Abstract
Autophagy is a major degradation pathway that engulfs, removes and recycles unwanted cytoplasmic material including damaged organelles and toxic protein aggregates. One type of autophagy, macroautophagy, is a tightly regulated process facilitated by autophagy-related (Atg) proteins that must communicate effectively and act in concert to enable the de novo formation of the phagophore, its maturation into an autophagosome and its subsequent targeting and fusion with the lysosome or the vacuole. Autophagy plays a significant role in physiology and its deregulation has been linked to several diseases, which include certain cancers, cardiomyopathies, and neurodegenerative diseases. Here, we summarize the key processes and the proteins that make up the macroautophagy machinery. We also briefly highlight recently uncovered molecular mechanisms specific to neurons allowing them to uniquely regulate this catabolic process to accommodate their complicated architecture and non-dividing state. Overall, these distinct mechanisms establish a conceptual framework addressing how macroautophagic dysfunction could result in maladies of the nervous system, providing possible therapeutic avenues to explore with a goal of preventing or curing such diseases.
Keywords: Autophagy, Macroautophagy, Neurons
Introduction
Cellular homeostasis is crucial to support life. The ability of cells to effectively and efficiently maintain the regulation and modulation of protein biogenesis and turnover play a key role in cellular growth, function and development. Cells must be able to maintain an efficient equilibrium between the different processes that govern protein homeostasis, which include synthesis, post-translational modifications, folding, localization and degradation. When such a delicate balance is compromised, cellular integrity may be detrimentally affected and could lead to the presence of aggregates comprised of misfolded proteins, or the accumulation of dysfunctional organelles—the hallmarks of many neurodegenerative diseases [1–3]. In many cases, pathology arises from mutated versions of a single protein. Examples include Parkinson and Huntington diseases wherein mutations in the proteins SNCA/α-synuclein and HTT (huntingtin), respectively, elicit the diseased state [4–6]. In recent studies, great emphasis has been placed on linking the deregulation of autophagy to the onset of such diseases brought about by toxic aggregates (previously reviewed in [1–3, 7–9]). Macroautophagy is one of the major degradative pathways that cells utilize to achieve proteostatic balance. Macroautophagy utilizes a conserved, eukaryotic molecular machinery that involves the sequestration of target materials and their subsequent delivery to and breakdown by the lysosome/vacuole [10, 11]. Macroautophagy occurs on a basal level but is upregulated when cells are exposed to extreme conditions (such as nutrient starvation). Even though it is primarily deemed as a protective process that allows cells to survive in response to various stressors, insufficient or excessive degradation from improperly controlled macroautophagy can lead to various degrees of damage to the stability and integrity of cells. In this review, we focus on understanding how macroautophagy functions and how the logistics of this highly complex process are accommodated within the complicatedly elongated structure of a neuron.
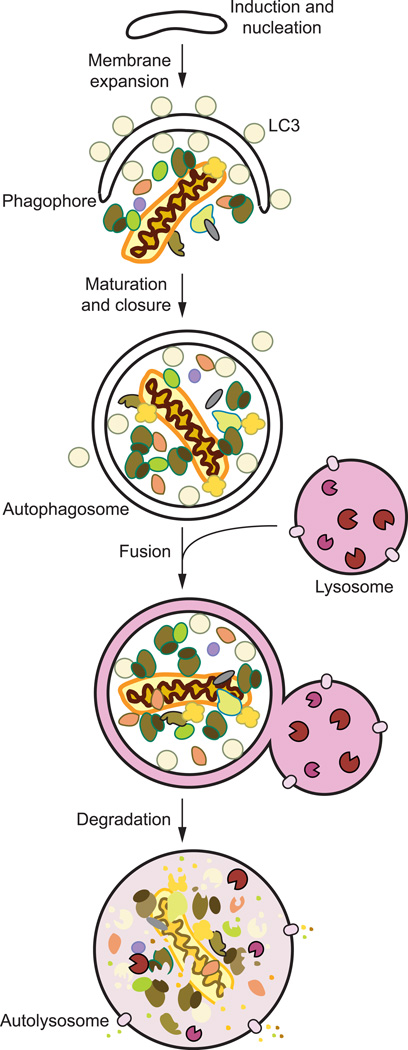
There are many types of autophagy—including microautophagy, chaperone-mediated autophagy and macroautophagy—all of which are highly dynamic processes, catalogued based on their substrate, selectivity, and their method of delivery to the lysosome/vacuole [10]. Briefly, microautophagy involves the transport of degradation targets by means of direct invagination or protrusion of the lysosomal/vacuolar membrane [12]. In chaperone-mediated autophagy, cytoplasmic chaperones identify degradation substrates in a highly specific manner by recognizing and binding a targeting motif, with the consensus KFERQ [13]. Lysosomal delivery of CMA cargo is achieved through direct translocation across the lysosomal membrane thereby allowing substrates to be degraded by lysosomal proteases. The most well characterized type of autophagy is macroautophagy (hereafter identified as autophagy). Macroautophagy involves the removal and recycling of cytoplasmic components by means of sequestration through a transient membrane compartment called the phagophore (Fig. 1). This compartment is expanded to envelop varying categories of substrates, providing immense cargo capacity that includes not only proteins, but also whole organelles. Once phagophore expansion and substrate engulfment is concluded, the membrane is sealed forming the double-membrane vesicular body termed the autophagosome. The autophagosome consequently fuses with the lysosome/vacuole where its cargo becomes exposed to and broken down by degradative hydrolases; the resulting macromolecules are subsequently released back into the cytosol for reuse [14].
Fig. 1.
Overview of autophagy. Autophagy occurs constitutively but at low levels; however, it is dramatically stimulated when cells experience stress brought about by nutrient starvation, lack of oxygen, or the presence of pathogens, among others. Once autophagy is induced, multiple autophagy-related proteins are then recruited to nucleate and assemble the double-membrane autophagosome precursor, called the phagophore. As it expands and elongates, the phagophore begins to enclose its cargo, either specifically or randomly. The phagophore becomes sealed, leading to a mature autophagosome that can dock onto and fuse with the vacuole. Upon engulfment, these vesicles along with their contents are broken down by resident vacuolar hydrolases. The resulting molecular components such as amino acids are recycled back into the cytosol for further use.
Macroautophagy, like microautophagy, can either selectively or nonselectively target cargo for degradation. In selective autophagy, specific targets are delivered to expanding autophagosome through the use of cargo receptors. These receptors include SQSTM1/p62, CALCOCO2/NDP52, NBR1 and OPTN, which mediate the sequestration of damaged mitochondria, toxic aggregates and invading pathogens [10, 14].
The de novo formation of an autophagosome is a remarkable feat and requires exquisite spatial and temporal control. For example, phagophore assembly and expansion must be entirely concluded before the autophagosome is completely sealed and formed, thus preventing inefficient cargo sequestration. Furthermore, key regulatory checkpoints must exist to allow only fully completed autophagosomes—and not immature phagophores—to fuse with the lysosome/vacuole, which would prevent the delivery of the cargo into the lumen. Albeit an already complicated processes, an additional layer of difficulty needs to be considered when dealing with autophagy in neuronal cells and their post-mitotic nature. Dendrites and axons, extensions projecting from the soma, can be quite long. Axonal extensions in human nerve cells can be close to 1 meter long. Dendrites, although much shorter than axons, can still achieve dimensions up to 2000 microns long. Thus, neurons face regulatory issues of facilitating autophagy at extreme distances. In a later section of this review, we briefly summarize recently uncovered mechanisms of autophagy regulation that occur in neurons.
The Core Autophagic Machinery
For the purposes of discussion, autophagy can be divided into several distinct stages [11, 15]: induction, nucleation of the phagophore, expansion and maturation into an autophagosome; docking and fusion with the lysosome/vacuole; and degradation and efflux (Fig. 1). These steps are carried out by proteins encoded by a group of autophagy-related (ATG) genes. The ATG proteins are grouped based on their respective functions into the following categories [16, 17]: (1) ULK1 kinase complex (ULK, ATG13, ATG101 and RB1CC1/FIP200); (2) the class III phosphatidylinositol (PtdIns) 3-kinase (PtdIns3K) complex (PIK3C3/Vps34, BECN1, PIK3R4/Vps15, and either ATG14 or UVRAG-SH3GLB1); (3) the membrane expansion complex (ATG9, ATG2-WIPI2); (4) the conjugation systems (ATG12–ATG5-ATG16L1 and LC3/GABARAP); and (5) components involved in lysosomal degradation and macromolecular efflux. Current knowledge of these steps in both yeast and mammalian systems are summarized below. Although the core machinery remains conserved, it is important to note that observations regarding mammalian autophagy presented below are derived from different cell types and differences in its regulation may exist from one cell type to another.
Autophagic Induction
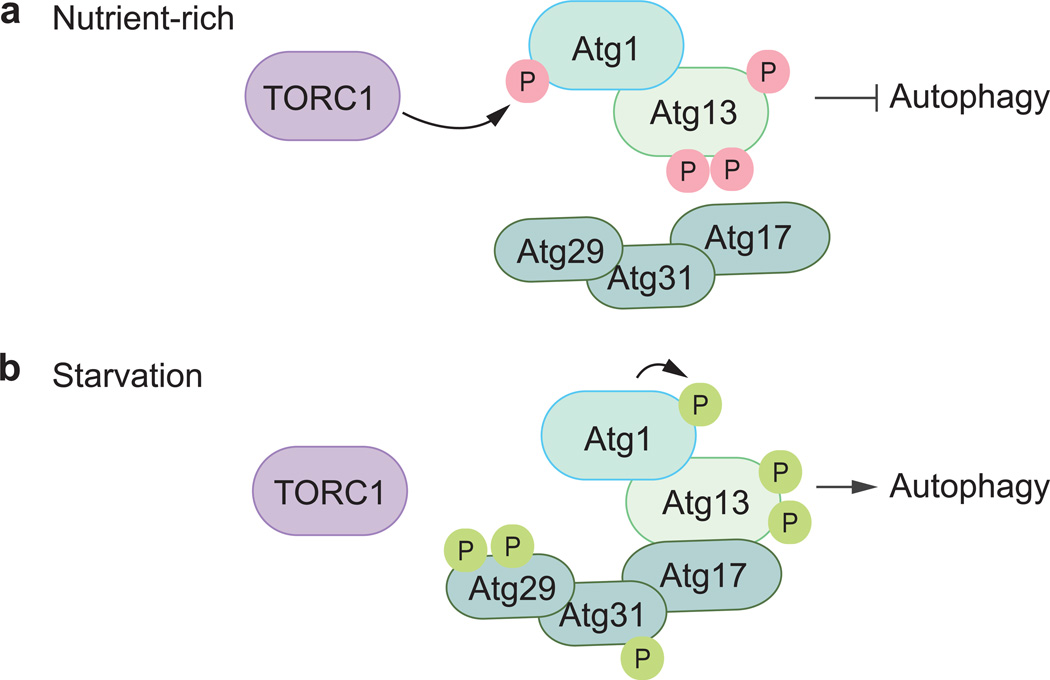
In yeasts, the formation of an autophagosome starts in a perivacuolar structure known as the phagophore assembly site (PAS). Initiation is carried out by the Atg1 kinase complex, which is composed of the Ser/Thr kinase Atg1 (Fig. 2), its regulatory subunit Atg13 and the Atg17-Atg31-Atg29 scaffolding complex [18–20]. For autophagy to commence, stimuli (e.g., amino acid deprivation) are detected by nutrient-sensing pathways, such as the target of rapamycin kinase complex (TORC1) [18]. Under normal conditions, the TORC1 and other nutrient-sensing kinases including protein kinase A maintain autophagy at a basal level by actively phosphorylating certain proteins including Atg1 and Atg13 (Fig. 2a) [18]. During starvation conditions, TORC1 and protein kinase A are inhibited resulting in the rapid dephosphorylation of Atg1 and Atg13. Atg1 becomes stimulated and auto-phosphorylation takes place resulting in its enhanced kinase activity, a functionality that is crucial for the recruitment and activation of other autophagy proteins, allowing them to be localized to the PAS and begin phagophore nucleation and assembly (Fig. 2b) [21–23].
Fig. 2.
The yeast Atg1 induction machinery. (a) Under nutrient-rich conditions, TORC1 actively phosphorylates Atg13, thereby inhibiting Atg1 activity. (b) During nitrogen starvation, TORC1’s kinase function is suppressed, allowing for Atg13 to become partially dephosphorylated resulting in the autophosphorylation of Atg1. In this induced form, Atg1 serves as a master regulator and cues in other Atg proteins to localize to the growing phagophore. Inhibitory and stimulatory phosphorylations are shown in pink and green, respectively.
In mammals, autophagy initiation is governed by the corresponding ULK kinase complex [24, 25]. Atg1 has two known counterparts in mammals: ULK1 and ULK2 (unc-51 like autophagy activating kinase 1/2); these proteins stably interact with RB1CC1, the homolog of Atg17, mammalian ATG13 and the novel accessory protein ATG101 [26–28]. During nutrient-rich conditions, the mechanistic target of rapamycin (serine/threonine kinase) complex 1 (MTORC1) principally binds to ULK1/2, directly inhibiting the latter’s auto-phosphorylation by phosporylating both ATG13 and ULK1/2. To facilitate autophagy (during starvation conditions or upon rapamycin treatment), MTORC1 is rapidly removed from the ULK kinase complex resulting in the dephosphorylation of both proteins and leading to the activation of ULK1/2’s kinase function [29, 30]. In this triggered state, ULK1/2 directly phosphorylates ATG13, RB1CC1 and itself thus allowing for the next stages in the autophagic process to proceed [29].
Membrane nucleation
Once induced, the nucleation and assembly of the phagophore is propagated by a set of proteins that catalyze the conversion of PtdIns to phosphatidylinositol 3-phosphate (PtdIns3P) (Fig. 3) [31]. This molecule then serves as a signal that recruits many other ATG proteins that recognize and preferentially bind PtdIns3P at the nucleation site. In yeasts, nucleation takes place at the PAS. In more complex eukaryotes, although a definitive PAS is lacking, it is thought that nucleation also occurs in specific subcellular locations in the cell. The class III PtdIns3K complex carries out the membrane modification involving the generation of PtdIns3P. There are two class III PtdIns3K complexes in yeast that function in either autophagy or the vacuolar protein sorting pathway, and at least three in mammals; the latter function at different stages of autophagy, and one of the complexes is inhibitory [32–34].
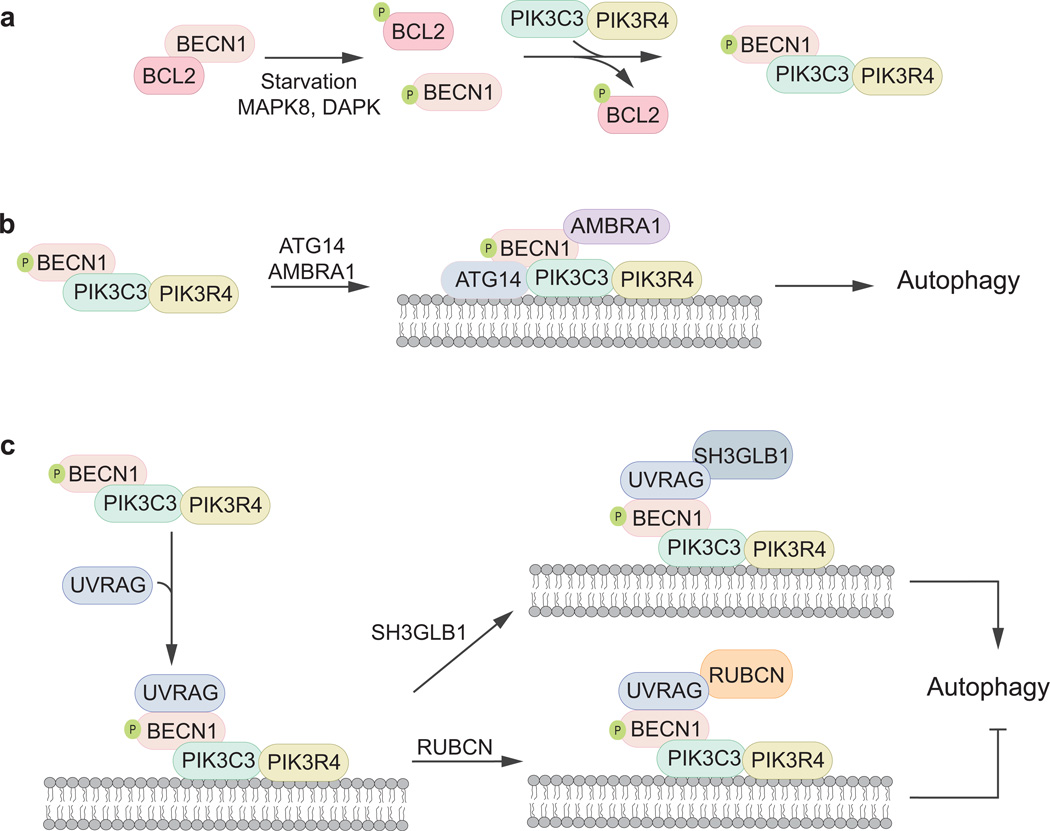
Fig. 3.
The class III PtdIns3K complex and the regulation of autophagosome nucleation. PtdIns3P is an important signaling molecule that allows for the nucleation and expansion of the phagophore. BECN1 is a key regulatory player in the activation of PtdIns3P synthesis carried out by PIK3C3 and its regulatory partner protein, PIK3R4. (a) Under non-inducing conditions, BECN1 is associated with BCL2 preventing its localization to the PAS. Upon nitrogen starvation, BECN1 and BCL2 become phosphorylated by DAPK and MAPK8/JNK1, respectively, allowing them to dissociate. (b, c) When autophagy is induced BECN1 is released from BCL2 and is then free to bind to a wide array of proteins that can suppress or stimulate autophagy. BECN1 bound to ATG14, AMBRA1 and UVRAG-SH3GLB1 result in autophagy activation whereas when bound to UVRAG-RUBCN, autophagy is inhibited.
The PtdIns3P produced by the Vps34/PIK3C3 complex serves as a PAS-recruiting particle that allows for several Atg proteins to localize at the nucleation site. These include Atg18 and Atg21 (WIPI1 and WIPI2 in mammals), both of which have high affinities for PtdIns3P [35, 36]. Although these proteins play a significant role in autophagy, their precise function is relatively unknown.
The BECN1-BCL2 interaction complex represents another important checkpoint that negatively regulates autophagy (Fig. 3a). During nutrient-rich conditions, BCL2 directly inhibits autophagy by binding to and preventing BECN1 from associating with the PtdIns3K complex [37, 38]. In contrast, during starvation, BCL2 dissociates from BECN1, allowing the latter to perform its function in PtdIn3P formation. BECN1’s different modes of interaction are controlled through post-translational modification. Specifically, phosphorylation serves as a molecular switch: BCL2 phosphorylation by MAPK8/JNK1 and BECN1 phosphorylation by DAPK, cause BECN1 and BCL2 to dissociate from one another.
Multiple studies have shown that BECN1 can serve as a molecular adapter that allows the ternary PtdIns3K complex to associate with various proteins forming different functional forms that can either suppress or upregulate autophagosome formation (Fig. 3). These BECN1 binding partners include, but are not limited to, ATG14 (the mammalian homolog of Atg14), AMBRA1 (autophagy/beclin-1 regulator 1), UVRAG (UV radiation resistance associated), and the anti-apoptopic BCL2 family of proteins [10, 39–47]. ATG14 and UVRAG bind to the PtdIns3K complex in a mutually exclusive fashion through their respective coiled-coil domains and they each allow the targeting of the nucleation machinery to different regions in the cells: ATG14 targets the complex to the omegasome (a precursor to the autophagosome, similar or equivalent to the PAS) and UVRAG to the cell surface and endosomal membranes. The ULK1-mediated release of AMBRA1 from DYNLL1 (dynein light chain LC8-type 1) and its subsequent binding to BECN1 is also required for the complex to localize to omegasomes in the ER [45, 46]. Overall, genetic and biochemical evidence show that the ATG14-BECN1-AMBRA1-PIK3C3-PIK3R4 complex plays an active role in the formation of the phagophore and therefore, promotes the onset of autophagosome formation (Fig. 3b) [41–45]. Conversely, the UVRAG-containing PtdIns3K complex can either downregulate or stimulate the onset of autophagy depending on its association with either SH3GLB1/Bif-1 (SH3-domain GRB2-like endophilin B1) [47] or RUBCN/Rubicon [44], respectively (Fig. 3c).
Phagophore expansion
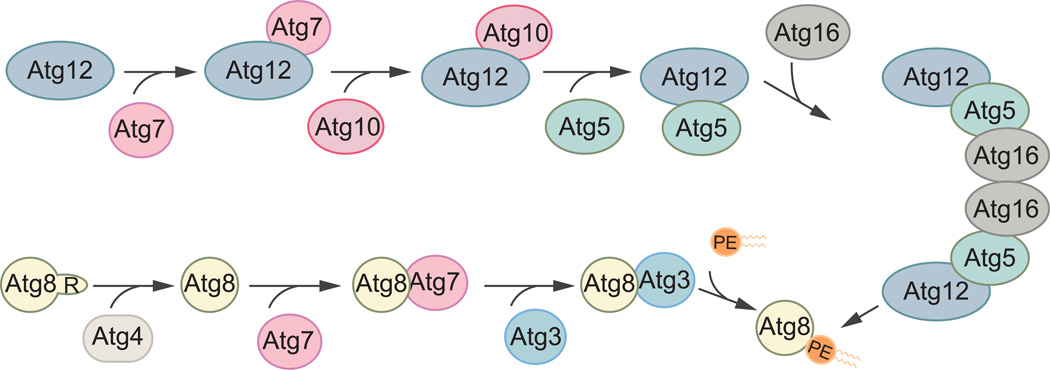
Autophagy employs the use of two ubiquitin-like conjugation systems to extend and elongate the phagophore (Fig. 4). In yeasts, these involve two ubiquitin-like proteins: Atg8 and Atg12; and the respective enzymes that allow for subsequent covalent modification to generate their respective conjugated forms [48–50]. Atg12 irreversibly associates with Atg5, a reaction dependent on its respective E1 activating and E2 conjugating enzymes, Atg7 and Atg10 [48, 51]. The Atg12–Atg5 conjugate and Atg16 then form a complex that serves as the E3 ligase facilitating the second ubiquitin-like conjugation reaction [52], the lipidation of Atg8 to form Atg8–phosphatidylenolamine (Atg8–PE) [53]. In order to covalently attach a PE molecule to Atg8, the latter’s terminal arginine residue must first be cleaved off by the Atg4 protease to expose a glycine residue at the C terminus. The proteolyzed protein is then recognized and processed by the same E1-activating enzyme, Atg7, and its own E2-like enzyme, Atg3. As mentioned earlier, the final ligation of Atg8 to PE is facilitated by the E3-like Atg12–Atg5-Atg16 complex [50].
Fig. 4.
The ubiquitin-like conjugation systems in autophagy. There are two ubiquitin-like proteins in yeasts, Atg12 and Atg8, both of which play crucial roles in the expansion of the phagophore and its subsequent maturation to become an autophagosome. In the first systems, Atg12 becomes conjugated to Atg5 in a reaction dependent on the E1 activating enzyme Atg7, and the E2-like Atg10. The Atg12–Atg5 conjugate then associates with Atg16 and act as the E3 ligase for the conjugation of Atg8 to PE. Atg8 undergoes a similar modification but must first be processed by Atg4, a cysteine protease, to remove its C-terminal arginine. Then, Atg7, Atg3 and Atg12–Atg5-Atg16 facilitate the conjugation of Atg8 to PE.
In more complex organisms, these ubiquitin-like systems are conserved [54, 55]. The mammalian counterparts function similarly to those in yeast. A key distinction, however, is the presence of numerous homologs of Atg8 (LC3 and GABARAP) and Atg4 (ATG4A, ATG4B, ATG4C and ATG4D). Both LC3 and GABARAP undergo the same covalent modification, however, LC3 is thought to function at earlier stages of phagophore assembly, whereas GABARAP acts further downstream of the autophagic pathway [56].
Although it is well-established that these ubiquitin-like conjugation systems are essential for autophagy, a comprehensive mechanistic understanding of how these covalent-modification reactions function towards phagophore expansion and maturation is incomplete. For example, the amount of Atg8 can regulate the size of autophagosomes [57, 58], however, the mechanistic details for how this is achieved are lacking. Atg8 proteins that are localized to the concave face of the phagophore, but not those on the convex surface, interact with cargo during selective autophagy. Furthermore, in the final stages of autophagy, Atg8 can be deconjugated from the membrane in an Atg4-dependent manner. How these various roles of Atg8 are separated, and how the interactions of Atg8, cargo receptors and Atg4 are regulated remains unclear.
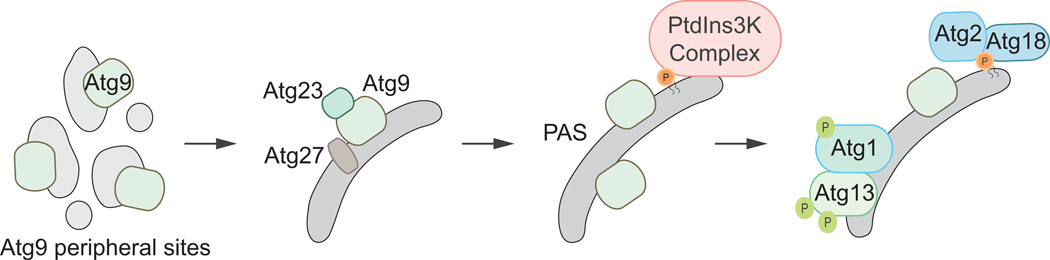
To further expand the phagophore, membrane must be delivered from various locations in the cell, as lipid synthesis does not occur at the PAS or the phagophore. This may not be the case with regard to the omegasome, which is thought to originate directly from the ER membrane and therefore may receive newly synthesized lipids by lateral movement. The putative membrane-recruiting component of autophagy involves the multi-pass integral membrane protein Atg9 [59, 60]. In yeast, the shuttling of Atg9 between peripheral sites (also termed Atg9 reservoirs, tubulovesicular structures and peripheral structures) and the phagophore is hypothesized to represent the transit and delivery of membranes (Fig. 5) [59]. Atg9 can oligomerize [61] and form a complex with other Atg proteins [62–65] that allows for its movement to and from peripheral sites and the PAS. The movement of Atg9 from the peripheral sites to the PAS is dependent on Atg11, Atg23 and Atg27 [62–65].
Fig. 5.
Atg9 cycling. Atg9 is a transmembrane protein that travels between the PAS and peripheral sites. The transit of Atg9 to the PAS from the peripheral sites requires Atg11, Atg23 and Atg27.
Mammalian ATG9 displays a similar localization pattern as its fungal counterpart and is thought to have the equivalent membrane transport functionality [66, 67]. ATG9 mainly localizes to the trans-Golgi network and late endosomes during nutrient-rich conditions [66]; however, during starvation conditions, ATG9 transits to the site of autophagy assembly as evidenced by its colocalization with autophagosomal markers [66, 67]. ATG9 movement to and from autophagosomes requires ULK1, PtdIns3K activity, and WIPI2. The regulation of ATG9 trafficking and its purported membrane transport activity remain poorly understood.
Fusion
The successful fusion of autophagosomes with the target degradative organelle results in the formation of autophagic bodies (yeast) and autolysosomes (complex eukaryotes). Although a clear-cut mechanism for this fusion process remains largely unknown [68], proteins that facilitate this step have largely been identified [69–74]. Interestingly, these proteins are also members of cellular machineries already known to be involved in other transport mechanisms that also end at the lysosome/vacuole. These include SNARE proteins; the class C Vps/homotypic fusion and vacuolar protein sorting (HOPS) complex [74]; and members of the ESCRT family of proteins.
Degradation and efflux
Once fusion with the lysosome/vacuole takes place, the inner membrane of the autophagosome and its contents are degraded by various hydrolases. Metabolites generated in these proteolytic processes, which include amino acids, are actively pumped out of these organelles back into the cytosol and are reutilized by the cell.
Autophagy in Neurons
In order to effectively process and transmit information, neurons have adapted a uniquely polarized cellular architecture that utilizes highly specialized structures not found in other cell types. These structures include the soma, dendrites, axons and synapses (Fig. 6). Due to the extreme polarization and daunting length of neuronal cells, their integrity relies heavily on the efficient and active transport of critical macromolecules, such as protein and lipids, to and from the cell body and synapses across axons and dendrites. Thus, neurons can be intensely susceptible to the accumulation of damaged organelles and harmful cytoplasmic aggregates that can easily lead to numerous neurodegenerative diseases [1, 3, 75]. For instance, mice lacking ATG7 in the central nervous system exhibit substantial neurodegeneration evidenced by behavioral defects and the presence of inclusion bodies [75]. Therefore, the cytoprotective properties of autophagy are extremely critical in maintaining cellular homeostasis in the central nervous system.
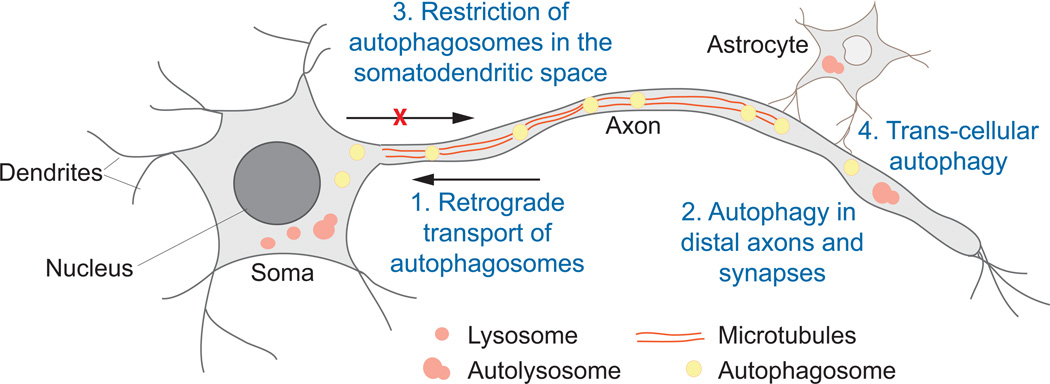
Fig. 6.
Autophagy in neurons. Neurons have a complex elongated shape composed of unique architectural features that include the soma (or the cell body), dendrites and axons. Four basic methods of autophagic clearance have been identified in neuronal cells. (1) Autophagosomes that form in axons can travel to the soma through motor proteins along microtubules; (2) autophagy is carried out at extreme boundaries of a neuron; (3) axonal autophagosomes (once they enter the soma) and somatic autophagosomes become restricted to the somatodendritic space; and (4) neighboring cells (in particular, astrocytes) aid in the degradation of damaged organelles in a transcellular, autophagy-dependent manner.
As in other cells, autophagy occurs at a constitutive basal level in neurons [75]. However, nutrient-starved neuronal cells show considerably lower levels of LC3 conjugated to PE (LC3-II) when compared to nutrient-starved non-polarized cell types [76]. This could be a consequence of faster autophagosome turnover in neurons. Alternatively, neuronal cells could simply respond differently to nutrient starvation—the comparatively lower amount of autophagosomes found in neurons may also be a consequence of a more tightly regulated mechanism of induction and formation—because neurons may not require a substantial level of autophagy and may be more susceptible to autophagic flux imbalance as a consequence of their post-mitotic nature and synaptic function [2]. Indeed, it was recently shown that neither drug-induced MTORC1 inhibition nor amino acid deprivation could produce a marked increase in detectable autophagy in axons [77]. Thus, a proper level of autophagy is essential in neurons to maintain cellular homeostasis as evidenced by numerous neurodegenerative diseases that have been linked to defective autophagy (summarized in [1–4, 7, 9]). Even seemingly nuanced mutations can lead to neuronal dysfunction. For example, a single mutation in a core member of the autophagic machinery has been identified as the cause of ataxia in patients with severe neurological abnormalities [78, 79]; an E122D mutation in ATG5 results in ineffective conjugation to ATG12 and, thus, overall decreases the level of autophagy in both mammalian and yeast model systems [79].
Spatial regulation of neuronal autophagy
Autophagy in neurons must be finely tuned, requiring exquisite spatial and temporal regulation necessary to accommodate an extended amount of cytoplasmic space. To accomplish this, neurons seemingly employ creative modes of autophagy control to account for their distinctive shape and length. Recent work shows that neurons employ at least four distinguishing mechanisms to execute autophagy as it traverses through the cellular milieu between the cell body and across distal axons and dendrites (Fig. 6). First, autophagosomes can form at the extreme boundaries of the cell, particularly in axons all the way to the synapse, and can be transported along microtubules towards the cell body, fusing with lysosomes along the way [80–82]. Second, autophagy can be carried out completely on the periphery of the cell and can be observed in remote axons [83]. Third, autophagy is spatially compartmentalized in neurons. Finally, the axon may enforce degradation of cargo, in particular, mitochondria, in a trans-cellular fashion with the aid of a neighboring cell [84]. Note that these events may not necessarily be occurring simultaneously and such mechanisms may be employed by the neuron to accommodate various developmental stimuli or stress (such as nutrient-deprivation and disease). These four mechanisms are further discussed in detail below.
Long-distance transport of autophagosomes
It has been long established that microtubules facilitate the retrograde transit of autophagosomes across long distances [82, 85, 86]. This highly processive, unidirectional movement dictates exquisite spatial and temporal control in order to overcome the challenges levied by the nature, shape and length of a neuron. An added complication is effected by the observation that distal axonal autophagosomes interact with both dynein and kinesin [82], motor proteins that facilitate retrograde and anterograde movement, respectively. Thus, if autophagosomes indeed remain associated with both cellular transit machineries, then their robust unidirectional movement towards the cell body must be carefully regulated. In recent studies [80, 81], the dynamic motor scaffold MAPK8IP1/JIP1 has been identified as the chief regulatory protein that controls the retrograde transport of axonal autophagosomes wherein anterograde movement through the motor kinesin becomes inhibited. Using knockdowns and mutational analyses, fluorescence microscopy assays show that MAPK8IP1 employs a multi-faceted strategy to facilitate the dynamic unidirectional movement of autophagosomes along an axon towards the soma. In distal axons, newly formed autophagosomes exhibit loosely regulated, bidirectional motion as they can bind both dynein and kinesin. As autophagosomes mature, MAPK8IP1, through its LC3-interacting region, is recruited to the autophagic membrane by associating with LC3-II. The presence of MAPK8IP1 results in the dissociation of kinesin’s heavy chain (KIF5) from the autophagosome and consequently causes the inhibition of anterograde flow, allowing the autophagosome to quickly and processively traverse the axon and enter the soma. The direction of movement along microtubules is also governed through the posttranslational modification of MAPK8IP1, which can be phosphorylated at residue S421. Indeed, when endogenous MAPK8IP1 is knocked down and replaced with its phosphomimetic version, a reduction in the retrograde movement of autophagosomes is observed, which stems from the inactivation of KIF5 [80]. Furthermore, a drastic increase in the number of either immobile, bidirectional and anterograde-moving autophagosomes is highly evident. As validation, this biochemical switch was subsequently reversed with a phospho-deficient MAPK8IP1 mutant; when MAPK8IP1 S421 is replaced by alanine, retrograde movement is restored. Overall, this multi-layered mechanism presents a simple yet effective way by which autophagy takes place in neurons: autophagosomes can act similar to a “garbage truck” and move along millimeters of axons, or “streets,” in a retrograde manner to collect “trash,” and deliver this cargo to the lysosome, the cell’s “garbage disposal and recycling center.”
The logistics of how ATG proteins are transported to and gathered at distal ends of the axon away from the cell body (the site of mRNA transcription) is poorly understood. Axons contain pools of inactive mRNAs that could be translationally activated in response to the appropriate stimuli [87]. However, it has also been established that the neuron can introduce new proteins to the axon terminal by means of a dynamic, long-range transport from the soma [87]. The latter seems to be the case for ATG9. It was recently shown that ATG9-containing vesicles can localize to the ends of the axons, near presynaptic regions, in an UNC-104/KIF1A-dependent manner [88]. Direct interference of ATG-9/ATG9 transport results in a dramatic reduction of autophagosome biogenesis in these developing neurons [88]. How other ATG proteins are delivered to these sites remains to be discovered.
Rapid autophagy in distal regions of the neuron
Although the retrograde hauling of autophagosomes across axons presents an elegant manner in which neurons have overcome longitudinal barriers to facilitate autophagy, certain situations require a rapid and relatively more efficient way to clean up surplus proteins and organelles in locations distal from the cell body. It was recently shown that, when called for, autophagy could be initiated in the edges of a neuron, without the need for long-distance transport of autophagosomes [83]. Using a genetically encoded and light-inducible photosensitizing agent to target the mitochondria (mitochondrially-targeted Killer Red, mt-KR), mitochondrial damage can be induced in cultured neuronal cells upon exposure to 555-nm light. This method is quite advantageous as damage can be site-specifically aimed at a small region of an axon, limiting autophagy induction to a substantially minute, outlying region of a neuron. As an alternative means of inducing mitochondrial damage, hippocampal neurons can be isolated using microfluidics, which allows local, focused treatment with antimycin A. Upon activation of mt-KR or addition of antimycin A, mitochondria are severely damaged as evidenced by their fragmentation, swelling and depolarization. Furthermore, these damaged mitochondria become selectively engulfed by autophagosomes (a process called mitophagy) that subsequently fuse with axonal lysosomes as evidenced by the colocalization of RFP-LC3 and LAMP1-YFP. This highly localized, axonal version of mitophagy is also regulated by the known mitochondria damage sensing and mitophagy-inducing proteins PARK2 and PINK1. Indeed, PARK2 is rapidly recruited to the mitochondria damaged by mt-KR or antimycin A. Furthermore, axonal mitophagy is clearly dependent on the Park2 or Pink1 genes [83]; axonal autophagosomes bearing functionally compromised mitochondria are not observed in cells that lack Park2 or Pink1. In summary, once induced, the initiation, nucleation, assembly and completion of autophagosomes can be accomplished in relatively far-flung regions away from a neuron’s cell body. This mechanism definitively illustrates how cells, in particular neurons, gain spatial and temporal advantage in preventing further neuronal damage that can be caused by reactive oxygen species in these damaged mitochondria.
Compartmentalized neuronal autophagy
Interestingly, in vivo investigations suggest a muted role for PINK1 and PARK2 in maintaining mitochondrial integrity in the axons of living Drosophila larvae. Although the axonal transport of mitochondria is impaired in the absence of PINK1, mitochondrial density in both the axon and soma remains relatively unchanged [89]. Conversely, in PARK2-deficient cells, reduced mitochondrial flux along axons mainly stems from a decrease in the number of axonally located mitochondria and not diminished transport [90]. Furthermore, in both cases, abnormal mitochondrial morphology is only observed in the cell body but not in axons. These results suggest that neurons handle mitochondrial quality control through PARK2-PINK1-dependent mitophagy in a compartment-specific manner [89, 90]. Such compartmentalization of neuronal autophagy is further demonstrated by a recent inquiry into the dynamics of bulk autophagy, following the formation and maturation of autophagosomes within various regions of the neuron. It was shown that autophagosomes formed within the soma are restricted to the somatodendritic space and are not permitted to travel to the axon [77]. Furthermore, axonally generated autophagosomes, upon entering the soma, remain confined to the cell body and are incapable of returning to the axon [77]. The regulatory factors that govern the gating of these locally- or distally-formed autophagosomes at the junctions between the axon and soma remain unclear.
Trans-cellular autophagy
Recently, the degradation of axonal mitochondria by a neighboring, adjacent cell has been observed [84]. This novel phenomenon, termed transmitophagy, provides another attractive mechanism as to how neurons have evolved pathways to protect themselves from oxidative stress and damage, in a quick and efficient manner. Trans-cellular mitophagy was tracked using serial block-face scanning electron microscopy in retinal ganglion cells. Multiple mitochondria appear gathered together in regions of axons that are adjacent to neighboring cells, in particular, astrocytes. Over time, these clustered mitochondria form protrusions that eventually separate and split from axons and become subsequently enveloped by a nearby astrocyte. These mitochondria are degraded in these neighboring cells as evidenced by the use of tandem fluorescent protein markers fused to a mitochondrial resident protein, COX8A (cytochrome c oxidase subunit 8A). When dually tagged with low pH-sensitive EGFP and low pH-stable mCherry, COX8A serves as a convenient marker that distinguishes the location of the mitochondria in these retinal ganglion cells. Solitary mCherry puncta persist only when the mitochondria associate with the lysosome and when they colocalize with the cytoplasm of the neighboring astrocyte. In a similar MitoFISH experiment, the fate of axonal mitochondrial DNA was also tracked. Corroboratively, these labeled nucleic acids trans-cellularly migrate to adjacent astrocytes.
Conclusion
Autophagy is a highly dynamic and complex process. Coupled with the challenges evoked by a neuron’s distinctive morphology and length, autophagy in this cell type exhibits an added layer of tremendous and rigorous kinetic and spatial regulation, wherein effective inter-and intracellular signaling and communication is indispensable. The recently uncovered mechanisms of how autophagy transpires in neurons provide an insight into how these cells have ingeniously adapted to overcome the temporal and geographical complexities imposed by its characteristic structure, size and shape. Moreover, these newly uncovered molecular strategies and pathways provide an attractive conceptual framework that will further address the inner workings of neuronal autophagy and how they can be exploited to address questions regarding disease prevention and cure.
Acknowledgments
This work was supported by NIH grant GM053396 to D.J.K.
References
- 1.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J Clin Invest. 2015;125:65–74. doi: 10.1172/JCI73944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Z, Yang B, Mo X, Xiao H. Mechanism and regulation of autophagy and its role in neuronal diseases. Mol Neurobiol. 2015;52:1190–1209. doi: 10.1007/s12035-014-8921-4. [DOI] [PubMed] [Google Scholar]
- 3.Kiriyama Y, Nochi H. The function of autophagy in neurodegenerative diseases. Int J Mol Sci. 2015;16:26797–26812. doi: 10.3390/ijms161125990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortes CJ, La Spada AR. Autophagy in polyglutamine disease: Imposing order on disorder or contributing to the chaos? Mol Cell Neurosci. 2015;66:53–61. doi: 10.1016/j.mcn.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyson T, Steiner JA, Brundin P. Sorting out release, uptake and processing of alpha-synuclein during prion-like spread of pathology. J Neurochem. 2015 doi: 10.1111/jnc.13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch-Day MA, Mao K, Wang K, et al. The role of autophagy in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2:a009357. doi: 10.1101/cshperspect.a009357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikoletopoulou V, Papandreou M-E, Tavernarakis N. Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ. 2015;22:398–407. doi: 10.1038/cdd.2014.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen D-N, Zhang L-H, Wei E-Q, Yang Y. Autophagy in synaptic development, function, and pathology. Neurosci Bull. 2015;31:416–426. doi: 10.1007/s12264-015-1536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatica D, Chiong M, Lavandero S, Klionsky DJ. Molecular mechanisms of autophagy in the cardiovascular system. Circ Res. 2015;116:456–467. doi: 10.1161/CIRCRESAHA.114.303788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W-W, Li J, Bao J-K. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xilouri M, Stefanis L. Chaperone mediated autophagy to the rescue: A new-fangled target for the treatment of neurodegenerative diseases. Mol Cell Neurosci. 2015;66:29–36. doi: 10.1016/j.mcn.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noda NN, Ohsumi Y, Inagaki F. ATG systems from the protein structural point of view. Chem Rev. 2009;109:1587–1598. doi: 10.1021/cr800459r. [DOI] [PubMed] [Google Scholar]
- 17.Popelka H, Klionsky DJ. One step closer to understanding mammalian macroautophagy initiation: Interplay of 2 HORMA architectures in the ULK1 complex. Autophagy. 2015;11:1953–1955. doi: 10.1080/15548627.2015.1087635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamada Y, Yoshino K-I, Kondo C, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabeya Y, Kamada Y, Baba M, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabeya Y, Noda NN, Fujioka Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 21.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh Y-Y, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–882. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J, Kuroyanagi H, Kuroiwa A, et al. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun. 1998;246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, Kuroyanagi H, Tomemori T, et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 26.Hosokawa N, Sasaki T, Iemura S-I, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 27.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 28.Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung CH, Jun CB, Ro S-H, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petiot A, Ogier-Denis E, Blommaart EF, et al. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 32.Furuya N, Yu J, Byfield M, et al. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 33.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 34.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polson HEJ, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 37.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Pattingre S, Sinha S, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salminen A, Kaarniranta K, Kauppinen A. Beclin 1 interactome controls the crosstalk between apoptosis, autophagy and inflammasome activation: impact on the aging process. Ageing Res Rev. 2013;12:520–534. doi: 10.1016/j.arr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–U69. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 44.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–U14. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 46.Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanida I, Mizushima N, Kiyooka M, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Dalton VM, Eggerton KP, et al. Apg7p/Cvt2p is required for the cytoplasmto-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 51.Shintani T, Mizushima N, Ogawa Y, et al. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabeya Y, Mizushima N, Yamamoto A, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 56.Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin M, Klionsky DJ. Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett. 2014;588:2457–2463. doi: 10.1016/j.febslet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mari M, Griffith J, Rieter E, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Backues SK, Orban DP, Bernard A, et al. Atg23 and Atg27 Act at the Early Stages of Atg9 Trafficking in S. cerevisiae. Traffic. 2014 doi: 10.1111/tra.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 64.Yen W-L, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Legakis JE, Yen W-L, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 66.Young ARJ, Chan EYW, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 67.Webber JL, Young ARJ, Tooze SA. Atg9 trafficking in mammalian cells. Autophagy. 2007;3:54–56. doi: 10.4161/auto.3419. [DOI] [PubMed] [Google Scholar]
- 68.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abeliovich H, Darsow T, Emr SD. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X, Mao K, Yu AYH, et al. The Atg17-Atg31-Atg29 complex coordinates with Atg11 to recruit the Vam7 SNARE and mediate autophagosome-vacuole fusion. Curr Biol. 2016;26:150–160. doi: 10.1016/j.cub.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C-W, Stromhaug PE, Kauffman EJ, et al. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 76.Yu WH, Kumar A, Peterhoff C, et al. Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for beta-amyloid peptide overproduction and localization in Alzheimer's disease. Int J Biochem Cell Biol. 2004;36:2531–2540. doi: 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 77.Maday S, Holzbaur ELF. Compartment-Specific Regulation of Autophagy in Primary Neurons. J Neurosci. 2016;36:5933–5945. doi: 10.1523/JNEUROSCI.4401-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yapici Z, Eraksoy M. Non-progressive congenital ataxia with cerebellar hypoplasia in three families. Acta Paediatr. 2005;94:248–253. doi: 10.1111/j.1651-2227.2005.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 79.Kim M, Sandford E, Gatica D, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife. 2016 doi: 10.7554/eLife.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu M-M, Holzbaur ELF. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J Cell Biol. 2013;202:495–508. doi: 10.1083/jcb.201302078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu M-M, Holzbaur ELF. MAPK8IP1/JIP1 regulates the trafficking of autophagosomes in neurons. Autophagy. 2014;10:2079–2081. doi: 10.4161/auto.34451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maday S, Wallace KE, Holzbaur ELF. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206:655–670. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis C-HO, Kim K-Y, Bushong EA, et al. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci USA. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol. 1993;121:305–315. doi: 10.1083/jcb.121.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci. 2011;31:7817–7830. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim E, Jung H. Local protein synthesis in neuronal axons: why and how we study. BMB Rep. 2015;48:139–146. doi: 10.5483/BMBRep.2015.48.3.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stavoe AKH, Hill SE, Hall DH, Colón-Ramos DA. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev Cell. 2016;38:171–185. doi: 10.1016/j.devcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Devireddy S, Liu A, Lampe T, Hollenbeck PJ. The organization of mitochondrial quality control and life cycle in the nervous system in vivo in the absence of PINK1. J Neurosci. 2015;35:9391–9401. doi: 10.1523/JNEUROSCI.1198-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sung H, Tandarich LC, Nguyen K, Hollenbeck PJ. Compartmentalized regulation of Parkin-mediated mitochondrial quality control in the Drosophila nervous system in vivo. J Neurosci. 2016;36:7375–7391. doi: 10.1523/JNEUROSCI.0633-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]