Abstract
Regulatory T (Treg) cells, expressing abundant amounts of the IL-2 receptor (IL-2R), are reliant on IL-2 produced by activated T cells. This feature implied a key role for a simple network based on IL-2 consumption by Treg cells in their suppressor function. However, congenital deficiency in IL-2R results in reduced expression of the Treg cell lineage specification factor Foxp3, confounding experimental efforts to understand the role of IL-2R expression and signaling in Treg suppressor function. Using genetic gain and loss of function approaches, we demonstrate that IL-2 capture is dispensable for control of CD4+ T cells, but is important for limiting CD8+ T cell activation, and that IL-2R dependent STAT5 transcription factor activation plays an essential role in Treg cell suppressor function separable from T cell receptor signaling.
Regulatory T (Treg) cells expressing the transcription factor Foxp3 restrain immune responses to self and foreign antigens1-3. Treg cells express abundant amounts of the interleukin 2 receptor α-chain (IL-2Rα; CD25), but are unable to produce IL-2. IL-2 binds with low affinity to IL-2Rα or the common γ -chain (γ c)-IL-2Rβ heterodimers, but receptor affinity increases ~1,000 fold when these three subunits together interact with IL-24. IL-2 and STAT5, a key IL-2R downstream target, are indispensable for Foxp3 induction and differentiation of Treg cells in the thymus5-11. IL-2Rβ and γ c are shared with the IL-15 receptor, whose signaling can also contribute to the induction of Foxp312. IL-2, in cooperation with the cytokine TGF-β, is also required for extrathymic Treg cell differentiation13.
While the role for IL-2R signaling in the induction of Foxp3 expression and Treg cell differentiation in the thymus has been well established by previous studies, the significance of IL-2R expression in mature Treg cells is not well understood. Although the deficiency in STAT5 abolishes Foxp3 expression, it can be rescued by increased amounts of the anti-apoptotic molecule Bcl2. This finding raised a possibility that a primary role for IL-2 is in the survival of differentiating Treg cells or their precursors14. It was also reported that ablation of the proapoptotic protein Bim can rescue Treg cells or their precursors from apoptosis associated with IL-2 or IL-2R deficiency and restore Treg cell numbers, but it did not prevent fatal autoimmunity15. However, a profound effect of a congenital deficiency in IL-2, Bcl2 and Bim on differentiation and selection of Treg and self-reactive effector T (Teff) cells confounds interpretation of this observation. Antibody-mediated neutralization of IL-2 in thymectomized mice reduces Treg cell numbers and Foxp3 expression in Treg cells16, 17. Thus, IL-2 supports Treg cell lineage stability after differentiation18, 19. However, expression of a transgene encoding IL-2Rβ chain exclusively in thymocytes was reported to rescue the lethal autoimmune disease in Il2rb−/− mice, suggesting that IL-2R expression is dispensable in peripheral Treg cells7, 11. Thus, a role for IL-2R expression and signaling in peripheral Treg cells remains uncertain. Hypothetically, a role for IL-2R in peripheral Treg cells could be threefold: 1) guidance for Treg cells to sense their targets – activated self-reactive T cells, which serve as a source of IL-2; 2) Treg cell-mediated deprivation of IL-2 as a mechanism of suppression, and 3) cell-intrinsic IL-2 signaling in differentiated Treg cells to support their maintenance, proliferation, or function due to triggering of JAK–STAT5, PI3K–Akt, or Ras–ERK signaling pathways. Previous studies primarily focused on the induction or maintenance of Foxp3, while other aspects of IL-2R function have not been firmly established due to aforementioned limitations.
Despite their high reliance on IL-2 for the maintenance of Foxp3 expression, Treg cells are unable to produce IL-2. The reason for the inhibition of autologous activation of STAT5 in Treg cells, and potential biological significance of this IL-2-based Treg-Teff cell regulatory loop, also remain unknown. It has been suggested that repression of IL-2 is required to maintain the ‘unbound’ state of high affinity IL-2R on Treg cells, and unbound IL-2R serves a key role in Treg cell-mediated suppression by depriving Teff cells of IL-220-24, however, whether this mechanism has a non-redundant role in suppression in vivo is unknown.
To address the role of IL-2R and downstream signaling pathways in differentiated Treg cells, we ablated IL-2Rα, IL-2Rβ, and STAT5 in Foxp3-expressing cells. By simultaneously inducing expression of an active form of STAT5, we assessed the differential requirements for IL-2R expression and IL-2 signaling for Treg cell homeostasis vs. suppressor activity.
Results
IL-2R is indispensable for Treg cell function
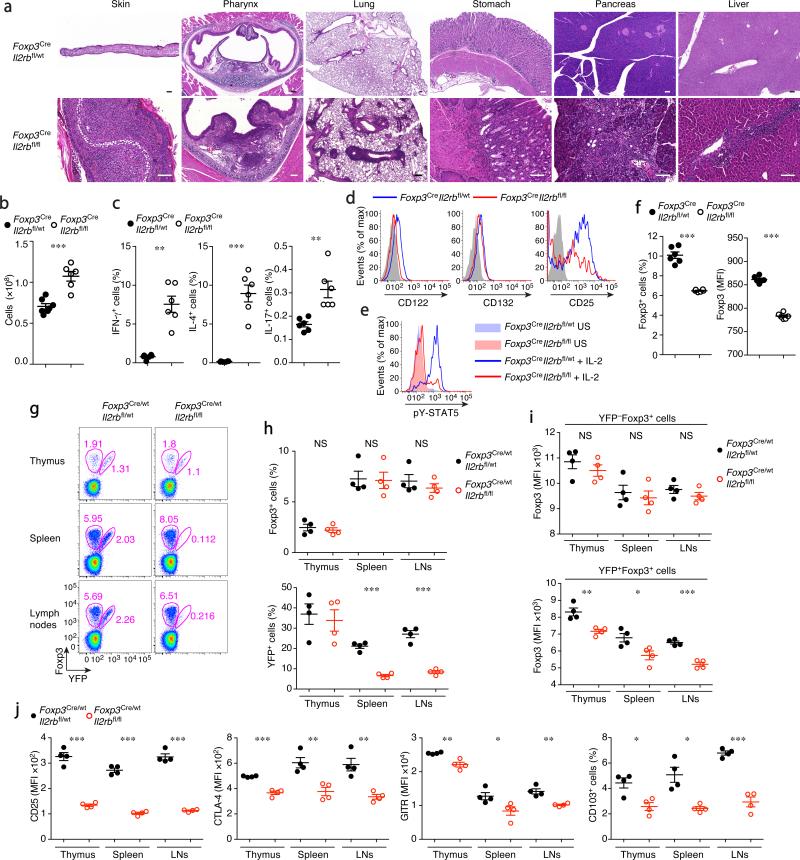
To definitively establish a role for IL-2R in Treg cell function in vivo, we generated a Treg cell-specific IL-2Rβ conditional knockout mice using Cre recombinase driven by the endogenous Foxp3 locus (Foxp3Cre), in which a loxP-flanked Il2rb allele (Il2rbfl/fl) was deleted in Treg cells after Foxp3 was expressed. Il2rbfl/flFoxp3Cre mice developed systemic fatal autoimmune inflammatory lesions and lymphoproliferation, albeit somewhat milder than that observed in Foxp3− mice (Fig. 1a–c). IL-2Rα expression was diminished in peripheral IL-2Rβ-deficient Treg cells (Fig. 1d), and tyrosine phosphorylation of STAT5 in response to IL-2 was lacking (Fig. 1e). The frequency of Foxp3+ cells among CD4+ T cells and the expression of Foxp3 on a per-cell basis were both diminished (Fig. 1f). In healthy heterozygous Il2rbfl/flFoxp3Cre/wt females, where both IL-2Rβ-sufficient (YFP−) and -deficient (YFP+) Treg cells co-exist due to random X-chromosome inactivation, IL-2Rβ-deficient Treg cells were underrepresented (Fig. 1g, h). It has been suggested that IL-2 is selectively required for the maintenance of CD62LhiCD44lo Treg cells, but is dispensable for CD62LloCD44hi Treg cells25. However, we found both CD62LhiCD44lo and CD62LloCD44hi Treg cells to be significantly reduced in the absence of IL-2Rβ in healthy heterozygous females. In these mice, IL-2Rβ-deficient Treg cells expressed reduced amounts of Foxp3 and Treg-cell ‘signature’ molecules IL-2Rα chain (CD25), CTLA-4, GITR, and CD103 regardless of CD62L and CD44 expression (Fig. 1i, j and Supplementary Fig. 1a). Although in diseased Il2rbfl/flFoxp3Cre mice, a majority of Treg cells were CD62LloCD44hi, this was likely a consequence of severe inflammation, because Treg cell frequencies were also markedly reduced at sites where CD62LloCD44hi cells were prevalent, i.e., the small and large intestines (Supplementary Fig. 1b). Accordingly, many characteristic Treg cell markers, except for CD25 and Foxp3, were upregulated as the result of Treg cell activation in Il2rbfl/flFoxp3Cre mice (Supplementary Fig. 1c). These observations suggested that both CD62LhiCD44lo and CD62LloCD44hi Treg cell subsets, including those residing in the non-lymphoid tissues, are dependent on IL-2, though under inflammatory conditions the latter can be sustained to some extent by IL-2R-independent signals. Despite the upregulation of CTLA-4, GITR, ICOS, and CD103, the ‘activated’ IL-2Rβ-deficient Treg cells from Il2rbfl/flFoxp3Cre mice were still incapable of controlling inflammation in the diseased mice and were not suppressive when co-transferred with Teff cells into lymphopenic recipients (data not shown).
Figure 1.
IL-2Rβ is indispensable for Treg cell function. (a) Histopathology of indicated organs of Foxp3CreIl2rbfl/wt and Foxp3CreIl2rbfl/fl mice. Scale bar, 100 μm. (b) Lymph node (LN) cellularity of indicated mice. (c) Cytokine production by splenic CD4+Foxp3− cells stimulated for 5 hr with anti-CD3/CD28. (d) IL-2R subunit expression by CD4+Foxp3+ cells from Foxp3CreIl2rbfl/wt (blue) and Foxp3CreIl2rbfl/fl (red) mice. (e) Intracellular phospho-STAT5 levels in Treg cells from the indicated mice unstimulated (US) or in vitro stimulated with rmIL-2 (1,000 U/ml) for 20 min. (f) The frequencies of Treg cells among LN CD3+CD4+ cells (left) and Foxp3 expression levels (MFI: mean fluorescence intensity) in the CD3+CD4+ Foxp3+ cells (right). (g-j) The analysis of healthy heterozygous female Foxp3Cre/wt mice. (g) YFP (Cre) expression and intracellular Foxp3 staining identify Treg cells with or without YFP-Cre expression. Gates shown are for CD3+CD4+ cells. (h) The frequencies of Foxp3+ cells among CD3+CD4+ cells (upper panel) and of Cre expressing cells among Foxp3+ cells (lower panel) in the indicated organs of Foxp3Cre/wtIl2rbfl/wt (black) and Foxp3Cre/wtIl2rbfl/fl (red) mice. (i) Foxp3 expression levels (MFI) in YFP−Foxp3+ (upper panel) and YFP+Foxp3+ (lower panel) cells. (j) The expression of indicated markers in YFP+Foxp3+ cells. Cells were analyzed by flow cytometry (b–j). 3–5 wk-old sex and age matched mice were analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (two-tailed unpaired Student's t test). Data are from one experiment representative of three independent experiments with similar results with three or more mice per group in each (b, c, f, h, i, j; each dot represents a single mouse; mean ± s.e.m.) or representative data of more than five (a) or ten (d, e, g) mice per group analyzed are shown.
Our findings raised the question whether ablation of IL-2Rα, which, in addition to facilitating IL-2 signaling, enables its sequestration from Teff cells, would result in a similar Treg cell deficiency and disease compared to those in Il2rbfl/flFoxp3Cre mice. Thus, we generated a loxP-flanked Il2ra allele (J.D.F. manuscript in preparation) and similarly induced its conditional ablation in Treg cells. We found that Treg cell-specific IL-2Rα deficiency resulted in a disease with comparable early onset and severity to those observed upon IL-2Rβ ablation (Supplementary Fig. 1d–f). Of note, germ-line deficiency of either Il2ra or Il2rb in mice on the same C57BL6/J as our conditional knockout mice resulted in a considerably less aggressive disease with a delayed onset, likely due to a role for IL-2R signaling in Teff cells (data not shown). Our findings also indicate that IL-15 was unable to effectively compensate for the loss of IL-2 signaling in differentiated Treg cells because in Foxp3CreIl2rafl/fl mice, Treg cells lacked only IL-2 signaling, whereas in Il2rbfl/flFoxp3Cre mice, they lacked both IL-2 and IL-15 signaling yet were similarly affected. This was in contrast to Treg cell differentiation in the thymus where IL-15 can contribute in part to Foxp3 induction12. Since IL-2R activates PI3K–Akt, MAPK, and JAK–STAT5 signaling pathways, we next sought to assess a role for STAT5 activation downstream of IL-2R signaling in Treg cells. We found that STAT5 ablation similarly impaired Treg cell function and Foxp3CreStat5a/bfl/fl mice were similarly affected by fatal autoimmunity as were mice harboring IL-2R deficient Treg cells (Supplementary Fig. 1g–k). Thus, in agreement with IL-2 neutralization studies, IL-2R signaling is required for Treg cell fitness in a cell-intrinsic manner.
STAT5b-CA partially rescues IL-2R deficient Treg function
The above findings implied that STAT5 activation downstream of IL-2R is continuously required for Treg cell function. However, a marked decrease in IL-2R observed in STAT5-deficient Treg cells (Supplementary Fig. 1g) made it impossible to separate a loss of STAT5 from impairment in all IL-2R functions, i.e., detection of IL-2, transduction of STAT5-dependent and -independent signals, and consumption and deprivation of IL-2, as a key contributor to the observed severe Treg cell dysfunction.
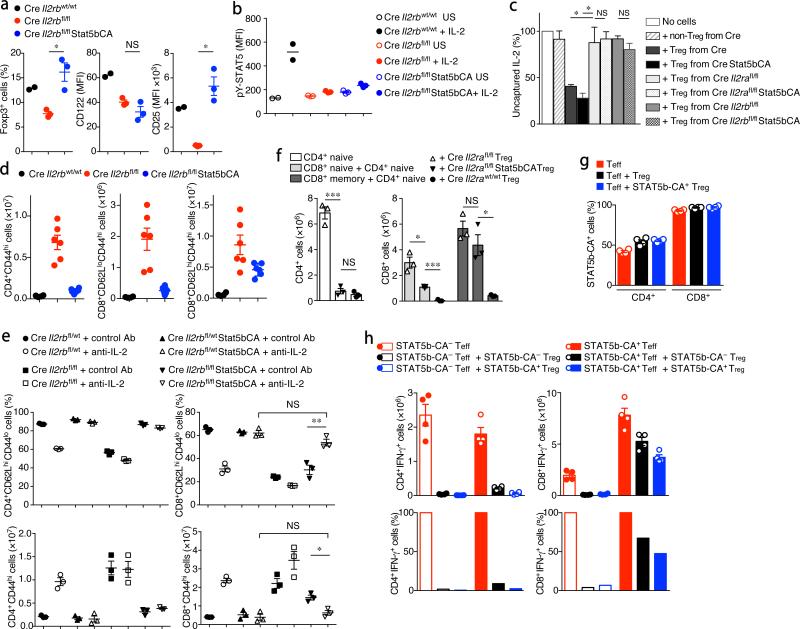
To address this major caveat and to understand a role for STAT5 vs. IL-2R, we asked whether expression of a gain-of-function form of STAT5b can rescue Treg cell function in the absence of IL-2R. A previous study using a transgene encoding a constitutively active form of STAT5b (STAT5b-CA) driven by the proximal lck promoter in the absence of IL-2Rβ showed rescue of Treg cell differentiation in the thymus, but not lymphoproliferative syndrome9. However, the expression of this transgene early during thymopoiesis leads to leukemic lymphoproliferation, which complicated the interpretation of these findings. In addition, both the activity of the proximal lck promoter and the expression of the transgene diminish in peripheral T cells in these mice9. Therefore, we generated a gene-targeted mouse strain utilizing the Rosa26 ‘gene trap’ locus26, where a ‘CAG’ promoter driven STAT5b-CA27 is preceded by a loxP-flanked STOP cassette (Supplementary Fig. 2a). In the resulting Rosa26Stat5bCA mice, STAT5b-CA is expressed only when the loxP sites undergo Cre mediated recombination. Introduction of the Rosa26Stat5bCA allele into Il2rbfl/flFoxp3Cre mice and the consequent expression of STAT5b-CA in IL-2Rβ-deficient Treg cells rescued the systemic inflammation and early fatal disease (Supplementary Fig. 2b). In these mice, Treg cell frequencies and numbers were comparable to or even surpassed their levels in IL-2R sufficient Foxp3Cre mice (Fig. 2a). Notably, the expression of IL-2Rα chain was increased despite the absence of IL-2Rβ chain (Fig. 2a), suggesting the expression of IL-2Rα on Treg cells is primarily controlled by STAT5-dependent, but not by STAT5-independent signaling. Importantly, these IL-2Rβ-deficient Treg cells with heightened IL-2Rα expression remained unresponsive to IL-2 (Fig. 2b).
Figure 2.
Restoration of the suppressor activity of IL-2R-deficient Treg cells in the presence of an active form of STAT5. (a) The frequencies of Foxp3+ cells among CD3+CD4+ cells and CD122 and CD25 expression by CD3+CD4+Foxp3+ cells. (b) Intracellular phospho-STAT5 levels in LN Treg cells from the indicated mice unstimulated (US) or in vitro stimulated with rmIL-2 (1,000 U/ml) for 20 min. (c) In vitro IL-2 capture assay. Treg and non-Treg cells from the indicated mice were sorted and cultured for 2 hr with recombinant human IL-2 (hIL-2). The amount of residual hIL-2 in the media after 2 hr was measured using flow cytometry-based bead array analysis and shown as percent value. (d) The cell numbers of indicated CD4+ and CD8+ cell subset (both CD3+Foxp3−) in the LNs of indicated mice (2 wk old). (e) The frequencies of naive (CD62LhiCD44lo) T cells among CD3+CD4+Foxp3− and CD3+CD8+Foxp3− cells (upper two panels) and the numbers of CD44hi activated CD3+CD4+Foxp3− and CD3+CD8+Foxp3− cells (lower two panels) in the LNs of indicated mice treated with IL-2 neutralizing antibodies or control IgG for 2 wks starting from day 5–7 after birth. (f) Analysis of the ability of IL-2R-sufficient and -deficient Treg cells to suppress the expansion of naive and activated/memory CD4+ and CD8+ T cells. CD4+Foxp3−CD62LhiCD44lo (CD4+ naive), CD8+Foxp3−CD62LhiCD44lo (CD8+ naive), and CD8+Foxp3−CD62LhiCD44hi (CD8+ memory) T cells were sorted from Foxp3Cre mice and adoptively transferred (1 × 106 cells each) into T cell-deficient (Tcrb−/−Tcrd−/−) mice together with Treg cells (2 × 105 cells) sorted from the indicated mice. CD4+Foxp3− and CD8+Foxp3− T cell numbers in the LNs of recipients 3 wks after transfer are shown. (g, h) Analysis of susceptibility of CD4+ and CD8+ T cells expressing an active form of STAT5 to Treg mediated suppression. (g) The frequencies of STAT5b-CA-expressing CD4+ and CD8+ Teff cells within total CD4+ and CD8+ Teff cells 3 wks after a transfer of in vitro TAT-Cre recombinase treated CD4+Foxp3− and CD8+Foxp3− T cells sorted from Foxp3CreRosa26Stat5bCA mice and transferred (1 × 106 cells each) into Tcrb−/−Tcrd−/− recipients. (h) The numbers and proportion (%) of IFN-γ-producing CD4+ and CD8+ T cells in the recipients without a co-transfer of Treg cells (red bars) or with 2 × 105 control (black bars) or STAT5b-CA-expressing Treg cells (blue bars) sorted from Foxp3Cre or Foxp3CreRosa26Stat5bCA mice, respectively. As a control, CD4+Foxp3− and CD8+Foxp3− T cells sorted from Foxp3CreRosa26wt mice were similarly treated with TAT-Cre and transferred to assess the susceptibility of STAT5b-CA− Teff cells to Treg mediated suppression (open bars). *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (two-tailed unpaired Student's t test). Data are from one experiment representative of two (b, c, e, f) or three (a, d, g, h) independent experiments with similar results with two or more (a, b) or three or more (c, d, e, f, g, h) mice per group in each experiment (each dot represents a single mouse; mean ± s.e.m.).
The observed restoration of the suppressor function of IL-2Rβ-deficient Treg cells and rescue of the early fatal disease upon STAT5b-CA expression raised the possibility that the reintroduced high IL-2Rα levels were responsible for these effects. However, the expression of STAT5b-CA similarly rescued the early fatal disease in Foxp3CreIl2rafl/fl mice (Supplementary Fig. 2c–h). Importantly, although the impaired capacity of Treg cells in both Il2rbfl/flFoxp3Cre and Foxp3CreIl2rafl/fl mice to capture and consume IL-2 was not rescued upon STAT5b-CA expression (Fig. 2c), CD4+ T cell reactivity was fully controlled in these mice (Fig. 2d and Supplementary Fig. 2d–h). These results suggested that the ability to capture and compete for IL-2 is dispensable for Treg cell mediated suppression of CD4+ T cell responses. To the contrary, however, expansion of CD8+ T cells, in particular, of activated CD62LhiCD44hi CD8+ T cells, was only marginally restrained in these mice (Fig. 2d and Supplementary Fig. 2f, h). Although the expansion of CD8+CD62LloCD44hi subset was relatively well, albeit not perfectly, controlled in neonatal mice (Fig. 2d and Supplementary Fig. 2f), this subset also gradually started to expand in these mice as early as 3 wks after birth (Supplementary Fig. 2i). Although both Il2rbfl/flFoxp3CreRosa26Stat5bCA and Il2rafl/flFoxp3CreRosa26Stat5bCA mice were rescued from premature death and showed significantly improved clinical status comparable to healthy controls, they gradually failed to thrive and started to succumb to disease accompanied by massively expanded activated CD62LhiCD44hi and CD62LloCD44hi CD8+ T cell subsets in LNs and tissues by approximately 12 wks of age (Supplementary Fig. 2i, j). These findings raised a possibility that IL-2 consumption by Treg cells, while dispensable for control of CD4+ T cells, is important for the restraint of CD8+ T cells.
Treg cells suppress CD8+ T cell responses via IL-2 depletion
To test if the impairment in consumption of IL-2 by Treg cells can account for the proliferation of CD8+ T cells in Foxp3CreIl2rbfl/flRosa26Stat5bCA mice, we administered IL-2 neutralizing antibodies to these and control mice starting from 5–7 days of age (Fig. 2e and Supplementary Fig. 3a). As IL-2 supports the differentiation of Treg cells in the thymus, IL-2 neutralization reduced the frequencies of Treg cells in all groups of mice and induced immunoactivation in control Il2rbfl/wtFoxp3Cre mice. In Il2rbfl/flFoxp3Cre mice, which spontaneously develop disease, the production of TH2 cytokines IL-4 and IL-13 by CD4+ T cells was significantly reduced by IL-2 neutralization; however, the activation of CD4+ and CD8+ T cells was at best only marginally reduced or unaffected. In contrast, the activation and proliferation of CD8+ T cells observed in Il2rbfl/flFoxp3CreRosa26Stat5bCA mice were almost completely suppressed by the treatment.
The relative reduction in CD8+CD62LloCD44hi and more pronounced proliferation of CD8+CD62LhiCD44hi T cell subset in Il2rbfl/flFoxp3CreRosa26Stat5bCA and Il2rafl/flFoxp3CreRosa26Stat5bCA mice raised a possibility that a loss of IL-2-consumption by Treg cells might selectively impair their suppression for memory CD8+ T cell expansion, but not the recruitment of naive CD8+ T cells into the effector cell pool. We tested this idea by adoptive transfer of CD4+ and CD8+ cell subsets into lymphopenic recipients (Fig. 2f). Consistent with the observation in Foxp3Cre mice, the impaired suppression of CD4+ T cell expansion and activation by IL-2R-deficient Treg cells was completely rescued by STAT5b-CA; in contrast, their ability to suppress memory CD8+ T cells was not restored, whereas suppression of naive CD8+ T cell expansion and expansion was only partially recovered. Thus, IL-2 consumption by Treg cells appears to have a non-redundant role in suppressing the expansion and activation of both naive and memory CD8+ T cell subsets, although this mechanism appears to be particularly prominent in control of the latter subset.
Although the majority of activated CD8+ T cells in Il2rbfl/flFoxp3Cre and Il2rbfl/flFoxp3CreRosa26Stat5bCA mice did not express detectable levels of IL-2Rα (Supplementary Fig. 3a), these cells could activate STAT5 in response to IL-2, albeit to a lesser extent than that observed in cells expressing IL-2Rα (Supplementary Fig. 3b). A small proportion of activated CD4+ T cells with undetectable IL-2Rα expression also responded to IL-2, but the majority of them did not. CD8+ T naive cells also responded to IL-2, while CD4+ T naive cells did not. Thus, both naive and activated CD8+ T cells appeared to be more sensitive to IL-2 than CD4+ T cells, and IL-2 consumption by Treg cells may markedly affect their activation. A corollary to this notion was that STAT5 activation in CD8+, but not CD4+ T cells may render the former resistant to Treg cell mediated suppression. Thus, we tested the effect of STAT5 activation on the proliferation of CD4+ and CD8+ T cells in the presence of Treg cells. For this purpose, we sorted CD4+Foxp3− and CD8+Foxp3− T cells from Foxp3CreRosa26Stat5bCA mice and induced the expression of STAT5b-CA in these cells by treating them with a recombinant Cre protein containing a membrane permeable TAT (trans-activating transcriptional activator from HIV virus) peptide (TAT-Cre). We adoptively transferred the treated cells into lymphopenic recipients with or without Treg cells. Although TAT-Cre treatment initially induced STAT5b-CA expression in approximately 30% of the treated CD4+ and CD8+ T cells, more than 95% of CD8+ T cells expressed STAT5b-CA three weeks after the transfer; whereas STAT5b-CA expressing CD4+ T cells expanded to 40-50% (Fig. 2g). Notably, STAT5b-CA+CD8+ T cells robustly expanded in the presence of either control (Foxp3Cre) or STAT5b-CA+ Treg cells (Fig. 2g, h). Although some degree of suppression of STAT5b-CA+CD8+ T cells by Treg cells was still observed, it was very mild compared to the suppression of STAT5b-CA−CD8+ T cells (Fig. 2h). In contrast, proliferation of and cytokine production by activated CD4+ T cells, regardless of the expression of STAT5b-CA, were well controlled by Treg cells (Fig. 2h). These observations suggest that STAT5 activation in CD8+, but not in CD4+ T cells prompts robust expansion of cells and confers pronounced resistance to Treg cell mediated suppression. Consistent with these findings, gain-of-function experiments where IL-2 was provided in the form of IL-2/ IL-2 immune complexes showed expansion of CD8+ T and CD4+ Treg, but not of CD4+ T cells28. Thus, while the ability to capture and compete for IL-2 is dispensable for Treg cell mediated suppression of CD4+ T cell responses, this mode of suppression appears essential for control of CD8+ T cells, which respond to excessive IL-2 more robustly than CD4+ T cells.
STAT5 activation in Tregcells boosts immunosuppression
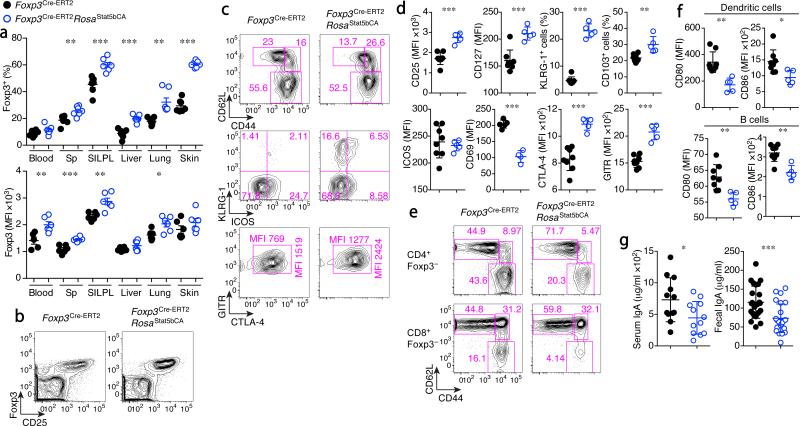
The lack of detectable STAT5 activation in response to IL-2 and of STAT5b-CA-driven expansion of IL-2R-sufficient Treg cells that escaped from Cre-mediated recombination (counterselection) in both Il2rbfl/flFoxp3CreRosa26Stat5bCA and Il2rafl/flFoxp3CreRosa26Stat5bCA mice indicated that the expression of an active form of STAT5 relieved Treg cells from their dependence on IL-2 signaling. This finding offered a unique opportunity to explore the biological significance of the aforementioned IL-2-dependent Treg–Teff cell regulatory network by uncoupling Treg cell function from IL-2 production by Teff cells. To address this question, we generated Rosa26Stat5bCAFoxp3Cre-ERT2 mice, which enabled tamoxifen-inducible expression of STAT5b-CA in differentiated Treg cells17. Induction of STAT5b-CA expression in ~20–30% of Treg cells upon a single tamoxifen administration was followed by their rapid increase in numbers at the expense of Treg cells with a non-recombined Rosa26Stat5bCA allele (Supplementary Fig. 4a, b). It is noteworthy that these cells exhibited a highly diverse TCR Vβ usage similar to that in control mice (Supplementary Fig. 4c).The experimental Foxp3Cre-ERT2Rosa26Stat5bCA mice remained healthy (Supplementary Fig. 4d, e). In these mice, the proliferated STAT5b-CA+ Treg cell population exhibited increased amounts of Foxp3, CD25, CTLA4, and GITR and an increased proportion of CD62LhiCD44hi vs. CD62LhiCD44lo cells, indicative of a STAT5b-CA impressed biasing of the Treg cell population towards an activated or memory cell state (Fig. 3a–d and Supplementary Fig. 4f). Consistent with the latter possibility, surface expression of IL-7R, KLRG1, and CD103 were increased (Fig. 3d). Notably, in lymph nodes (LNs) and Peyer's patches (PPs), Treg cells were not numerically increased despite the predominance of STAT5b-CA+ Treg cells (Supplementary Fig. 4b, g), suggesting that Treg cells with activated STAT5 preferentially distribute in non-lymphoid tissues. CD8+Foxp3+ cells were also increased upon induction of STAT5b-CA (Supplementary Fig. 4h). The ‘autonomous’ Treg cells, expressing active STAT5 showed heightened in vitro suppressor activity (Supplementary Fig. 4i) and effectively suppressed the basal state of activation and proliferative activity of CD4+ and CD8+ T cells in vivo, as well, as evidenced by the decreased numbers of Ki-67+ cells and CD62LloCD44hi Teff cells and a markedly increased CD62LhiCD44lo T naive cell pool (Fig. 3e and Supplementary Fig. 5a, b). Accordingly, CD4+ T cell production of pro-inflammatory cytokines, most prominently IL-4, and expression of CD80 and CD86 by B cells and dendritic cells (DCs) were reduced (Supplementary Fig. 5c and Fig. 3f). These results indicate that expression of STAT5-CA confers increased suppressor function to Treg cells. Previously, Treg cells were proposed to promote systemic TH17 type responses and IgA class switching in the gut29, 30. However, we found that serum and fecal IgA as well as TH17 responses in secondary lymphoid organs were reduced, rather than increased in the presence of STAT5b-CA+ Treg cells (Fig. 3g and Supplementary Fig. 5c). Serum IgM and IgE also showed a tendency towards a decrease, but this was not statistically significant (Supplementary Fig. 5d). These results were in agreement with an increase in TH17 responses and in both TH2- and TH1-type Ig class switch observed upon acute Treg cell ablation31. Since altered intestinal immune responses have been implicated in promoting colonic carcinogenesis, we explored an effect of a gain in Treg cell suppressor function afforded by activated STAT5 in an ApcMin model of colorectal cancer. Mice harboring the ApcMin mutation develop multiple adenomatous polyps in the small intestine32. ApcMinFoxp3Cre-ERT2Rosa26Stat5bCA mice developed a comparable or fewer numbers of polyps, but the average polyp size was increased (Supplementary Fig. 5e). These results were consistent with the idea that suppression of inflammation by Treg cells in tumor microenvironments promotes the growth of tumors once tumors or pre-cancerous lesions are already formed. However, the early stages of colonic carcinogenesis appeared not to be promoted but were potentially suppressed by Treg cells with augmented suppressor activity.
Figure 3.
Increased proliferative and suppressor activity of Treg cells expressing an active form of STAT5. (a–g) Analysis of Foxp3Cre-ERT2 (black dots) and Foxp3Cre-ERT2Rosa26Stat5bCA (blue dots) mice three months after a single tamoxifen treatment. (a) The frequencies of Foxp3+ cells among CD3+CD4+ cells (upper graph) and the expression levels of Foxp3 in CD3+CD4+Foxp3+ cells (lower graph) in the indicated organs. Sp: spleen, SILPL: small intestine lamina propria lymphocytes. (b) Analysis of splenic CD4+ T cells for the expression of CD25 and Foxp3. (c) Analysis of splenic CD4+Foxp3+ Treg cells. (d) The expression of indicated markers in splenic CD4+Foxp3+ Treg cells. (e) Analysis of splenic CD3+CD4+Foxp3− (upper panels) and CD3+CD8+Foxp3− (lower panels) cells. (f) The expression of CD80 and CD86 on DCs (CD11c+MHC class IIhi) and B cells (B220+CD11c−) in the LNs. (g) Serum and fecal IgA levels as determined by ELISA. Sex and age matched mice were analyzed. Cells were analyzed by flow cytometry (a–f). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired Student's t test). Data are from one experiment representative of two (a, d, f, g) independent experiments with similar results with five or more mice per group in each experiment (a, d, f, g; each dot represents a single mouse; mean ± s.e.m.) or representative data of more than ten mice per group analyzed are shown (b, c, e).
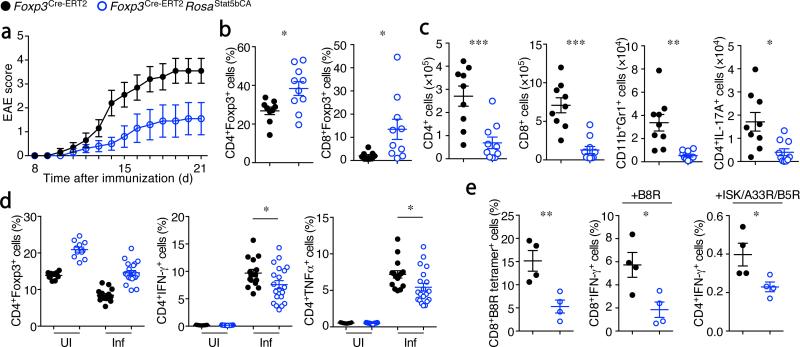
In addition to restraining the basal immune reactivity in physiological settings and modulating colon carcinoma development, ‘autonomous’ Treg cells afforded superior protection against autoantigen-induced autoimmunity. We found that Foxp3Cre-ERT2Rosa26Stat5bCA mice were highly resistant to experimental autoimmune encephalomyelitis (EAE) (Fig. 4a–c). The frequencies of CD4+Foxp3+ cells were significantly increased in the brain and spinal cord of these mice (Fig. 4b), and infiltration of inflammatory cells, including neutrophils and IL-17-producing CD4+ TH17 cells into these organs, was significantly reduced (Fig. 4c). Pathogen-specific responses were also diminished in Foxp3Cre-ERT2Rosa26Stat5bCA mice. Although Listeria-specific TH1 responses were only modestly suppressed (Fig. 4d), vaccinia virus-specific CD8+ T cell responses were markedly reduced in the presence of STAT5b-CA+ Treg cells (Fig. 4e). Our observation of diminished responses to infectious agents and modulation of cancer progression may provide a rationale as to why Treg cells lack IL-2 production and autonomous activation of STAT5, and instead are reliant on activated T cells as a source of IL-2.
Figure 4.
Potent suppressor function of Treg cells expressing an active form of STAT5. (a–c) Analysis of EAE induced upon immunization with MOG peptide in CFA. (a) Average disease scores (n=10 each). (b) The frequencies Foxp3+ cells among brain-infiltrating CD3+CD4+ (left) and CD3+CD8+ (right) cells in mice shown in (a). (c) The numbers of the indicated brain-infiltrating cells in mice shown in (a). (d) T cell responses against Listeria monocytogenes in uninfected (UI) or infected (Inf; day 8 after infection) mice. The frequencies of Foxp3+ Treg cells among CD3+CD4+ cells (left). The frequencies of IFN-γ (middle) and TNFα (right) producing CD4+TCRβ+Foxp3− cells after 5 hr in vitro re-stimulation with heat-killed Listeria in the presence of DCs. (e) Anti-viral T cell responses in mice on day 8 after infection with non-replicating vaccinia virus. The frequencies of vaccinia B8R peptide-specific CD8+ T cells detected using H-2Kb-B8R tetramer (left), IFN-γ production by CD8+Foxp3− (middle) and CD4+Foxp3− (right) cells after a 5 hr in vitro stimulation with B8R peptide or a mixture of three vaccinia virus-specific peptides (ISK, A33R, and B5R). Cells were analyzed by flow cytometry (b–e). Sex and age matched Foxp3Cre-ERT2 (black) and Foxp3Cre-ERT2Rosa26Stat5bCA (blue) mice were challenged or infected as indicated 2-3 months after a single tamoxifen treatment. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired Student's t test). Data are from one experiment representative of two independent experiments with similar results with ten (a–c) or four (e) mice per group in each experiment, or pooled from four independent experiments with n=11 (UI), n=15 (Inf, control), or n=20 (Inf, Rosa26Stat5bCA) mice per group are shown (d). Each dot represents a single mouse (b–e). mean ± s.e.m. (a–e).
A distinct role for STAT5 activation in Treg cells
Next, we sought to address the question of how sustained STAT5 signaling may potentiate Treg cells’ suppressive ability. In genetic loss- and gain-of-function studies, STAT5 activity in Treg cells correlated with their proliferative capacity and expression of IL-2Rα and Foxp3. However, the in vitro suppression assays above, as well as the reduction in immune activation in LNs and PPs of Foxp3Cre-ERT2Rosa26Stat5bCA mice, where fewer Treg cells were found than in control mice suggested that the enhanced immunosuppression observed in Foxp3Cre-ERT2Rosa26Stat5bCA mice was not simply due to a numerical increase of Treg cells, but that their suppressor activity on a per cell basis was also augmented. It was also unlikely that mild upregulation of Foxp3 in the presence of STAT5b-CA could account for the increased suppressor activity of Treg cells as we found that genome-wide Foxp3 binding does not change upon activation of Treg cells, which lead to an increase in Foxp3 expression more pronounced than the one caused by STAT5b-CA33. The increase in Foxp3 protein in STAT5b-CA+ Treg cells compared to control was particularly noticeable in the CD25lo Treg cell subset (Fig. 3b; Average fold changes in Foxp3 MFI in Foxp3+ Treg cells from Foxp3Cre-ERT2Rosa26Stat5bCA mice vs. those from Foxp3Cre-ERT2 mice; CD25hi = 1.06 fold, CD25lo = 1.36 fold (n = 6)), consistent with the observation that STAT5b-CA+ Treg cells were relieved from their dependence on IL-2. Nevertheless, STAT5b-CA+ Treg cells exhibited a more potent suppressor function than CD25hiFoxp3hi Treg cells from control mice when co-transferred with Teff cells into lymphopenic recipients despite comparably high expression of Foxp3 (data not shown). Thus, the increased suppressor activity of STAT5b-CA+ Treg cells was unlikely to be due to the increased Foxp3.
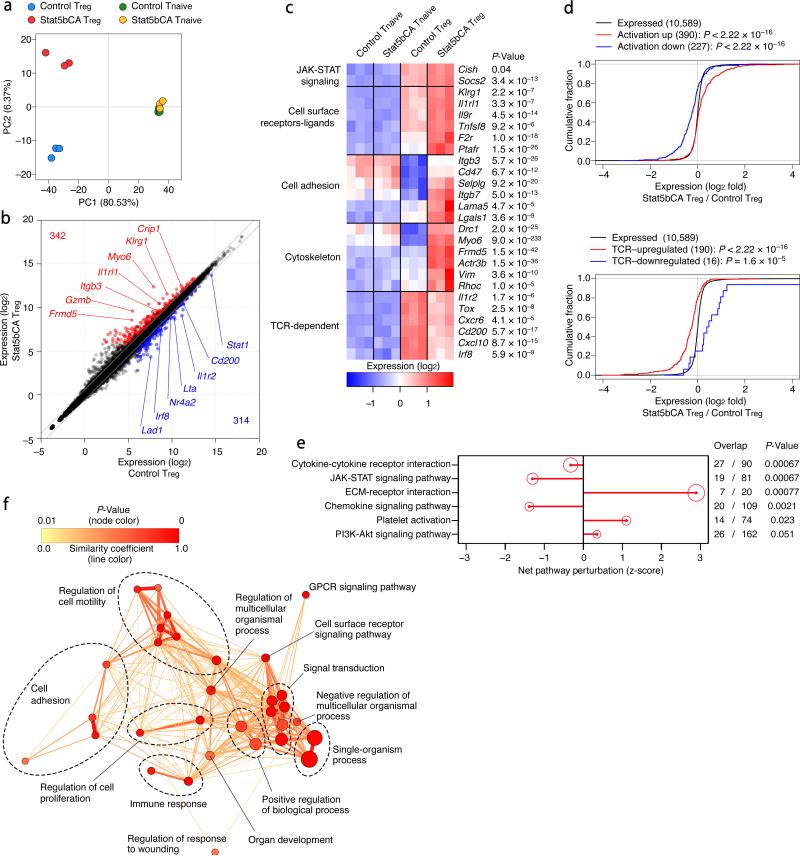
To gain insight into the potential mechanisms underlying the heightened suppressor function conferred by sustained STAT5 activation, we sorted mature Treg cells from Foxp3Cre-ERT2 and Foxp3Cre-ERT2Rosa26Stat5bCA mice that expressed comparable levels of Foxp3 and analyzed gene expression in these cells using RNA-seq. While the gene expression profiles of CD4+ T naive cells from both groups of mice were nearly identical, Treg cell gene expression was markedly affected by the active form of STAT5 (Fig. 5a and Supplementary Fig. 6a). Among all expressed genes (~11,000) in either Treg or CD4+ T naive cell populations analyzed, 342 genes were upregulated and 314 genes were downregulated in STAT5b-CA+ Treg cells compared to control cells (Fig. 5b and Supplementary Fig. 6b). The gene set upregulated in STAT5b-CA+ Treg cells encoded various cell surface molecules and receptors involved in cell adhesion, migration, and cytoskeletal reorganization (Fig. 5c). Several genes that were upregulated or downregulated in control Treg cells compared to T naive cells showed opposite trends in STAT5b-CA+ Treg cells, suggesting that STAT5b-CA does not simply reinforce the Treg cell signature. A previous study showed that exposure of Treg cells to inflammation induced upon transient Treg cell depletion leads to a marked change in their gene expression and a potent increase in their suppressor function33. Consistent with the heightened suppressor function of STAT5b-CA+ Treg cells, we found that the gene expression changes in these cells conferred by an active form of STAT5 correlated with those found in highly activated Treg cells in inflammatory settings (Fig. 5d). TCR signaling is required for the ability of Treg cells to exert their suppressor function34, 35. Thus, it was possible that TCR and STAT5 dependent signaling pathways in Treg cells are acting upon a largely overlapping set of genes whose expression they jointly regulate to potentiate Treg cell suppressor activity. However, our analysis revealed that the gene set affected by the active form of STAT5 was distinct from that expressed in Treg cells in a TCR-dependent manner (Fig. 5d). Thus, both TCR and STAT5 signaling pathways play an indispensable role in Treg cell suppressor activity in vivo by controlling largely distinct sets of genes and likely distinct aspects of Treg cell suppressor activity.
Figure 5.
RNA-seq analysis of Treg cells expressing an active form of STAT5. (a) Principal component analysis of RNA-seq datasets, using the top 15% of genes with the highest variance. Each dot represents an RNA sample from a single mouse. Note that “Stat5bCA T naive” cells are T naive cells sorted from tamoxifen-treated Foxp3Cre-ERT2Rosa26Stat5bCA mice and do not express STAT5b-CA. Only “Stat5bCA Treg” cells express STAT5b-CA in the four groups of cells presented. (b) Plot of gene expression (as log2 normalized read count) in control vs. STAT5b-CA expressing Treg cells. The diagonal lines indicate fold change of at least 1.5× or 0.67× fold. Significantly up- and down-regulated genes (defined as genes with at least 1.5× or 0.67× fold change, adjusted P-value ≤ 0.05, and expression above a minimal threshold based on the distribution of all genes) are colored red or blue, respectively, and their numbers are shown. (c) Heat map of selected genes. Three replicates are shown in order. P-values are for control Treg vs. STAT5b-CA expressing Treg cells. (d) The empirical cumulative distribution function (ECDF) for the log2 fold change of all expressed genes in STAT5b-CA vs. control Treg cells, is plotted along with ECDFs for the subsets of genes up- or down-regulated by inflammatory activation in Treg cells33 (upper graph), or the subsets of genes up- or down-regulated in a TCR-dependent manner in CD44hi Treg cells34 (lower graph). (e) Signaling Pathway Impact Analysis (SPIA) of KEGG pathways. The 6 most statistically significant pathways that show enrichment among differentially expressed (DE) genes in STAT5b-CA vs. control Treg cells are shown. The net pathway perturbation indicates the status of the pathway (positive = activated; negative = inhibited) based on the activating or inhibitory relationships of DE genes within the pathway. The size of the red circle is proportional to the degree of enrichment, and the FDR-adjusted global P-value reflecting both enrichment and perturbation is shown. (f) Network analysis of GO term enrichment among significantly upregulated genes in STAT5b-CA vs. control Treg cells. Upregulated genes were analyzed for over-represented GO terms using BiNGO in Cytoscape, and the resulting network was calculated and visualized using EnrichmentMap. Groups of similar GO terms (Supplementary Table 1) were manually circled. Line thickness and color are proportional to the similarity coefficient between connected nodes. Node color is proportional to the FDR-adjusted P-value of the enrichment. Node size is proportional to gene set size.
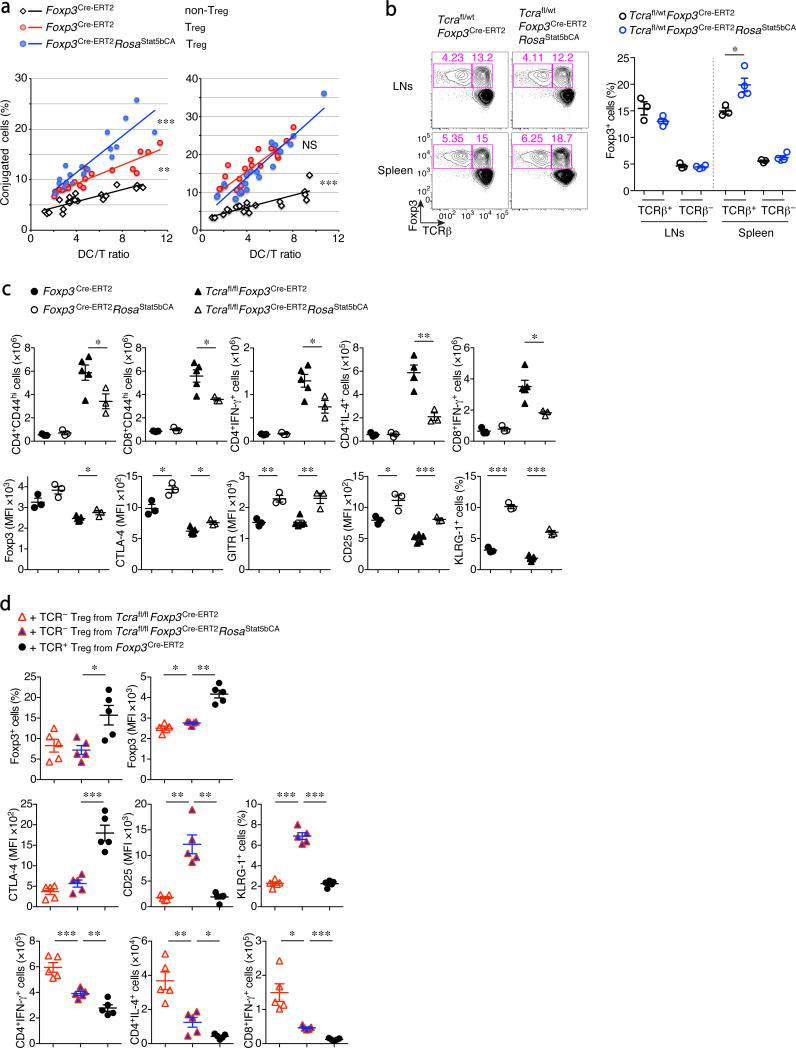
To better understand aspects of Treg cell function potentiated by STAT5 activation, we performed signaling pathway and molecular function enrichment analyses, which revealed overrepresentation of gene sets involved in cell-cell and extracellular matrix interactions, cell adhesion, and cellular locomotion among genes differentially expressed in STAT5b-CA+ Treg cells (Fig. 5e, f, Supplementary Fig. 6c). This result suggested that in Treg cells, STAT5 activation might potentiate their interactions with the target cells. Since intravital imaging of Treg cells in vivo had previously revealed their stable interactions with DCs36, we assessed the potential effect of constitutively active STAT5 expression in Treg cells on their ability to form conjugates with DCs in vitro. In agreement with the gene set enrichment analysis, we found that STAT5b-CA expression in Treg cells promotes conjugate formation between Treg and DCs (Fig. 6a). Heightened interactions of STAT5b-CA+ Treg cells with DCs in vitro were consistent with the decreased expression of co-stimulatory molecules by DCs observed in tamoxifen-treated Foxp3Cre-ERT2Rosa26Stat5bCA mice.
Figure 6.
Augmented STAT5 signaling in Treg cells increases the conjugate formation between Treg cells and DCs and potentiates suppressor function in a TCR independent manner. (a) Analysis of in vitro conjugate formation between T cells and DCs. CFSE-labeled Treg and non-Treg cells from the indicated mice were co-cultured with graded numbers of CellTrace Violet-labeled CD11c+ DCs from C57BL/6J mice for 720 min in the absence (left panel) or presence (right panel) of rmIL-2 (100 IU/ml). Each dot represents a flow cytometric analysis of conjugate formation in a single well. (b) Expression of Foxp3 and TCRβ by CD4+ cells in the LNs and spleen of Tcrafl/wt heterozygous mice treated with tamoxifen for 2 wks. The frequencies of TCR-sufficient and -deficient Foxp3+ cells among CD4+ cells are summarized in the right panel. (c) T cell activation and pro-inflammatory cytokine production in the LNs of indicated mice treated with tamoxifen for 2 wks. The lower five panels show the expression of the indicated molecules in LN Treg cells.
(d) The analysis of T cells transferred into Tcrb−/−Tcrd−/− recipients. WT CD4+Foxp3− and CD8+Foxp3− T cells (5 × 105 cells each) were transferred together with TCR-ablated (TCRβloCD3lo) or TCR-sufficient Treg cells (3 × 105 cells) sorted from the indicated mice that had been treated with tamoxifen for 2 wks. The frequencies of Treg cells and Foxp3 expression levels in Treg cells (upper two panels), the expression of the indicated molecules in Treg cells (middle three), and the numbers of cytokine producing CD4+ and CD8+ T cells (lower three) in the LNs of recipient mice 3 wks after the transfer are shown. Sex and age matched mice were analyzed. Cells were analyzed by flow cytometry (a–d). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (modified ANCOVA in Prism software (a) or two-tailed unpaired Student's t test (b, c, d)). Data are from one experiment representative of three (a) or two (b, d) or four (c) independent experiments with similar results with three or more (b, c) or five or more (d) mice per group in each experiment (b, c, d; each dot represents a single mouse; mean ± s.e.m.).
These findings raised a question whether STAT5 activation can potentiate the suppressor function of Treg cells in a TCR-independent manner. To test this notion, we analyzed Foxp3Cre-ERT2Rosa26Stat5bCA mice expressing a conditional Tcra allele. As we reported previously, tamoxifen-inducible Cre-mediated TCR ablation in Treg cells is highly efficient in these mice and resulted in immune activation resulting from impaired suppressor function34. In heterozygous Foxp3Cre-ERT2Tcrafl/wt mice, Cre mediated recombination can theoretically result in ablation of TCR in up to a half of Treg cells due to the allelic exclusion at the Tcra locus. We observed a small proportion of TCR-deficient Treg cells after 2-wks of tamoxifen administration in these mice (Fig. 6b). Although the expression of the active form of STAT5 was observed in ~50% of TCR-sufficient and -deficient Treg cells in Foxp3Cre-ERT2Tcrafl/wtRosa26Stat5bCA mice, the proportion of only TCR-sufficient, but not -deficient STAT5b-CA expressing Treg cells was increased. The marked increase in T cell activation and pro-inflammatory cytokine production was mitigated in part upon expression of the active form of STAT5 in tamoxifen-treated Foxp3Cre-ERT2Tcrafl/flRosa26Stat5bCA mice (Fig. 6c). This partial recovery of Treg cell suppressor function by the active form of STAT5 in TCR-ablated Treg cells was also confirmed in experiments where flow-sorted TCR-deficient STAT5b-CA+ Treg cells and Teff cells were adoptively transferred into lymphopenic recipients (Fig. 6d). Although the rescue was incomplete, these results suggested that enhanced STAT5 signaling could potentiate Treg cell suppressor activity in the absence of contemporaneous TCR-dependent signals. Indeed, some features of Treg cells that had been observed in TCR-sufficient STAT5b-CA+ Treg cells were still present in TCR-ablated STAT5b-CA+ Treg cells (Fig. 6c, d). It should be noted, however, that STAT5b-CA expression failed to rescue suppressor function in Foxp3CreTcrafl/flRosa26Stat5bCA mice where TCR deletion occurred immediately after the induction of Foxp3. It was previously shown that TCR signaling is required for Treg cells to acquire an activated, antigen-experienced phenotype and suppressor function34. Thus, our results suggest that activation of STAT5 potentiates TCR-independent suppressor function in mature Treg cells that have already undergone TCR-dependent maturation. This observation is reminiscent of the sequential requirement for these two signals, TCR and IL-2R, in the differentiation of Treg cells in the thymus where STAT5 signal promotes differentiation of Treg precursors that have experienced permissive TCR signaling37.
Discussion
Previous analysis of mice with germ-line deficiency in IL-2 and IL-2R subunits demonstrated that IL-2 is a key cytokine required for the induction of Foxp3 and the differentiation of Treg cells in the thymus5-11 . Furthermore, antibody-mediated IL-2 neutralization and provision of IL-2-complexes, as well as genetic dissection of regulatory elements within the Foxp3 locus, revealed an important role for IL-2 in the maintenance of mature Treg cells and in stabilization of Foxp3 expression during their extrathymic differentiation16, 28, 38. These findings raised a question of whether IL-2R signaling can also directly promote Treg cell suppressor capacity and, therefore, serve as a critical nexus linking differentiation and maintenance of Treg cells with their suppressor function. An early in vitro study proposed a role for IL-2 signaling based on indirect evidence21. In addition, IL-2 consumption by Treg cells was suggested to play an essential role in Treg cell suppressor function by causing death of activated CD4+ T cells due to IL-2 deprivation20-24. On the other hand, other reports suggested that IL-2R is dispensable for the ability of Treg cells to suppress effector T cell proliferation8, 39. Furthermore, the rescue of the disease in Il2ra−/− and Il2rb−/− mice observed upon adoptive transfer of wild-type Treg cells suggested the existence of major mechanisms of Treg cell-mediated suppression independent of IL-2-deprivation6, 7. However, the latter studies left open a major question as to whether IL-2 consumption by Treg cells is essential for suppression of IL-2R-sufficient Teff cells since IL-2 is likely a major driver of autoimmune disease in the absence of functional Treg cells.
A major limiting factor in efforts to experimentally assess a role for IL-2R signaling in, and IL-2 consumption by Treg cells in their function in vivo has been the lack of adequate genetic tools. We addressed this issue through generation of conditional Il2ra and Il2rb alleles and their ablation in Treg cells in combination with the induced expression of an active form of STAT5. These new genetic tools enabled us to unequivocally demonstrate that IL-2R signaling has a cell intrinsic, non-redundant role not only in the maintenance of mature Treg cells and their fitness, but also in their suppressor function. Furthermore, we found that STAT5 deficiency in Treg cells results in a similar loss of suppressor function and that expression of an active form of STAT5 can rescue fatal disease resulting from IL-2R deficiency. These results suggest a key role of IL-2R–STAT5 signaling in linking differentiation and maintenance of Treg cells and their function. STAT5 binds to the Foxp3 promoter and the intronic Foxp3 regulatory element CNS2 and is involved in Foxp3 induction and maintenance38. Runx-CBFβ complexes also bind to CNS2 and the Foxp3 promoter and affect Foxp3 expression levels40. Although both CNS2- and CBFβ-deficient Treg cells do exhibit reduced Foxp3 expression resembling that of STAT5- or IL-2R-deficient Treg cells, the impairment of suppressor function in the latter was much more severe. Thus, the decrease in Foxp3 expression alone cannot account for a severe loss of Treg cell suppressor function in the absence of STAT5 or IL-2R. Indeed, our analysis of gene expression and functional features conferred upon expression of the active form of STAT5 pointed to a heightened ability of Treg cells to bind to DC and suppress their activation. Furthermore, expression of an active form of STAT5 partially rescued the near-complete loss of Treg suppressor function in the absence of TCR signaling34, 35. These results may appear at odds with the previous finding that STAT5b-CA transgene driven by the proximal lck promoter and Eμenhancer failed to curtail fatal lymphoproliferative disease in Il2rb−/− mice despite restoring Foxp3 expression and Treg cell differentiation in the thymus9. However, the interpretation of the latter result is problematic due to a massive expansion of pre-leukemic T and B cells and reduced expression of the STAT5b-CA transgene in peripheral Treg cells.
Our studies clearly demonstrated that IL-2-deprivation by Treg cells was fully dispensable for suppression of IL-2R-sufficient CD4+ T cells even though IL-2R signaling was required. However, IL-2R dependent IL-2 consumption by Treg cells was indispensable for suppression of CD8+ T cell responses. The latter seemingly unexpected finding makes sense in light of the observed exquisite sensitivity of both naive and activated CD8+ T cells to IL-2 induced stimulation. Furthermore, IL-2 is produced upon activation of both naive CD4+ and CD8+ T cells within hours after TCR engagement in contrast to effector cytokines such as IL-4 and IFN-γ whose production requires T naive cell differentiation into Teff cells on a much longer time scale41. These distinguishing features provide a likely explanation for a need for a distinct mechanism of control of CD8+ T cell responses by Treg cells through IL-2 consumption.
It has been suggested that sensing of local IL-2 production by Treg cells enables ‘licensing’ of their suppressor function21. However, the rescue of suppression of CD4+ T cell responses by IL-2R-deficient Treg cells expressing an active form of STAT5 suggests that activated Treg cells can suppress autoimmunity without identifying the cellular source of IL-2. Thus, while IL-2 is a booster for Treg cell suppressor function, it may not play an indispensable role as a cue for specific targeting.
Genetically modified T cells are emerging as a potent means of therapy in some forms of cancer. The observed enhanced suppressor activity of Treg cells expressing an active form of STAT5 and significantly reduced severity of organ-specific autoimmunity in their presence suggest that such a modification of Treg cells may hold promise for an optimal design of Treg cell-based therapies for a variety of autoimmune and inflammatory disorders and in organ transplantation.
Our findings highlight the central role of IL-2 receptor signaling driven STAT5 activation in supporting and boosting suppressor function of differentiated Treg cells. In this regard, it is noteworthy that although a Foxp3 ortholog has not been identified in birds, chicken and duck CD4+ T cell subsets expressing high amounts of IL-2Rα chain possess in vitro suppressor activity suggesting the importance of evolutionary conservation of IL-2Rα function in suppressive T cells42, 43.
Methods
Mice
Foxp3Cre and Foxp3Cre-ERT2 mice were described previously17, 44. Il2rafl mice were generated by J.D.F. Stat5a/bfl mice were provided by L Henninghausen. ApcMin mice were purchased from the Jackson Laboratory. The targeting strategies to generate Il2rbfl (generated by U.K.) and Rosa26Stat5bCA alleles are shown in Supplementary Fig. 7. Tcrafl mice were described previously34. The experimental mice were either generated on or backcrossed onto a C57BL/6J (B6) background, bred and housed in the specific pathogen-free animal facility at Memorial Sloan Kettering Cancer Center (MSKCC). All animal experiments were approved by institutional animal care and use committee at MSKCC and were performed in accordance with the institutional guidelines. For survival analysis, mice were monitored daily and unhealthy mice were euthanized once they are found lethargic and counted as non-survivors. For tamoxifen treatment, tamoxifen (Sigma-Aldrich) was dissolved in olive oil at a concentration of 40 mg/ml. Mice were given oral gavage of 100 μl of tamoxifen emulsion per treatment. In EAE and infection experiments, mice were challenged 2 to 3 months after a single tamoxifen gavage and assessed as described previously38.
Flow cytometry and cell sorting
Cells were stained with fluorescently tagged antibodies purchased from eBioscience, BD Biosciences, Tonbo Bioscience, or BioLegend (Supplementary Table 2) and analyzed using a BD LSR II flow cytometer. Flow cytometry data were analyzed using FlowJo software (TreeStar). For intracellular cytokine staining, cells were stimulated for 5 hr with CD3 and CD28 antibodies (5 μg/ml each) in the presence of brefeldin A or monensin, harvested and stained with eBioscience Fixation Permeabilization kit. For intracellular phosphorylated STAT5 staining, cells were stimulated with or without rmIL-2 for 20 min, fixed and permeabilized with 4% PFA followed by 90% methanol, and stained with anti-pY-STAT5 antibody (BD Biosciences). Cell sorting of Foxp3+ and Foxp3− cells was performed based on YFP or GFP expression using a BD FACSAria II cell sorter.
Listeria and Vaccinia infection
Mice were intravenously injected into the tail vein with Listeria monocytogenes (LM10403S; 2000 cells/mouse) on day 0 and analyzed on day 8. For the detection of Listeria-specific immune responses, splenic DCs from unchallenged B6 mice sorted using CD11c microbeads (Miltenyi) were cultured in wells of a 96 well U-bottom plate (2 × 104 cells/well) with heat-killed Listeria monocytogenes (2 × 107 cells/well) for 6 hr prior to the analysis. The cells were then co-cultured with splenic T cells obtained from Listeria-infected mice (1 × 105 cells/well) for 5 hr in the presence of brefeldin A, and cytokine producing T cells were detected by flow cytometry. For vaccinia virus infection, mice were intraperitoneally injected with non-replicating virus (5 × 107 PFU/mouse) on day 0 and analyzed on day 8. Splenocytes were re-stimulated with several vaccinia virus derived antigenic peptides (1 μg/ml) for 5 hr in the presence of brefeldin A, and cytokine producing T cells were detected by flow cytometry.
In vivo IL-2 neutralization
Mice were i.p. injected with a cocktail of two different anti-IL-2 monoclonal antibodies JES6-1 and S4B6-1 (BioXcell) or isotype matched control antibody (rat IgG2a, 2A3; BioXcell), 200 μg each, twice a week, starting from 5–7 days after birth.
TAT-Cre protein treatment of T cells
For the induction of STAT5b-CA expression in non-Treg cells, 1 × 107 CD4+Foxp3− or CD8+Foxp3− T cells sorted from the LNs and spleens of Foxp3Cre and Foxp3CreRosa26Stat5bCA mice were resuspended in 2 ml of serum-free RPMI media containing a TAT-Cre recombinase (Millipore; 50 μg/ml) and incubated at 37°C for 45 min. The cells were washed with RPMI containing 10% FCS, resuspended in PBS, and injected into T cell-deficient (Tcrb−/−Tcrd−/−) mice together with or without separately sorted Treg cells for in vivo suppression assay.
In vitro IL-2 capture assay
Pooled cells from LNs and spleens were depleted of B cells and accessary cells by panning and T cells were enriched. The cells were stained with anti-CD8 and anti-B220 Abs, and CD4+ Treg cells were sorted on the basis of GFP (YFP) expression alone in CD8-negative population. The sorted cells were divided among 8 wells of a 96-well V-bottomed plate (2 × 105 cells/well) in 25 μl RPMI medium (10% FCS) with or without increasing doses of recombinant human IL-2 (0.016 to 12 U/ml), followed by incubation for 2 h at 37 °C. Depletion of IL-2 from the medium was assessed with the BD Cytometric Bead Array and Human IL-2 Enhanced Sensitivity Flex Set according to the manufacturer's instructions (BD Biosciences).
In vitro T–DC conjugation assay
Treg cells and non-Treg cells were sorted in the same manner as IL-2 capture assay. Splenic CD11c+ DCs were isolated by MACS from B6 mice injected with Flt3L-secreting B16 melanoma cells. Treg and non-Treg cells were stained with CFSE. DCs were stained with CellTrace Violet (Molecular Probes). 1 × 104 Treg or non-Treg cells were cultured together with graded numbers of DCs (1 × 104 to 1 × 105) in a 96-well round-bottomed plate for 720 min in the presence or absence of rmIL-2 (100 IU/ml). Frequencies of Treg cells conjugated with DCs (% CTV+CFSE+/CFSE+) were analyzed by flow cytometry.
In vitro suppression assay
Naive CD4+ T cells (responder cells) and Treg cells were FACS purified and stained with CellTrace Violet (CTV). 4 × 104 naive CD4+ T cells were cultured with graded numbers of Treg cells in the presence of 1 × 105 irradiated, T-cell-depleted, CFSE-stained splenocytes and 1 μg/ml anti-CD3 antibody in a 96 round-bottom plate for 80 hr. Cell proliferation of responder T cells and Treg cells (live CFSE−CD4+Foxp3− and Foxp3+) was determined by flow cytometry based on the dilution of fluorescence intensity of CTV of the gated cells.
Measurements of serum and fecal immunoglobulin levels
Serum IgM, IgG1, IgG2a, IgG2b, IgG2c, IgG3 and IgA levels were determined by ELISA using SBA Clonotyping System (Southern Biotech). IgE ELISA was performed using biotinylated anti-IgE antibody (BD Biosciences) and HRP-conjugated streptavidin. For measurement of fecal IgA levels, fresh fecal pellets were collected and dissolved in extraction buffer (7 μl per mg pellet) containing 50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 1mM EDTA, 1 mM DTT, and protease inhibitor cocktail (Complete mini; Roche). Supernatants were collected after centrifugation, titrated, and IgA levels were measured by ELISA.
Statistical Analysis for Animal Experiments
Each mouse was tagged with a unique identification number, and researchers were blinded to the genotypes of mice except for adjusting sample size included in a single experiment and after data analysis is completed. Wild type mice with suspected congenital anomalies were excluded from the study. Cell samples that showed less than 70% cell vitality after preparation or after in vitro stimulation were excluded from the study. Statistical analyses were performed using Prism software with two-tailed unpaired Student's t test. Welch's correction was applied when F test was positive. P values < 0.05 were considered significant.
RNA sequencing
Male 8-wk-old Foxp3Cre-ERT2Rosa26Stat5bCA (STAT5b-CA) and Foxp3Cre-ERT2 (control) mice, nine mice for each experimental group, received a single dose (4 mg) of tamoxifen by oral gavage 4 months before isolation. Splenic CD4+Foxp3(YFP/GFP)+GITRhiCD25hi Treg and CD4+Foxp3(YFP/GFP)−CD62LhiCD44lo naive T cells were double sorted using a BD FACSAria II cell sorter, and a total of 12 samples were generated. Spleen T cell subsets isolated from three individual mice in the same experimental group were pooled into one biological replicate; three biological replicates were subjected to RNA-seq analysis for each experimental group. Total RNA was extracted and used for poly(A) selection and Illumina TruSeq paired-end library preparation following manufacturer's protocols. Samples were sequenced on the Illumina HiSeq 2500 to an average depth of 27.5 million 50-bp read pairs per sample. All samples were processed at a same time and sequenced on the same lane to avoid batch effects.
Read alignment and processing followed the method previously described45. Briefly, raw reads were trimmed using Trimmomatic v0.32 with standard settings to remove low-quality reads and adaptor contamination46. The trimmed reads were then aligned to the mouse genome (Ensembl assembly GRCm38) using TopHat2 v2.0.11 implementing Bowtie2 v2.2.2 with default settings. Read alignments were sorted with SAMtools v0.1.19 before being counted to genomic features using HTSeq v0.6.1p1. The overall read alignment rate across all samples was 74.5%. Differential gene expression was analyzed using DESeq2 1.6.3 in R version 3.1.047.
Bioinformatic analyses for RNA-seq
The distribution of read counts across all genes was bimodal. The assumption that this corresponded to “expressed” and “non-expressed” genes was supported by examination of marker genes known to be expressed or not expressed in Treg and T naive cells. The local minimum between the two peaks was chosen to be the threshold for expression. Using this threshold of ~60 normalized reads, 10,589 out of 39,179 genes were called as present. Significantly up- (342 genes) and down-regulated (314) genes between STAT5b-CA versus control Treg cells were defined as expressed genes with fold changes of at least 1.5× or 0.67×, respectively, and FDR-adjusted P-value ≤ 0.05.
TCR-upregulated (i.e., TCR-dependent) genes were defined as genes downregulated (at least 0.57× fold change) in TCR-deficient compared to TCR-sufficient CD44hi Treg cells, while TCR-downregulated genes are upregulated (at least 1.75×, Padj ≤ 0.001) in TCR-deficient CD44hi Treg cells (GSE61077)34. Activation-upregulated genes are genes upregulated (2× fold change, Padj ≤ 0.01) in Treg cells from Foxp3DTR mice recovering from punctual Treg cell depletion (GSE55753)33.
Signaling Pathway Impact Analysis (SPIA) was performed using the R package of the same name48. Significantly up- and downregulated genes, and their fold changes, were analyzed as one set for enrichment and perturbation of 90 Mus musculus KEGG pathways accessed on October 5, 2015. The net pathway perturbation Z-score was calculated using the observed net perturbation accumulation, and the mean and SD of the null distribution of net perturbation accumulations. Global P-values were calculated using the normal inversion method with Bonferroni correction.
Biological process (BP) gene ontology (GO) term over-representation was calculated using BiNGO v3.0.349 in Cytoscape v3.2.1, employing the hypergeometric test and applying a significance cutoff of FDR-adjusted P-value ≤ 0.05. The 10,589 expressed genes were entered as the reference set, and the GO ontology and annotation files used were downloaded on Oct. 25, 2015 (Supplementary Table 1). The output from BiNGO was imported into EnrichmentMap v2.0.150 in Cytoscape to cluster redundant GO terms and visualize the results. An EnrichmentMap was generated using a Jaccard similarity coefficient cutoff of 0.2, a P-value cutoff of 0.001, an FDR-adjusted cutoff of 0.005, and excluding gene sets with fewer than 10 genes. The network was visualized using a prefuse force-directed layout with default settings and 500 iterations. Groups of similar GO terms were manually circled.
Supplementary Material
Acknowledgments
We thank T. Kitamura (The University of Tokyo) for mSTAT5b-CA vector (pMX-STAT5B(1*6)) and K. Rajewsky (Max Delbrück Center) for Rosa26 construct, L. Henninghausen (NIH) for Stat5a/b conditional allele. Supported by the Japan Society for the Promotion of Science, Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation (T.C.), Lucille Castori Center for Microbes, Inflammation & Cancer (T.C.), NIH Medical Scientist Training Program grant T32GM07739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program (A.G.L. and X.F.), the US National Institutes of Health (P30 CA008748 to MSKCC Core Facilities, R37 AI034206 to A.Y.R.), the Ludwig Center at Memorial Sloan Kettering Cancer Center (A.Y.R.), Hilton-Ludwig Cancer Prevention Initiative funded by the Conrad N. Hilton Foundation and Ludwig Cancer Research (A.Y.R.), and the Howard Hughes Medical Institute (A.Y.R.).
Footnotes
Author contributions
T.C., J.D.F. and A.Y.R. conceived the project and designed the experiments; T.C., A.K.K., A.G.L., X.F., Y.Z., G.G., and Y.F. conducted experiments; U.K. generated Il2rbfl allele; J.D.F. generated Il2rafl allele; T.C., J.D.F. and A.Y.R. wrote and edited the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Accession codes
References
- 1.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 2.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 3.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 4.Waldmann TA. The multi-subunit interleukin-2 receptor. Annual review of biochemistry. 1989;58:875–911. doi: 10.1146/annurev.bi.58.070189.004303. [DOI] [PubMed] [Google Scholar]
- 5.Furtado GC, Curotto de Lafaille MA, Kutchukhidze N, Lafaille JJ. Interleukin 2 signaling is required for CD4(+) regulatory T cell function. The Journal of experimental medicine. 2002;196:851–857. doi: 10.1084/jem.20020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida AR, Legrand N, Papiernik M, Freitas AA. Homeostasis of peripheral CD4+ T cells: IL-2R alpha and IL-2 shape a population of regulatory cells that controls CD4+ T cell numbers. J Immunol. 2002;169:4850–4860. doi: 10.4049/jimmunol.169.9.4850. [DOI] [PubMed] [Google Scholar]
- 7.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 8.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nature immunology. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 9.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 10.Yao Z, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nature reviews. Immunology. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 12.Vang KB, et al. IL-2, -7, and -15, but not thymic stromal lymphopoeitin, redundantly govern CD4+Foxp3+ regulatory T cell development. J Immunol. 2008;181:3285–3290. doi: 10.4049/jimmunol.181.5.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malin S, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nature immunology. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barron L, et al. Cutting edge: mechanisms of IL-2-dependent maintenance of functional regulatory T cells. J Immunol. 2010;185:6426–6430. doi: 10.4049/jimmunol.0903940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. The Journal of experimental medicine. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu N, et al. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hori S. Lineage stability and phenotypic plasticity of Foxp3(+) regulatory T cells. Immunological reviews. 2014;259:159–172. doi: 10.1111/imr.12175. [DOI] [PubMed] [Google Scholar]
- 20.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nature immunology. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 21.Thornton AM, Donovan EE, Piccirillo CA, Shevach EM. Cutting edge: IL-2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J Immunol. 2004;172:6519–6523. doi: 10.4049/jimmunol.172.11.6519. [DOI] [PubMed] [Google Scholar]
- 22.Barthlott T, et al. CD25+ CD4+ T cells compete with naive CD4+ T cells for IL-2 and exploit it for the induction of IL-10 production. International immunology. 2005;17:279–288. doi: 10.1093/intimm/dxh207. [DOI] [PubMed] [Google Scholar]
- 23.Busse D, et al. Competing feedback loops shape IL-2 signaling between helper and regulatory T lymphocytes in cellular microenvironments. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3058–3063. doi: 10.1073/pnas.0812851107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi T, et al. Construction of self-recognizing regulatory T cells from conventional T cells by controlling CTLA-4 and IL-2 expression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2116–2125. doi: 10.1073/pnas.1307185110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smigiel KS, et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. The Journal of experimental medicine. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zambrowicz BP, et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onishi M, et al. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Molecular and cellular biology. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, et al. Foxp3(+) regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity. 2011;34:409–421. doi: 10.1016/j.immuni.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature immunology. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 32.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 33.Arvey A, et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nature immunology. 2014;15:580–587. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nature immunology. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vahl JC, et al. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 2014;41:722–736. doi: 10.1016/j.immuni.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Tadokoro CE, et al. Regulatory T cells inhibit stable contacts between CD4+ T cells and dendritic cells in vivo. The Journal of experimental medicine. 2006;203:505–511. doi: 10.1084/jem.20050783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran DQ, et al. Analysis of adhesion molecules, target cells, and role of IL-2 in human FOXP3+ regulatory T cell suppressor function. J Immunol. 2009;182:2929–2938. doi: 10.4049/jimmunol.0803827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudra D, et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nature immunology. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J Immunol. 2004;172:6136–6143. doi: 10.4049/jimmunol.172.10.6136. [DOI] [PubMed] [Google Scholar]
- 42.Shanmugasundaram R, Selvaraj RK. Regulatory T cell properties of chicken CD4+CD25+ cells. J Immunol. 2011;186:1997–2002. doi: 10.4049/jimmunol.1002040. [DOI] [PubMed] [Google Scholar]
- 43.Andersen KG, Nissen JK, Betz AG. Comparative Genomics Reveals Key Gain-of-Function Events in Foxp3 during Regulatory T Cell Evolution. Frontiers in immunology. 2012;3:113. doi: 10.3389/fimmu.2012.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for methods
- 44.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature protocols. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarca AL, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 50.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.