290 | 8.6 | 91.4 | 290 | 43.4 | 56.6 | 290 | 43.1 | 56.9 |
| Oligoastrocytic tumors | 1506 | 87.9 | 12.1 | 515 | 9.5 | 90.5 | 515 | 47.8 | 52.2 | 515 | 49.3 | 50.7 |
| Ependymal tumors | 3002 | 49.5 | 50.5 | 1053 | 0.9 | 99.1 | 1053 | 0.3 | 99.7 | 1053 | 0.3 | 99.7 |
| Glioma malignant, NOS | 1059 | 40.2 | 59.8 | 361 | 2.8 | 97.2 | 361 | 6.4 | 93.6 | 361 | 6.4 | 93.6 |
| Choroid plexus tumors | 387 | 41.9 | 58.1 | – | – | – | – | – | – | – | – | – |
| Other neuro-epithelial tumors | 53 | 43.4 | 56.6 | 20 | 5.0 | 95.0 | 20 | 0.0 | 100.0 | 20 | 0.0 | 100.0 |
| Neuronal and mixed neuronal-glial tumors | 1703 | 52.4 | 47.6 | 118 | 0.8 | 99.2 | 118 | 7.6 | 92.4 | 118 | 7.6 | 92.4 |
| Embryonal tumors | 1825 | 49.9 | 50.1 | – | – | – | – | – | – | – | – | – |
| Medulloblastoma | 1058 | 57.3 | 42.7 | – | – | – | – | – | – | – | – | – |
| PNET | 365 | 43.8 | 56.2 | – | – | – | – | – | – | – | – | – |
| ATRT | 185 | 45.4 | 54.6 | – | – | – | – | – | – | – | – | – |
| All other embryonal | 217 | 28.1 | 71.9 | – | – | – | – | – | – | – | – | – |
| Nerve sheath tumors | 7811 | 11.6 | 88.4 | – | – | – | – | – | – | – | – | – |
| Meningioma | 28,058 | 52.3 | 47.7 | – | – | – | – | – | – | – | – | – |
| Mesenchymal tumors | 523 | 26.0 | 74.0 | – | – | – | – | – | – | – | – | – |
| Primary melanocytic lesions | 51 | 3.9 | 96.1 | – | – | – | – | – | – | – | – | – |
| Other neoplasms related to the meninges | 1371 | 24.7 | 75.3 | – | – | – | – | – | – | – | – | – |
| Germ cell tumors, cysts and heterotopias | 623 | 1.4 | 98.6 | – | – | – | – | – | – | – | – | – |
| Tumors of the pituitary | 15,409 | 1.5 | 98.5 | – | – | – | – | – | – | – | – | – |
| Craniopharyngioma | 1243 | 20.6 | 79.4 | – | – | – | – | – | – | – | – | – |
| Hemangioma | 869 | 2.2 | 97.8 | – | – | – | – | – | – | – | – | – |
| Neoplasm, unspecified | 720 | 5.7 | 94.3 | – | – | – | – | – | – | – | – | – |
| All other | 28 | 17.9 | 82.1 | – | – | – | – | – | – | – | – | – |
| Total | 101,116 | 46.5 | 53.5 | 14,115 | 9.0 | 91.0 | 14,115 | 9.5 | 90.5 | 14,115 | 9.6 | 90.4 |
Table 2. Frequency and percent complete by SEER registries by required site specific factors (SSF) (SEER, 2004–2012, varying).
| Variable | World Health Organization (WHO) grade classification | Methylation of O6-methyl-guanine-methyltransferase (MGMT) | Chromosome 1p: loss of heterozygosity (LOH) | Chromosome 19q: loss of heterozygosity (LOH) | ||||
|---|---|---|---|---|---|---|---|---|
| SSF | 1 | 4 | 5 | 6 | ||||
|
|
|
|
|
|||||
| SEER registry | Total | Complete (%) | Total | Complete (%) | Total | Complete (%) | Total | Complete (%) |
| Alaska natives | 118 | 40.7 | – | – | – | – | – | – |
| California | 41,758 | 44.9 | 5664 | 9.3 | 5664 | 8.8 | 5664 | 8.9 |
| San Francisco-Oakland SMSA | 5366 | 52.8 | 733 | 9.0 | 733 | 11.1 | 733 | 10.9 |
| California excluding SF/SJM/LA | 22,486 | 43.3 | 3,287 | 6.1 | 3287 | 9.0 | 3287 | 9.0 |
| Los Angeles | 10,857 | 42.0 | 1248 | 16.9 | 1,248 | 6.8 | 1,248 | 7.0 |
| San Jose-Monterey | 3049 | 52.3 | 396 | 13.1 | 396 | 9.8 | 396 | 9.8 |
| Connecticut | 4712 | 47.9 | 679 | 9.3 | 679 | 6.9 | 679 | 6.9 |
| Detroit (metropolitan) | 5560 | 48.9 | 697 | 20.1 | 697 | 14.1 | 697 | 14.3 |
| Georgia | 10,147 | 47.0 | 1449 | 8.7 | 1449 | 5.6 | 1449 | 6.0 |
| Atlanta (metropolitan) | 3467 | 44.2 | 462 | 12.3 | 462 | 6.5 | 462 | 6.9 |
| Rural Georgia | 150 | 48.0 | 25 | 8.0 | 25 | 0.0 | 25 | 0.0 |
| Greater Georgia | 6530 | 48.5 | 962 | 7.0 | 962 | 5.3 | 962 | 5.7 |
| Hawaii | 1322 | 51.3 | 137 | 2.9 | 137 | 6.6 | 137 | 6.6 |
| Iowa | 3957 | 47.1 | 629 | 0.8 | 629 | 7.0 | 629 | 7.0 |
| Kentucky | 5345 | 49.7 | 804 | 12.4 | 804 | 13.1 | 804 | 13.3 |
| Louisiana | 4849 | 36.7 | 716 | 1.0 | 716 | 4.7 | 716 | 4.7 |
| New Jersey | 11,128 | 47.2 | 1655 | 9.1 | 1655 | 5.6 | 1655 | 5.5 |
| New Mexico | 2195 | 50.8 | 280 | 4.3 | 280 | 10.0 | 280 | 9.6 |
| Seattle (puget sound) | 6527 | 50.9 | 921 | 14.2 | 921 | 28.0 | 921 | 28.0 |
| Utah | 3498 | 50.3 | 471 | 1.7 | 471 | 10.6 | 471 | 10.4 |
| Total | 101,116 | 46.5 | 14,115 | 9.0 | 14,115 | 9.5 | 14,115 | 9.6 |
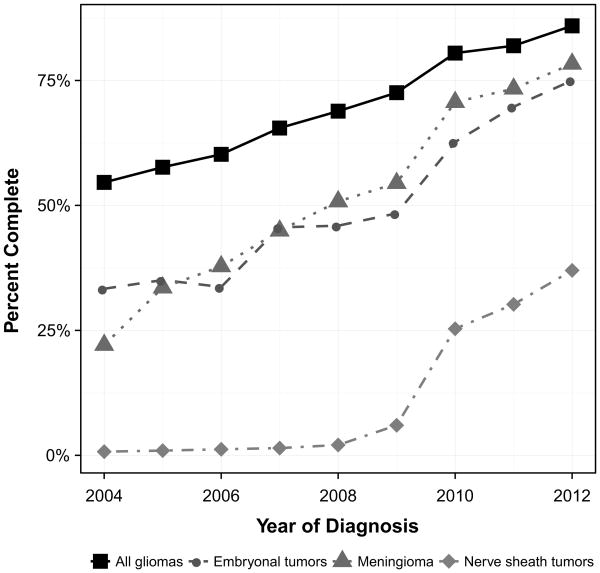
Fig. 1.
Percent of records with complete WHO grade by year of diagnosis, SEER 2004–2012
CS site-specific factor 1—World Health Organization (WHO) grade classification
Overall, WHO grade completeness steadily increased from 2004 to 2012, increasing from 30.3 to 63.9 %. Percent completeness by year is presented in Fig. 2. The tumors with the largest increase in completeness were the astrocytoma entities, increasing from 18.2 to 80.8 %. Other neuro-epithelial tumors decreased in completeness from 2004 to 2012, 44.4–18.2 %. From 2004 to 2012, the histologies with the highest average percentage of completion for WHO grade were anaplastic oligodendroglioma (87.9 %), oligoastrocytic tumors (87.9 %), and anaplastic astrocytoma (86.1 %). In 2012, the tumors with the highest percentage of completion were oligoastrocytic tumors with 98.1 % completeness. Completeness for WHO grade by registry ranged from 36.7 to 52.8 % (Table 2).
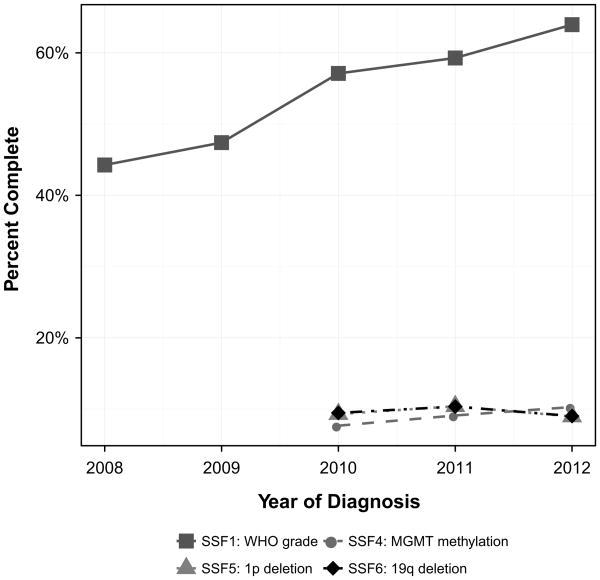
Fig. 2.
Percent Completeness trend for site specific factors (SSF): SSF1, SSF2, SSF3, SSF7, and SSF8 from SEER 2008–2012
CS site-specific factor 4—methylation of the O6-methylguanine-methyltransferase (MGMT) gene promoter
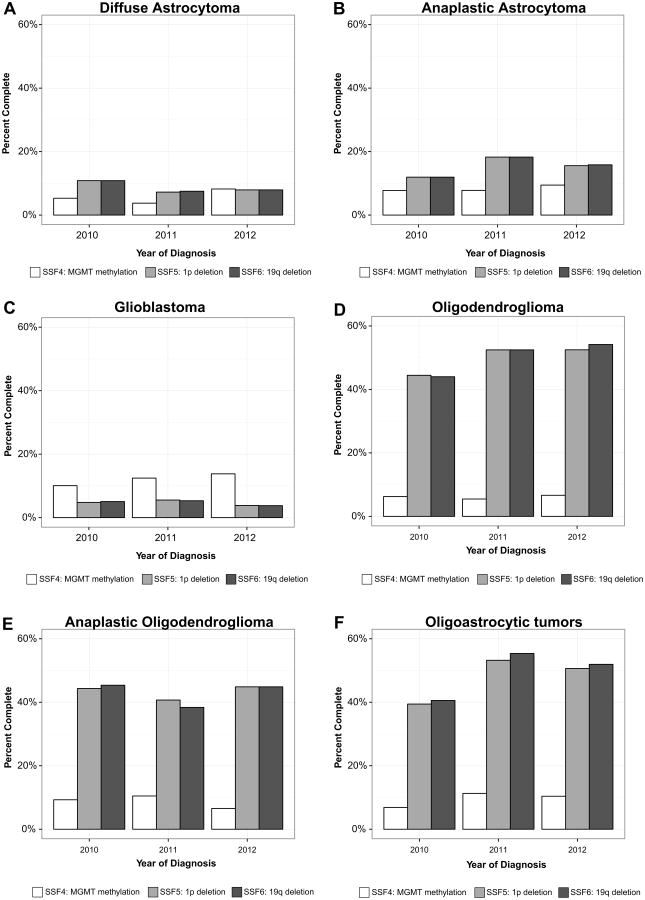
Overall, methylation of MGMT completeness increased slightly from 2010 to 2012, increasing from 7.7 to 10.3 %. Percent of record completeness by year for diffuse astrocytoma, anaplastic astrocytoma, glioblastoma, oligodendro-glioma, anaplastic oligodendroglioma, and oligoastrocytic tumors are presented in Fig. 3. The histology with the largest increase in completeness was oligoastrocytic tumors, increasing from 6.9 to 10.4 %. From 2010 to 2012, the histologies with the highest average percentage of completion for methylation of MGMT were glioblastoma (12.1 %), oligoastrocytic tumors (9.5 %), and anaplastic oligodendroglioma (8.8 %). In 2012, the histology with the highest percentage of completion was glioblastoma with 13.8 % completeness. Completeness for methylation of MGMT by registry ranged from 0.8 to 20.1 % (Table 2).
Fig. 3.
Percent of record complete by year for required site specific factors (SSF) 4–6 for (a) diffuse astrocytoma, (b) anaplastic astrocytoma, (c) glioblastoma, (d) oligodendroglioma, (e) anaplastic oligodendroglioma, and (f) oligoastrocytic tumors, SEER 2010–2012
CS site-specific factor 5—chromosome 1p: loss of heterozygosity (LOH)
Overall, 1p deletion (also known as loss of heterozygosity) completeness decreased negligibly from 2010 to 2012, decreasing from 9.3 to 9.0 %. Percent of record completeness by year for diffuse astrocytoma, anaplastic astrocytoma, glioblastoma, oligodendroglioma, anaplastic oligodendroglioma, and oligoastrocytic tumors are presented in Fig. 3. The histology with the largest measureable increase in completeness was oligoastrocytic tumors, increasing from 39.4 to 50.6 %. Several histologies experienced decreases in completeness from 2010 to 2012. The histology with the largest measureable decrease was diffuse astrocytoma, decreasing from 10.8 to 7.9 %. From 2010 to 2012, the average change in 1p deletion completeness was a 2.4 % increase. From 2010 to 2012, the histologies with the highest average percentage of completion were oligodendroglioma (49.8 %), oligoastrocytic tumors (47.8 %), and anaplastic oligodendroglioma (43.3 %). In 2012, the histology with the highest percentage of completion was oligodendroglioma with 52.5 %. Completeness for 1p deletion by registry ranged from 0.0 to 28.0 % (Table 2).
CS site-specific factor 6—chromosome 19q: loss of heterozygosity (LOH)
Overall, 19q deletion (also known has loss of heterozygosity) completeness slightly decreased from 2010 to 2012, decreasing from 9.5 to 9.0 %. Percent of record completeness by year for diffuse astrocytoma, anaplastic astrocytoma, glioblastoma, oligodendroglioma, anaplastic oligodendroglioma, and oligoastrocytic tumors are presented in Fig. 3. The histology with the largest increase in completeness was oligoastrocytic tumors, increasing from 40.6 to 51.9 %. Several histologies experienced decreases in completeness from 2010 to 2012. The histology with the largest decrease was diffuse astrocytoma, decreasing from 10.8 to 7.9 %. From 2010 to 2012, the histologies with the highest average percentage of completion for 19q deletion were oligodendroglioma (50.2 %), oligoastrocytic tumors (49.3 %), and anaplastic oligodendroglioma (42.9 %). In 2012, the histology with the highest percentage of completion was oligodendroglioma with 54.1 %. Completeness for 19q deletion by registry ranged from 0.0 to 28.0 % (Table 2).
Discussion
Completeness of SSF for brain tumors in the SEER 18 data varied significantly by SSF, year of diagnosis, and histologic type. There are many factors that could lead to this variation. Only 4 of the 8 described factors (see supplementary Table 1) are required by SEER for collection: SSF1, SFF4-6. Some of these factors are only (or mostly) relevant to specific histologic tumor types. SSF1 is potentially applicable to all brain tumor types and, correspondingly, has the highest completeness. SSF4-SSF6 components are mostly applicable to specific glioma subtypes and, therefore, it is expected that completeness of these factors would vary significantly by histology.
WHO grade is a critical factor for brain and CNS tumor prognosis. Previous analyses have examined the completeness of this variable, as well as the concordance (defined as being coded with a WHO grade that is appropriate for the histology code) for selected histologies [14]. Overall, when this variable is complete, it is generally coded with a value that is an accurate WHO grade for that histology. It is to be noted that some tumors that occur within the sites classified as brain and CNS do not have an assigned WHO grade. These include tumors that are classified as endocrine tumors (e.g. pituitary adenoma), or hematopoietic tumors (e.g. primary CNS lymphomas). Some brain and CNS tumors are not assigned a WHO grade in clinical practice (e.g. hemangioblastoma, and schwannoma) as these measures may not be clinically important. Both of these may contribute to lower completeness of WHO grade overall. While there is some correlation between the ICD-O-3 histology and behavior codes, WHO grade when assigned is widely used for treatment decisions for patients with brain tumors. As WHO grade is a factor that is considered to be prognostic [3], it is critical in relevant histologies to have high completeness of this variable for measurement of grade-specific survival patterns.
Methylation of the promoter of the MGMT gene has been validated as a significant predictor of improved survival in glioblastoma [15, 16]. Methylation of the gene promoter inhibits transcription of this gene, thus ‘silencing’ the gene. MGMT is a DNA repair protein, and it is assumed that the decreases in protein levels increase sensitivity to the alkylating chemotherapies (e.g. temozolomide) that are often used in the treatment of gliomas [17]. Studies have shown that tumors with methylated MGMT promoters are more sensitive to alkylating chemotherapy and radiotherapy [18], and the methylation status is often used to predict the likelihood of response in glioma patients to temozolomide [19]. However, methylation status of MGMT is not useful for many brain and CNS histologies, as MGMT activity varies significantly between histologies, as well as by WHO grade [20]. MGMT methylation has been found in only about 40 % of gliomas and very rarely in non-glial brain tumors [21].
Though methylation of the MGMT promoter can be found in lower grade gliomas, it only has significant clinical relevance in glioblastoma. There are many reasons why this test might not be ordered as part of standard clinical practice, contributing to low completeness. Some physicians may treat all patients with temozolomide regardless of MGMT methylation status, and as a result this test would not be relevant to determining an individual's treatment. One of the more common methods to measure the level of promoter methylation of MGMT uses a combination of real-time quantitative PCR and methylation-specific PCR, and many hospitals do not have the infrastructure to conduct these tests in a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory environment. Comparison of techniques has suggested that pyrosequencing may provide a more accurate assessment of methylation status [22], but this technique is significantly more expensive and may not be feasible for widespread implantation in the clinic. These analyses have also shown that methylation quantification for a particular tumor may vary depending on the test used.
Methylation of the MGMT promoter is quantified as the total number of the 17 CpG island sites within this region with an attached methyl group, although not all 17 sites are examined consistently. The point along this scale when the promoter is determined to be fully ‘silenced’ is not well-established and the results of these test may be difficult to interpret. As these tests are often not conducted within most hospitals, the long turnaround time for receiving results from an outside institution may deter pathologists from ordering these tests. Additionally, the test for MGMT methylation may not have been recommended by clinical practice guidelines consistently and, therefore, the reimbursement by third parties could be a limiting factor for ordering the test. Though this factor was first determined to be prognostic in 2005 [15], the National Comprehensive Cancer Network (NCCN; http://www.nccn.org/) guidelines for CNS tumors did not recommended this test during the time-period examined in this study. As a result the availability of this biomarker in SEER in this time frame (2010–2012) data is low (12 % of glioblastomas had reported MGMT test results). MGMT testing for glioblastoma has been incorporated in NCCN guidelines since 2013 which is expected to influence future test rates based on SEER data.
The utility of MGMT methylation as a prognostic marker may also be diminishing in light of recent molecular analyses. The results of the European Organisation for Research and Treatment of Cancer (EORTC) 26951 trial found that the prognostic outcome predicted by MGMT promoter methylation varied depending on the IDH1 mutation status of the tumor in grade III gliomas [23], and suggest use of this biomarker in the context of known IDH1 status. The German Oncology Working Group (NOA) 08 trial found MGMT methylation to have significant prognostic value in the elderly, where incidence of IDH1/2 mutations and the G-CIMP phenotype are rare compared to younger cohorts [24, 25]. Recent analyses of data generated by The Cancer Genome Atlas have confirmed the importance of methylation in predicting prognosis in glioma [26] with a strong survival benefit conferred by having a highly methylated phenotype [glioma-CpG island methylator phenotype (G-CIMP)] [27]. These phenotypes are correlated [28], though the G-CIMP phenotype is much rarer in glioblastoma than MGMT methylation. While this phenotype may be a better predictor of survival in glioma than promoter methylation of MGMT alone [29], genome-wide methylation assays present many of the same problems as methylation quantification at a specific locus.
Whole-arm deletions in the short arm of chromosome 1 and the long arm of chromosome 19 have been recognized as markers of prolonged median survival in oligodendroglioma since 1998 [30]. This marker significantly predicts positive response to chemotherapy and radiation treatment in oligodendroglioma and anaplastic oligodendroglioma [31–33], and is most relevant in lower grade gliomas as compared to glioblastoma. Recent analyses of data collected by the Cancer Genome Atlas have demonstrated that these alterations can precisely stratify low grade gliomas by prognosis when combined with mutation status in isocitrate dehydrogenase 1/2 (IDH1/2) [26, 34]. These results suggest that 1p/19q deleted glioma is a distinct entity with a separate process of gliomagenesis than gliomas that do not have 1p/19q deletion. With the 2016 revision to the WHO classification of tumors of the CNS, characterization of this feature is essential to diagnosing oligodendroglioma.
Testing for these markers has many of the same limitations as those found in testing MGMT methylation status. Some physicians may choose treatment that does not vary with their patient's status on these markers, and thus ordering these tests would not be relevant. These markers are most often evaluated using fluorescence in situ hybridization (FISH), and many hospitals may not have the equipment or personnel to conduct these tests. As a result, these tests may require sending tissue samples to other laboratories, causing a delay that may deter pathologists from ordering these tests. The results of FISH testing are somewhat subjective, as there may be significant interpatient variability in the extent to which these arms are deleted.
Testing for 1p loss and 19q loss was recommended by NCCN guidelines for tumors with an oligodendroglial component during the timeframe included in this study (2010–2012). Subsequently, the availability of this biomarker in the SEER data was the highest (around 50 % for relevant histologies). However, these data may not fully represent the testing rates or compliance with guidelines. Cancer registrars are predominantly hospital-based with varying access to the medical records of community oncology practices. If the test was ordered by and reported to a community provider the test results may not have been available to the cancer registrars for abstracting for SEER. SEER registries collect pathology reports on reportable cases as available. On average, 80 % of SEER cases do have one or more pathology reports associated with the case, though this number varies from registry to registry with some registries being close to 100 %. Thus, SEER registrars have the opportunity to capture the test if it was incorporated in the pathology report. The College of American Pathologists (CAP) recommends that all molecular tests are incorporated in the pathology report including tests that were performed at a different lab. However, adherence to this recommendation is not known, particularly for tests that were ordered by community oncologists.
Recent analyses of the lower grade gliomas, including WHO grade II and III (astrocytomas, oligodendrogliomas, and oligoastrocytomas) have found that these deletions are a critical prognostic factor that—along with mutations in IDH1/2—can be used to accurately stratify all grade II and III diffuse gliomas [34, 35]. In addition, codeletion of chromosome 1p/19q predicts a better outcome and a higher likelihood of temozolomide response in patients with such low grade gliomas [36]. Notably, the 2016 WHO classification of tumours of the CNS [37] includes these factors as diagnostic criteria for oligodendrogliomas and anaplastic oligodendrogliomas [3, 38, 39].
Molecular characterization of glioma has strongly established the importance of IDH1/2 mutation in subtyping these tumors. Though IDH1/2 mutation has been recognized as being a significant prognostic marker since 2009 [40], the strength of evidence related to this marker has increased in recent years. In addition to recent analyses of grade II and grade III diffuse gliomas, a recent analysis of grade II–IV glioma has demonstrated that this is the most critical marker for stratifying all glioma [26]. This variable is not a component of the AJCC system, and despite being assessed frequently in neuropathology, it is not collected in the cancer registration process. Given the increasing importance of 1p/19q codeletion and IDH1/2 mutations for cancer subtyping and prognosis it is imperative that cancer registries continue to collect 1p/19q codeletion data and begin to incorporate IDH1/2 mutation testing in cancer surveillance as soon as possible.
The SSFs for brain and CNS tumors include many variables that are strong prognostic factors and critical for use in generating cancer survival statistics relevant for clinical practice. Cancer registrars are dependent on neuropathologists, or sometimes other clinicians, to order these tests. They are also dependent on neurosurgeons, neuro-radiologists, and neuro-oncologists to include these data in their medical records as related to each individual patient's care. Increased attention to these factors (in particular SSF4, SSF5 and SSF6) in the scientific literature and clinical practice guidelines may be responsible for increased reporting on these factors, as well as encouraging their inclusion in the medical records and specifically in the pathology reports. Indeed, with the publication of the 2016 WHO classification of CNS tumors, pathologists will not be able to make particular diagnoses (e.g., “Oligodendroglioma, IDH-mutant and 1p/19q-codeleted”) in the absence of such molecular data.
The wide range of completeness values for each factor by SEER registry suggest that there is geographic variation that influences the completeness of these factors. While this can reflect differences in test utilization, it also can be partially explained with varying access to medical records and different abstracting practices. Each registry ranges in its completeness of each factor as well, with no one registry having the highest level of completeness in all factors. The type of institution where a patient is diagnosed may also determine whether these tests are ordered or included in the clinical records. Tertiary referral centers or teaching institutions may be more likely to order testing for these factors, though this dataset does not contain hospital-level information to know definitively.
Some of the SSF are not or have never been required information for cancer collection by SEER or by the individual SEER registries, and registrars may not have abstracted these variables as a result. In addition, since they are not required within SEER, they are not subject to SEER's quality control standards and would not be released by SEER to the general research community. All of these reasons may contribute to variation by registry. Insurance coverage for these tests may vary and may affect ordering and subsequent inclusion in medical records, thereby contributing to these lower reporting rates. Nonetheless, it is expected that the publication of the 2016 WHO classification of CNS tumors, with its incorporation of molecular parameters into specific diagnoses, will encourage third-party payers to cover these critical tests, and will in turn increase their completeness in tumor registries.
There are significant changes to glioma histology classification that are included in the 2016 WHO classification of CNS tumors [5]. Oligoastrocytoma has been long considered an entity that is distinct from astrocytoma and oligodendroglioma. In the 2016 WHO classification of CNS tumors, because recent molecular analyses have suggested that these tumors do not exist as a distinct entity [41] and can be more specifically diagnosed as astrocytoma and oligodendroglioma, the diagnosis of oligoastrocytoma is strongly discouraged and qualified with a “not otherwise specified” designation. Changes in diagnostic criteria have occurred over time for many brain tumor histologies, including previous changes in criteria for glioma and in embryonal tumors, which jeopardize long-term assessments of brain tumor incidence and survival trends. When using data collected prior to large changes in histologic criteria, it is critical to keep in mind the changes to the histology schema that have occurred over time in revisions to the WHO classification of CNS tumors. Investigators using these data would be well served by analyzing data collected before and after the implementation of major revisions separately prior to combining these data for analysis.
Cancer registries are invaluable resources for monitoring population-level patterns of cancer incidence and survival. However, there are limitations to these data. In particular, there is no central pathology review. and only a limited number of variables are collected This means that despite changes to the histology schema that may occur over time, it is not possible without additional variables to go back and re-classify any tumors based on new criteria. In addition to changes in histologic criteria over time, there is significant interrater variability in histopathological diagnosis of glioma [42, 43]. Due to these reasons, lack of central pathology review can limit the use of cancer registry data in many analyses. A recent analysis of data from EORTC 26951 and 26882 found that while consensus of diagnosis in glioma was 66 % overall, known molecular markers could be incorporated in a diagnostic algorithm along with histologic features to increase the rate of consensus [44].
With the recent revision to the WHO criteria for CNS tumors [5], IDH1/2 mutation and 1p/19q codeletion will become the primary factors by which gliomas are classified. Incorporation of these factors into cancer registration as soon as possible is critical to providing accurate and useful data for brain tumors within the cancer registry system. Including these molecular markers, which more accurately stratify gliomas than histopathology alone, will significantly increase the utility of these data for the monitoring of brain tumor incidence and survival. While other data sources, such as insurance billing records or hospital-based datasets, allow for more detailed interrogation of patterns of cancer treatment and outcomes, cancer registries are the only source for statistics that are truly generalizable to the US population. As technology continues to improve and cancer diagnostics become increasingly molecular, it is critical that these clinically relevant data are collected within the cancer registration process.
Conclusion
With the increasing focus on ‘precision medicine,’ it has become important to the brain tumor community that we measure tumor markers that affect brain tumor diagnosis, prognosis and predict the response to therapy at the population level. Survival after treatment with various therapies cannot be accurately evaluated without taking these critical pieces of information into account. Cancer registries are essential to measuring the burden of cancer on the US population, and the collection of new variables that are reflective of contemporary clinical practice is key to providing accurate and useful statistics to researchers, clinicians, patients, and their families.
Supplementary Material
Supplementary Table 1. Overview of SEER Site Specific Factors (SSF) 1-8, definitions of completeness and applicable years
Acknowledgments
Funding: QTO, HG, and JSB-S were supported by the National Cancer Institute Case Comprehensive Cancer Center Support Grant (P30CA043703). CBTRUS is honored to be included among the research grant recipients of the following organizations, which have contributed to the maintenance of the CBTRUS database: the Centers for Disease Control and Prevention (CDC) under Agreement 5U58DP00383, the Sontag Foundation, Genentech, Novocure, Inc., Celldex Therapeutics, Inc., AbbVie, Inc., along with the Musella Foundation, Voices Against Brain Cancer, Elekta, the Zelda Dorin Memorial Fund, as well as private and in kind donations.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-016-2217-7) contains supplementary material, which is available to authorized users.
Compliance with ethical standards: Conflict of interest: There are no conflicts of interest to report.
References
- 1.Surveillance Epidemiology and End Results (SEER) program required SEER site-specific factors for collaborative stage. http://seer.cancer.gov/tools/ssf/
- 2.Kleihues P, Cavenee W, editors. Tumours of the nervous system: World Health Organization Classification of tumours. IARC Press; Lyon: 2000. [Google Scholar]
- 3.Louis D, Wiestler O, Cavanee W, editors. WHO Classification of tumours of the central nervous system. International Agency for Research on Cancer; Lyon: 2007. [Google Scholar]
- 4.American Joint Committee on Cancer. Collaborative stage data collection system. 2015 http://www.cancerstaging.org/cstage/
- 5.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 6.Surveillance Epidemiology and End Results (SEER) program SEER*Stat database: incidence—SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2015 Sub (1973–2012 varying)—linked to county attributes—Total U S, 1969–2013 counties. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch; released April 2015, based on the November 2014 submission. [Google Scholar]
- 7.Surveillance Epidemiology and End Results (SEER) Program. Number of persons by race and hispanic ethnicity for SEER participants (2010 census data) 2015 [Google Scholar]
- 8.International Agency for Research on Cancer. International Classification of Diseases for Oncology, 3rd edn (ICD-O-3) 2000. [Google Scholar]
- 9.Collaborative Stage Data Collection System—CSv02.05 Cancer Schema. Brain: brain and cerebral meninges. Collaborative Stage Data Collectionn System—CSv02.05 Cancer Schema. 2013 http://web2.facs.org/cstage0205/brain/Brainschema.html.
- 10.Collaborative Stage Data Collection System—CSv02.05 Cancer Schema. CNSOther—other parts of central nervous system. 2013 http://web2.facs.org/cstage0205/cnsother/CNSOther-schema.html.
- 11.Collaborative Stage Data Collection System—CSv02.05 Cancer Schema. Intracranial gland: pituitary gland, craniopharyngeal duct, and pineal gland. Collaborative Stage Data Collection System—CSv02.05 Cancer Schema. 2013 http://web2.facs.org/cstage0205/intracranialgland/IntracranialGlandschema.html.
- 12.Surveillance Epidemiology and End Results (SEER) Program. SEER*Stat software version 8.2.1. National Cancer Institute, DCCPS, Surveillance Research Program; 2015. www.seer.cancer.gov/seerstat. [Google Scholar]
- 13.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16:iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lym RL, Ostrom QT, Kruchko C, Couce M, Brat DJ, Louis DN, Barnholtz-Sloan JS. Completeness and concordancy of WHO grade assignment for brain and central nervous system tumors in the United States, 2004–2011. J Neurooncol. 2015 doi: 10.1007/s11060-015-1775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 16.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 17.Stupp R, Hegi ME, Gilbert MR, Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol. 2007;25:4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- 18.Yin D, Xie D, Hofmann WK, Zhang W, Asotra K, Wong R, Black KL, Koeffler HP. DNA repair gene O6-methylguanine-DNA methyltransferase: promoter hypermethylation associated with decreased expression and G:C to A:T mutations of p53 in brain tumors. Mol Carcinog. 2003;36:23–31. doi: 10.1002/mc.10094. [DOI] [PubMed] [Google Scholar]
- 19.Kaina B, Margison GP, Christmann M. Targeting O(6)-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell Mol Life Sci. 2010;67:3663–3681. doi: 10.1007/s00018-010-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christmann M, Verbeek B, Roos WP, Kaina B. O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim Biophys Acta. 2011;1816:179–190. doi: 10.1016/j.bbcan.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura M, Watanabe T, Yonekawa Y, Kleihues P, Ohgaki H. Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:C→A:T mutations of the TP53 tumor suppressor gene. Carcinogenesis. 2001;22:1715–1719. doi: 10.1093/carcin/22.10.1715. [DOI] [PubMed] [Google Scholar]
- 22.Christians A, Hartmann C, Benner A, Meyer J, von Deimling A, Weller M, Wick W, Weiler M. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS One. 2012;7:e33449. doi: 10.1371/journal.pone.0033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wick W, Meisner C, Hentschel B, Platten M, Schilling A, Wiestler B, Sabel MC, Koeppen S, Ketter R, Weiler M, Tabatabai G, von Deimling A, Gramatzki D, Westphal M, Schackert G, Loeffler M, Simon M, Reifenberger G, Weller M. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81:1515–1522. doi: 10.1212/WNL.0b013e3182a95680. [DOI] [PubMed] [Google Scholar]
- 24.Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, Combs SE, Vesper J, Braun C, Meixensberger J, Ketter R, Mayer-Steinacker R, Reifenberger G, Weller M NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13:707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 25.Wiestler B, Claus R, Hartlieb SA, Schliesser MG, Weiss EK, Hielscher T, Platten M, Dittmann LM, Meisner C, Felsberg J, Happold C, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, Grimm C, Weichenhan D, Tews B, Reifenberger G, Capper D, Muller W, Plass C, Weller M, Wick W Neuro-oncology Working Group of the German Cancer Society. Malignant astrocytomas of elderly patients lack favorable molecular markers: an analysis of the NOA-08 study collective. Neuro Oncol. 2013;15:1017–1026. doi: 10.1093/neuonc/not043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG, Jr, Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH, Noushmehr H, Iavarone A, Verhaak RG TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Bent MJ, Erdem-Eraslan L, Idbaih A, de Rooi J, Eilers PH, Spliet WG, den Dunnen WF, Tijssen C, Wesseling P, Sillevis Smitt PA, Kros JM, Gorlia T, French PJ. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic oligodendrogliomas and oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19:5513–5522. doi: 10.1158/1078-0432.CCR-13-1157. [DOI] [PubMed] [Google Scholar]
- 29.van den Bent MJ, Gravendeel LA, Gorlia T, Kros JM, Lapre L, Wesseling P, Teepen JL, Idbaih A, Sanson M, Smitt PA, French PJ. A hypermethylated phenotype is a better predictor of survival than MGMT methylation in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. Clin Cancer Res. 2011;17:7148–7155. doi: 10.1158/1078-0432.CCR-11-1274. [DOI] [PubMed] [Google Scholar]
- 30.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 31.Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, Mehta M. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31:337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelbaum MA, Hu C, Peereboom DM, Macdonald DR, Giannini C, Suh JH, Jenkins RB, Laack NN, Brachman DG, Shrieve DC, Souhami L, Mehta MP. Phase II trial of pre-irradiation and concurrent temozolomide in patients with newly diagnosed anaplastic oligodendrogliomas and mixed anaplastic oligoastrocytomas: long term results of RTOG BR0131. J Neurooncol. 2015;124:413–420. doi: 10.1007/s11060-015-1845-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kou-wenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L, Enting RH, French PJ, Dinjens WN, Vecht CJ, Allgeier A, Lacombe D, Gorlia T, Hoang-Xuan K. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 34.The Cancer Genome Atlas Research Network. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O'Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, Yena P, Black A, Bowen J, Dicostanzo K, Gastier-Foster J, Leraas KM, Lichtenberg TM, Pierson CR, Ramirez NC, Taylor C, Weaver S, Wise L, Zmuda E, Davidsen T, Demchok JA, Eley G, Ferguson ML, Hutter CM, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Ayala B, Baboud J, Chudamani S, Jensen MA, Liu J, Pihl T, Raman R, Wan Y, Wu Y, Ally A, Auman JT, Balasundaram M, Balu S, Baylin SB, Beroukhim R, Bootwalla MS, Bowlby R, Bristow CA, Brooks D, Butterfield Y, Carlsen R, Carter S, Chin L, Chu A, Chuah E, Cibulskis K, Clarke A, Coetzee SG, Dhalla N, Fennell T, Fisher S, Gabriel S, Getz G, Gibbs R, Guin R, Hadjipanayis A, Hayes DN, Hinoue T, Hoadley K, Holt RA, Hoyle AP, Jefferys SR, Jones S, Jones CD, Kucherlapati R, Lai PH, Lander E, Lee S, Lichtenstein L, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, Mayo M, Meng S, Meyerson ML, Mieczkowski PA, Moore RA, Mose LE, Mungall AJ, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Protopopov A, Ren X, Roach J, Sabedot TS, Schein J, Schumacher SE, Seidman JG, Seth S, Shen H, Simons JV, Sipahimalani P, Soloway MG, Song X, Sun H, Tabak B, Tam A, Tan D, Tang J, Thiessen N, Triche T, Jr, Van Den Berg DJ, Veluvolu U, Waring S, Weisenberger DJ, Wilkerson MD, Wong T, Wu J, Xi L, Xu AW, Yang L, Zack TI, Zhang J, Aksoy BA, Arachchi H, Benz C, Bernard B, Carlin D, Cho J, DiCara D, Frazer S, Fuller GN, Gao J, Gehlenborg N, Haussler D, Heiman DI, Iype L, Jacobsen A, Ju Z, Katzman S, Kim H, Knijnenburg T, Kreisberg RB, Lawrence MS, Lee W, Leinonen K, Lin P, Ling S, Liu W, Liu Y, Liu Y, Lu Y, Mills G, Ng S, Noble MS, Paull E, Rao A, Reynolds S, Saksena G, Sanborn Z, Sander C, Schultz N, Senbabaoglu Y, Shen R, Shmulevich I, Sinha R, Stuart J, Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghunathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Calatozzolo C, Campos B, Carlotti CG, Jr, Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Finocchiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, McLendon R, McPherson C, Neder L, Nguyen P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rathmell WK, Rotin D, Scarpace L, Schilero C, Senecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Vallurupalli M, Wang Y, Warnick R, Williams F, Wolinsky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Zhang J. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, Sarkar G, Caron AA, Kollmeyer TM, Praska CE, Chada AR, Halder C, Hansen HM, McCoy LS, Bracci PM, Marshall R, Zheng S, Reis GF, Pico AR, O'Neill BP, Buckner JC, Giannini C, Huse JT, Perry A, Tihan T, Berger MS, Chang SM, Prados MD, Wiemels J, Wiencke JK, Wrensch MR, Jenkins RB. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaloshi G, Benouaich-Amiel A, Diakite F, Taillibert S, Lejeune J, Laigle-Donadey F, Renard MA, Iraqi W, Idbaih A, Paris S, Capelle L, Duffau H, Cornu P, Simon JM, Mokhtari K, Polivka M, Omuro A, Carpentier A, Sanson M, Delattre JY, Hoang- Xuan K. Temozolomide for low-grade gliomas: predictive impact of 1p/19q loss on response and outcome. Neurology. 2007;68:1831–1836. doi: 10.1212/01.wnl.0000262034.26310.a2. [DOI] [PubMed] [Google Scholar]
- 37.Louis DN, O H, Wiestler OD, Cavanee WK, editors. WHO classification of tumours of the central nervous system. International Agency for Research on Cancer; Lyon: 2016. [Google Scholar]
- 38.Ellison DW. Multiple molecular data sets and the classification of adult diffuse gliomas. N Engl J Med. 2015;372:2555–2557. doi: 10.1056/NEJMe1506813. [DOI] [PubMed] [Google Scholar]
- 39.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, Figarella-Branger D, Fuller GN, Giangaspero F, Giannini C, Hawkins C, Kleihues P, Korshunov A, Kros JM, Beatriz Lopes M, Ng HK, Ohgaki H, Paulus W, Pietsch T, Rosenblum M, Rushing E, Soy-lemezoglu F, Wiestler O, Wesseling P International Society Of N-H. International Society Of Neuropathology-Haarlem consensus guidelines for nervous system tumor Classification and grading. Brain Pathol. 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S, Jones DT, Pfister SM, Herold-Mende C, Wick W, Mueller W, Hartmann C, Paulus W, von Deimling A. Farewell to oligoastrocytoma: in situ molecular genetics favor Classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 2014;128:551–559. doi: 10.1007/s00401-014-1326-7. [DOI] [PubMed] [Google Scholar]
- 42.van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician's perspective. Acta Neuropathol. 2010;120:297–304. doi: 10.1007/s00401-010-0725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aldape K, Simmons ML, Davis RL, Miike R, Wiencke J, Barger G, Lee M, Chen P, Wrensch M. Discrepancies in diagnoses of neuroepithelial neoplasms: the San Francisco Bay Area Adult Glioma Study. Cancer. 2000;88:2342–2349. [PubMed] [Google Scholar]
- 44.Kros JM, Huizer K, Hernandez-Lain A, Marucci G, Michotte A, Pollo B, Rushing EJ, Ribalta T, French P, Jamine D, Bekka N, Lacombe D, van den Bent MJ, Gorlia T. Evidence-based diagnostic algorithm for glioma: analysis of the results of pathology panel review and molecular parameters of EORTC 26951 and 26882 trials. J Clin Oncol. 2015;33:1943–1950. doi: 10.1200/JCO.2014.59.0166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Overview of SEER Site Specific Factors (SSF) 1-8, definitions of completeness and applicable years