Abstract
Mitophagy is a selective form of macro-autophagy in which mitochondria are specifically targeted for autophagic degradation. Mitophagy plays an important role in cellular homeostasis by eliminating dysfunctional mitochondria and reducing mitochondrial mass as an adaptive response to stress. Cells execute mitophagy through several non-redundant mechanisms, including the PINK1/Parkin partnership that modulates turnover of depolarized mitochondria and stress-induced BNIP3, NIX and FUNDC1 molecular adaptors that interact directly with LC3 to promote mitophagy. These pathways are deregulated in human diseases, including cancer, neurodegeneration, metabolic disorders, muscle atrophy, ageing and inflammation, reflecting the importance of mitophagy as a cellular housekeeping function.
Keywords: Mitochondria, autophagy, Parkin, PINK1, BNIP3, NIX, FUNDC1, disease, ageing
Mechanisms of mitophagy
Mitochondrial quality control is key to maintaining properly functioning mitochondria that in turn are necessary for adaptive metabolism and survival in response to cellular stress. Mitochondrial-specific molecular chaperones (such as HSP60) and anti-oxidants (such as mitochondrial catalase) play a role in detoxifying the mitochondria in response to excess Reactive Oxygen Species (ROS) or the mitochondrial unfolded protein response. Additionally, mitochondrial fusion can promote mitochondrial well-being by distributing and diluting out damaged mtDNA, mitochondrial proteins or lipids amongst the wider mitochondrial reticulum. However, a major component of cellular control of mitochondrial integrity is the specialized form of macro-autophagy known as mitophagy.
There are multiple mechanisms by which mitochondria are targeted for degradation at the autophagosome but the best understood pathways are PINK1/Parkin induced mitophagy [1] and BNIP3- and NIX-dependent mitophagy [2]. In response to mitochondrial depolarization, PINK1 (PTEN-induced putative kinase 1) serine/threonine kinase accumulates at the outer mitochondrial membrane (OMM), where it phosphorylates ubiquitin on mitochondrial proteins to recruit autophagy cargo adaptors, such as OPTN and NDP52 that bind directly to LC3 at the autophagosome leading to mitochondrial degradation within autophagolysosomes [1]. PINK1 also recruits the E3 ubiquitin ligase Parkin that ubiquitinates specific substrates at the OMM, including VDAC, Miro and Mitofusin-2 to amplify the signal initiated by PINK1 activity [1]. In addition to Parkin, there are other mitochondrial E3 ubiquitin ligases with more restricted expression, such as Mul1 that promotes mitophagy in skeletal muscle.
In addition to clearing dysfunctional mitochondria, mitophagy also promotes a reduction in overall mitochondrial mass as an adaptive response to hypoxia, nutrient deprivation or developmental signals. The stress-responsive nature of mitophagy is explained in part by the control of expression and activity of key mitophagy adaptors by known stress response pathways. For example, BNIP3 is a transcriptional target of HIF-1, PPARγ, Rb/E2F, FoxO3, activated Ras and p53, while NIX (BNIP3L) is both HIF-1 and p53-regulated, and FUNDC1 is a HIF-1 target also. While evolutionarily related proteins, BNIP3 and NIX exhibit tissue-specificity, with BNIP3 mainly expressed in the heart, muscle and liver and NIX expressed strongly in hematopoietic tissues where it promotes mitochondrial clearance during red blood cell maturation. The mechanism of BNIP3- and NIX-mediated mitophagy differs from that of the Parkin/PINK1 pathway in that these proteins serve as direct adaptors targeting mitochondria to the autophagosome. In brief, induced BNIP3 and NIX (BNIP3L) integrate as homo-dimers into the OMM via C-terminal trans-membrane domains. The amino terminal regions of these proteins protrude into the cytosol where they interact with processed LC3-II at nascent phagophores via conserved LC3-interacting region (LIR) motifs. The interaction of BNIP3 with LC3 is modulated by phosphorylation on critical serine/threonine residues suggesting additional post-translational means to stress-induced regulation of mitophagy. Interestingly, both BNIP3 and NIX have previously been reported to function as pro-apoptotic BH3-only proteins although the BH3 domains are weakly conserved and redundant for function and the high levels of BNIP3 and NIX expression in liver, heart and red blood cells in the absence of cell death suggests that mitophagy, rather than cell death, is the primary function of these proteins. Similar to BNIP3 and NIX, FUNDC1 is a hypoxia-inducible mitochondrial adaptor that binds to LC3 directly through a LIR motif. Under conditions of hypoxia, ULK1 kinase, a key component of the autophagy preinitiation complex, translocates to mitochondria where it phosphorylates FUNDC1 to enhance the interaction of FUNDC1 with LC3 to promote mitophagy [3].
Mitophagy in human disease
Cancer
PARK2 (Parkin), BNIP3 and NIX (BNIP3L) are deleted or silenced in various different human cancers. Also consistent with a role for mitophagy in preventing cancer, expression of Parkin, BNIP3 and NIX (BNIP3L) is regulated by key tumor suppressors (p53, pRB) and oncogenes (NF-κB, FoxOs, HIF-1). PARK2 (Parkin) is frequently deleted in human breast, ovarian, bladder and lung cancers and expression of Parkin is undetectable in most cancer cell lines. In addition, Parkin null mice exhibit an increased incidence of spontaneous liver tumors and are sensitized to irradiation-induced lymphomas, supporting a tumor suppressive function for Parkin. Beyond its role in mitophagy, Parkin regulates levels of key cell cycle drivers, including Cyclin D1, Cyclin E, and CDK4 that may also contribute to its role as a tumor suppressor. Similar to Parkin, deregulated expression of BNIP3 and NIX (BNIP3L) has been linked to cancer in both patients and mouse models. BNIP3 and NIX (BNIP3L) are upregulated in ductal carcinoma in situ (DCIS) but expression is lost upon progression to invasive carcinoma, and is associated with increased proliferation and lymph node metastasis. Of interest, BNIP3 is frequently deleted in triple negative breast cancer (TNBC), an aggressive subtype of human breast cancer and BNIP3 loss is predictive of metastasis and poor prognosis for this cohort of patients [4]. BNIP3 is also epigenetically silenced in other tumor types, including lung, gastric, pancreatic and liver cancers, particularly upon progression to invasive and metastatic disease.
Neurodegenerative disease
Mutation of PARK2 (Parkin) and PARK6 (PINK1) have both been independently linked to familial cases of Parkinsons’ Disease (PD) linking mitophagy defects to degeneration of dopaminergic neurons [1]. Indeed, dopaminergic neurons from patients with either familial or sporadic PD appear to be sensitized to respiratory chain inhibitors and to have increased levels of mtDNA mutations leading to mitochondrial dysfunction. Other mutations linked to PD, such as those in α-synuclein that lead to accumulation of α-synuclein aggregates indirectly leads to a block in mitophagy through inhibition of general autophagy. Overall, these findings point to a key role for mitophagy in preventing PD.
Muscle Atrophy & Aging
Autophagy rates decrease with age in most tissues and thus turnover of mitochondria also decreases, leading to loss of mitochondrial function. This has been linked to reduced respiration, increased ROS production and age-related damage in various tissues, including cardiac muscle, skeletal muscle, pancreas and liver. Overall, any perturbation of mitochondrial quality control, such as reduced mitophagy, is predicted to promote aging in line with the “free radical theory of ageing” since mitochondria are the major source of ROS in the cell.
Metabolic Disease and Diabetes
Mitochondrial dysfunction has been linked to insulin resistance, metabolic syndrome and type 2 diabetes both due to inherited mutations and acquired mtDNA mutations as people age. Although specific mitophagy defects have not been linked per se to insulin resistance, age-dependent loss of mitochondrial function is strongly linked to onset of Type 2 diabetes in the elderly. Clearly, defects in mitochondrial metabolism arising due to reduced mitophagy, such as reduced fatty acid oxidation that cause lipids to accumulate in the liver or in muscle, would be predicted to alter insulin signaling and lead to diabetes.
Mitochondrial Anti-viral Signaling
Mitophagy plays an important role in limiting pro-inflammatory signals induced by either pathogen associated molecular patterns (PAMPs), such as viral DNA, or damage-associated molecular patterns (DAMPs), such as extracellular ATP. Both PAMPs and DAMPs induce mitochondrial membrane permeabilization, ROS production and leakage of mtDNA in turn activates inflammasome complexes in the cell and production of IL-1β and caspase-1 activity that can feed back to cause further mitochondrial damage. Mitophagy plays a key role in limiting inflammasome activation by eliminating damaged mitochondria and inhibition of mitophagy results in hyperactivation of inflammsomes and pro-inflammatory cell death [5].
Summary
Mitophagy defects and loss of mitochondrial function are linked to numerous disease states. In cancer, this has raised the possibility of targeting mitochondria for therapeutic purposes, since tumor cells have increased ROS levels and have rewired their metabolism to be more dependent on specific aspects of mitochondrial metabolism. This makes cancer cells more susceptible than normal cells to respiratory chain inhibitors and other mitochondrial poisons such as antibiotics that inhibit mitochondrial protein translation and could be re-purposed for cancer treatment. Clearly, this approach for cancer treatment would not be effective for other diseases, such as PD and inflammatory disease, where ideally the opposite approach of stimulating mitophagy would be preferred, although the means to do this needs to be developed. In summary, this is an emerging research area with significant potential for both fundamental discovery and therapeutic gain.
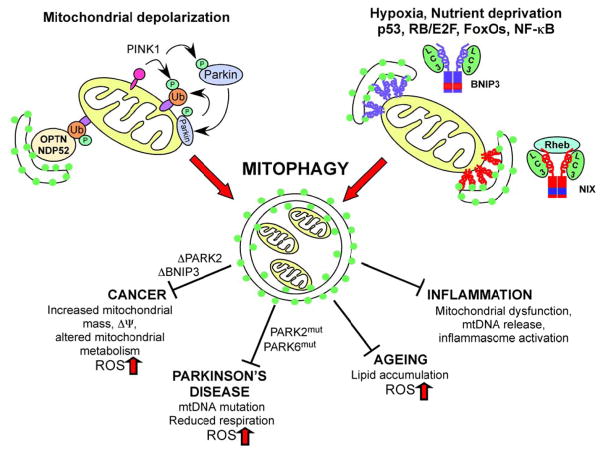
Figure 1. Schematic illustrating mechanisms and role of mitophagy in disease.
PINK1/Parkin-mediated mitophagy promotes elimination of depolarized mitochondria through the concerted action of PINK1 kinase on ubiquitin and the Parkin E3 ubiquitin ligase that is recruited by PINK1 to the outer mitochondrial membrane where it ubiquitinates substrates such as Mitofusin-2 and VDAC1. The actions of BNIP3 (blu ue) and NIX (red) are induced by stress signals downstream of hypoxia, nutrient deprivation and specific developmental cues. Both BNIP3 and NIX function as homo-dimers at the OMM and interact directly with processed LC3 at phagophore membranes to promote mitochondrial turnover. The interaction of NIX with LC3 and its mitophagy functions are stimulated by interaction with Rheb. Defects in mitophagy contribute to cancer, Parkinson’s disease, ageing and inflammation amongst other diseases as a result of mitochondrial dysfunction, increased ROS production, mtDNA mutation, reduced respiration, altered metabolism and other adverse consequences of the loss of this important mitochondrial quality control mechanism.
Acknowledgments
Grant support for this work includes NIH RO1 CA162405 to KFM.
T32 CA009594 supported MZS.
Footnotes
Conflict of Interest Statement: The authors declare that they have no conflicts of interest.
References
- 1.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chourasia AH, Boland ML, Macleod KF. Mitophagy & Cancer. Cancer Metab. 2015;3:1–11. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu WK, Tian W, Hu Z, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chourasia AH, Tracy K, Frankenberger C, et al. Mitophagy defects arising from BNip3 loss promote tumor progression to metastasis. EMBO Rep. 2015;16:1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, Inflammation, and Immunity: A Troika Governing Cancer and Its Treatment. Cell. 2016;166:288–298. doi: 10.1016/j.cell.2016.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]