Abstract
Triosephosphate isomerase (TIM; EC 5.3.1.1) is a key enzyme involved in glycolysis and gluconeogenesis. Glycolysis is one of the most regulated metabolic pathways, however little is known about the structural mechanisms for its regulation in non-model organisms, like crustaceans. To understand the structure and function of this enzyme in invertebrates, we obtained the crystal structure of triosephosphate isomerase from the marine Pacific whiteleg shrimp (Litopenaeus vannamei, LvTIM) in complex with its inhibitor 2-phosphogyceric acid (2-PG) at 1.7 Å resolution. LvTIM assembles as a homodimer with residues 166-176 covering the active site and residue Glu166 interacting with the inhibitor. We found that LvTIM is the least stable TIM characterized to date, with the lowest range of melting temperatures, and with the lowest activation enthalpy associated with the thermal unfolding process reported. In TIMs dimer stabilization is maintained by an interaction of loop 3 by a set of hydrophobic contacts between subunits. Within these contacts, the side chain of a hydrophobic residue of one subunit fits into a cavity created by a set of hydrophobic residues in the neighboring subunit, via a "ball and socket" interaction. LvTIM presents a Cys47 at the "ball" inter-subunit contact indicating that the character of this residue is responsible for the decrease in dimer stability. Mutational studies show that this residue plays a role in dimer stability but is not a solely determinant for dimer formation.
Keywords: triosephosphate isomerase, TIM, shrimp, prawn, Litopenaeus vannamei, dimer stabilization
1. INTRODUCTION
Triosephosphate isomerase (TIM) is an obligated dimer, with the exception of Archaea in which this protein assembles as a tetramer [1-4]. Each TIM subunit is comprised of approximately 250 residues and dimerization is thought to be necessary for the stability and proper assembly of the active site loops. Each TIM monomer assembles as a barrel comprised by eight β-loop-α motifs, forming a parallel eight-stranded β-barrel surrounded by eight α-helices on the outside of the monomer [5, 6]. TIM plays a critical role in the glycolytic pathway interconverting dihydroxyacetone phosphate and D-glyceraldehyde-3-phosphate [6]. Its catalytic properties and mechanism have been described in detail [6-8]. Its catalytic rate is diffusion-controlled, a property that identifies it as an “evolutionarily perfect enzyme” [7]; furthermore, neither cofactors nor metal ions are required for the reaction and no allostery or cooperativity has been found to date [9]. The critical role of TIM is that it serves as a crossroad between glycolysis, lipid metabolism, gluconeogenesis and pentose phosphate pathways. The TIM barrel fold is very conserved among enzymes and the crystal structure of triosephosphate isomerase has been determined in many species such as Gallus gallus [10], Saccharomyces cerevisiae [11], Trypanosoma brucei [12], Escherichia coli [13], Homo sapiens [5], Bacillus stearothermophilus [14], Plasmodium falciparum [15], Trichomonas vaginalis [16] and others.
The study of (β–α)8 barrel proteins and TIMs in particular as a putative drug targets has been thoroughly addressed [17]. The TIM dimer interface is a starting point towards rational drug design against diseases originated by protozoan parasites that heavily depend on glycolysis for energy [15]. Molecules that interact with exposed cysteines like methylmethane thiosulfonate (MMTS) selectively inhibit TIMs from Leishmania donovani [18], T. cruzi [19], T. brucei [20] and Giardia lamblia [21]. The strength of the inhibition is dependent on the degree of conjugation of a reactive cysteine (Cys15 in the case of T. cruzi) and the perturbation of the interactions at the homodimeric interface.
Thermodynamic studies indicate that in TIMs the energy necessary to dissociate the homodimer is higher that the energy needed to unfold the monomers [1, 22-24]. The exceptions to this fact are TIMs from Trichomonas vaginalis (TvTIMs). These TIMs only differ between them in 4 among 252 amino acids and the energy necessary to dissociate their dimers is lower than the energy to unfold its monomers [16, 25, 26]. In order to understand glycolysis regulation in crustaceans we characterized the triosephosphate isomerase from shrimp Litopenaeus vannamei (LvTIM) with focusing on its stability and the "ball and socket" model postulated for TvTIMs [16, 26].
2. MATERIALS AND METHODS
2.1 Reagents
All biochemical reagents, chemicals and enzymes were purchased from Sigma-Aldrich. HisTrap™ Fast Flow and a Superdex 75 100-300 and 200 100-300 columns were used for IMAC and size exclusion chromatography respectively. Columns were obtained from GE Healthcare Life Sciences Inc. (Piscataway, NJ USA). Gene synthesis and the expression vector pJetExpress404 (T5 promoter) were provided by DNA2.0 (Menlo Park, CA, USA). Protein concentration was estimated using the micro BCA (bicinchoninic acid) from Pierce-Thermo Fisher Scientific (Rockford IL, USA).
2.2 LvTIM overexpression and purification
The cDNA of LvTIM was deduced from the shrimp transcriptome [27] and confirmed by sequencing at the GATC Genomic Analysis and Technology Core Lab at The University of Arizona (Tucson AZ, USA). The sequence was deposited in GenBank under access code JX431295. The deduced amino acid sequence was optimized for its heterologous expression in E. coli, synthesized and cloned into the pJetExpress404 (T5 promoter, ampicillin resistance marker) for recombinant expression in bacteria. The construct was transformed into strain E. coli BL21(DE3). A single ampicillin-resistant transformed colony was used to inoculate 5 ml of Luria-Bertani (LB) media, supplemented with 100 μg ml−1 ampicillin; shaken for eight h at 37 °C. This pre-inoculum was used to inoculate 50 ml of LB media and finally to inoculate 1 l of LB media in a Fernbach flask. The culture was incubated at 37 °C and shaken at 200 RPM until an OD600= 0.6 was reached. Expression of LvTIM was induced by addition of isopropyl thiogalactoside (IPTG) to a final concentration of 0.4 mM. The culture was shaken for 18 h at 200 RPM at 16 °C and the bacterial biomass was harvested by centrifugation at 5000 × g at 4 °C for 15 min and stored at −80 °C until used. For bacterial lysis, 1 g of the bacterial pellet was resuspended in 5 ml of ice-cold lysis buffer (20 mM Tris-HCl, 0.5 M NaCl, 0.5 mM PMSF and 0.1 mg ml−1 lysozyme). The resuspended cell culture was incubated on ice for 20 min and then sonicated with 3 pulses of 30 s each with a Branson sonicator.
Bacterial lysate was clarified by centrifugation at 35000 × g for 30 min at 4 °C. The supernatant was passed through a 5 ml HisTrap™ Fast Flow column previously equilibrated with buffer A (20 mM Tris-HCl, 0.5 M NaCl). The column was washed with 10 volumes of buffer A and the elution of LvTIM was executed with a linear gradient of imidazole from 0 to 0.5 M in running buffer A. LvTIM eluted at 200-250 mM of imidazole in a final volume of 9 ml. The eluate was immediately dialyzed in 3 changes of 500 ml of dialysis buffer (0·1 M triethanolamine (TEA) buffer, pH 7.4, 50 mM NaCl.) at 4 °C for 12 h. A second purification step, also used as quaternary structure confirmation, was performed using a Superdex 200 size exclusion column previously calibrated with molecular mass standards in dialysis buffer. The molecular mass markers were albumin (66 kDa), ovalbumin (44 kDa) and trypsin inhibitor (21.5 kDa).
2.3 Enzyme kinetics and stability
Protein concentration was measured by the micro BCA method (Pierce), using dilutions of bovine serum albumin (BSA) as standard, and with the measured the absorbance at 280 nm using a calculated molecular coefficient of 35825 M−1 cm−1 [28]. The activity assay was done as previously reported [18], following the decrease in absorbance at 340 nm at 25 °C due to the oxidation of NADH. The assay was done in a 1 ml quartz cell containing 50 mM Tris pH 7.6, 1 mM glyceraldehyde 3-phosphate, 0.2 mM NADH and 1 U of α-glycerophosphate dehydrogenase [29]. The reaction was started by adding 5 μg ml−1 of LvTIM in the reaction mixture.
The optimum temperature was determined using a Peltier-temperature controlled UV spectrophotometer Cary 50 (Varian-Agilent Technologies, Santa Clara, CA, USA) varying the assay temperature from 20 to 65 °C. To determine the pH optimum of LvTIM, enzyme activity was measured in 50 mM reaction buffer, changing the pH from 4 to 10 units (pH 4.0–5.0, Acetate buffer; pH 6.0–7.0, Tris-HCl buffer; 7.0–10, Trizma base buffer) using the standard assay method described. For the determination of Km and Vmax the concentration range of glyceraldehyde 3-phosphate was from 0.05 to 2.4 mM.
2.4 Thermal denaturation profiles monitored by circular dichroism (CD)
The temperature-induced denaturation of LvTIM was assessed by monitoring the ellipticity changes at a wavelength of 220 nm, while the temperature of the sample was increased at a constant rate, usually 2 °C min−1, or as stated in the text. The temperature scans were between 25 and 80 °C. LvTIM was diluted in 20 mM Tris-HCl at pH 7.4. The experiments were performed at several protein concentrations (10, 15, 20, 50, and 100 μg mL−1). The assays were performed using a JASCO J-815 spectropolarimeter (Jasco Inc., Easton, MD) equipped with a PTC-348WI Peltier-type cell holder for temperature control and magnetic stirring using a cell with a path length of 1 cm. Actual temperatures within the cell were registered with the external cell holder probe. Cooling profiles were recorded after denaturation transitions had been completed. Both heating and cooling profiles were controlled through the Peltier accessory.
2.5 Denaturation Kinetics
The time-course denaturation of LvTIM was followed by monitoring changes in ellipticity at 220 nm. The 1.0 cm path length cell was filled up to 98% of its total volume (3.0 mL) with Tris buffer to equilibrate it at the temperature of the experiment, which was measured with the external probe of the Peltier accessory. Afterward, the necessary volume of concentrated LvTIM solution to complete the cell volume. Samples were vigorously stirred to promote rapid mixing and temperature equilibration. Under these conditions, the dead time of experiments was less than five seconds. Kinetic data were adjusted to a single decay equation (θt = θf + A exp [−kt]) where, θt is the signal measured at time t, θf is the final signal value, A, represents the amplitude of each phase, and, k, is the unfolding rate constant for the reaction. The effect of temperature on the rate constants of the reaction, observed by changes in the secondary structure for the denaturation of LvTIM was analyzed by plotting ln (k/T) vs. 1/T, corresponding to the well-known Eyring’s equation:
| (1) |
where, k is the rate constant of an elementary reaction; kB and ħ are the Boltzman’s and Planck’s constants, respectively; ΔH# and ΔS# are the activation enthalpy and entropy, correspondingly.
2.6. Crystal Structure
Purified LvTIM was dialyzed in Tris-HCl pH 7.5, 200 mM NaCl and concentrated to 8 mg mL−1. Initial crystallization experiments were performed by the hanging drop method using an incomplete factorial screen of 192 conditions at 23 °C. As the initial crystallization experiments were unsuccessful, a second round of crystallization experiments was performed with the addition of 2-phosphoglyceric acid (2-PG) at a final concentration of 15 mM to the LvTIM protein. Needle-like crystals appeared in a reservoir solution containing 100 mM Tris-HCl pH 8.5, 30% w/v polyethylene glycol 4,000 and 200 mM lithium sulfate monohydrate. Protein crystals grew for one-week reaching dimensions of 400 × 50 × 50 μm. Protein crystals were transferred to a cryoprotectant solution containing 80% of the reservoir and 20% glycerol and were flash-frozen in liquid nitrogen.
Diffraction and data collection was carried on beamline X6A of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL), USA, using a CCD-ADSC Quantum 270 detector. A total of 360 images were collected at 0.979 Å. The data set was integrated and scaled using XDS and XSCALE respectively [30]. Phases were solved by molecular replacement method (MR) using PHASER [31] and the ligand modeled with the coordinates from RCSB ligand entry 2PG (http://www.rcsb.org/pdb/ligand/ligandsummary.do?hetId=2PG). The model used for MR was the three-dimensional structure of Tenebrio molitor TIM, with PDB accession code 2I9E [32]. Refinement was carried out with the program PHENIX [33] and rebuilding with COOT [34]. Figures were made with CCP4mg [35] and PyMOL [36].
3. Results and discussion
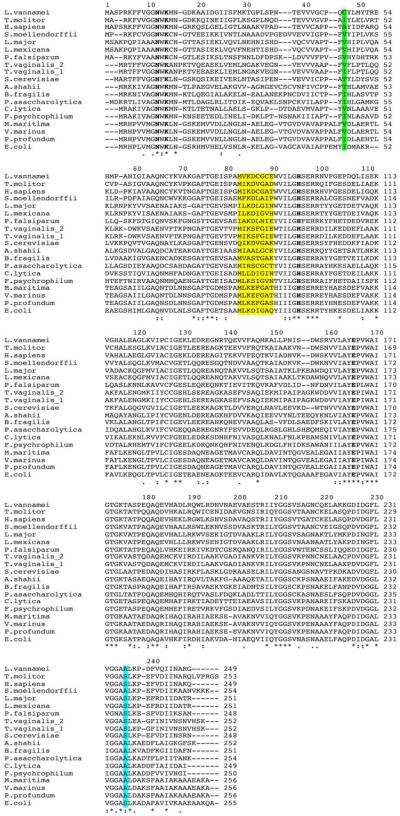
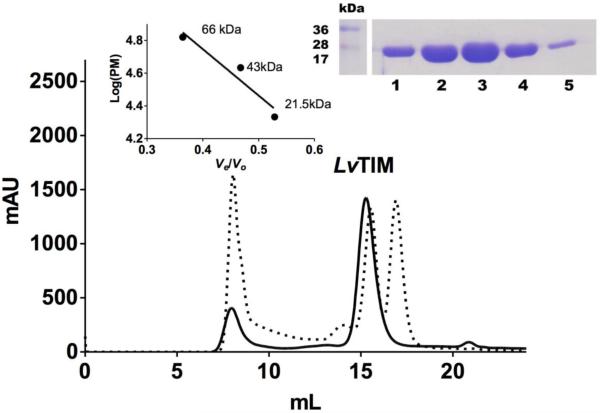
The alignment of the LvTIM amino acid sequence with TIMs from other species (Fig. 1) indicate that LvTIM contains the consensus active site signature [AYEPVWAIGTG] (PROSITE access number: PS00171), where Glu 166 is the catalytic residue that participates as a proton acceptor. The sequence alignment also illustrates the presence of other well-conserved catalytic amino acid residues that are involved in catalysis as His 96, and the substrate-binding residues Asn 12 and Lys 14 (Fig. 1). To further characterize LvTIM, we overexpressed and purified the recombinant protein in E. coli using IMAC and gel filtration. Heterologously expressed LvTIM reach a purity above 95% after IMAC and gel filtration (Fig. 2 inset). The oligomeric state of LvTIM was estimated to be a dimer by gel filtration on an S200 Gel filtration column (GE Healthcare). The dimeric nature of LvTIM was assessed by measuring its retention time in comparison to molecular mass standards; the data indicates that LvTIM was retained as a protein with a Stokes radius corresponding to a weight of 54 kDa, indicating that this protein is a dimer in solution (Fig. 2).
Figure 1.
Multiple sequence alignment of LvTIM and other organism. Both mesophilic and psychrophilic TIM sequences were included. Mesophylic: L. major, L. mexicana, P. falciparum, T. molitor, H. sapiens, S. cerevisiae, T. vaginalis, E. coli, Selaginella moellendorffii and Porphyromonas asaccharolytica and psychrophilic: V. marinus, Flavobacterium psychrophilum, Photobacterium profundum, Alistipes shahii, Bacteroides fragilis, P. asaccharolytica, Cellulophaga lytica, Moritella marina. The ball and socket is show as green shadow (ball) and yellow shadow (socket). Also amino acid residue in loop 8 associated to psychrophilic TIMs is show as light blue shadow. Active site residues are bold.
Figure 2.
Size-exclusion chromatography of LvTIM. Solid line corresponds to the LvTIM protein sample with Blue Dextran and the dotted line corresponds to bovine serum albumin (66 kDa), ovalbumin (45 kDa) and soybean trypsin inhibitor (21 kDa) with blue dextran. The eluted protein fractions are observed in the inset.
3.1 Enzyme kinetics
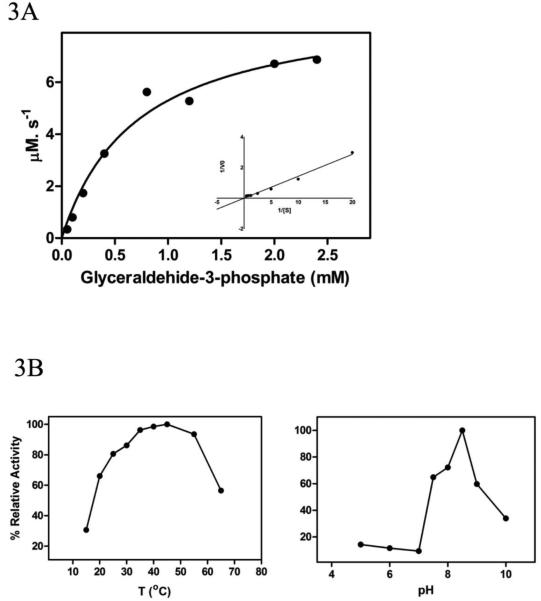
The recombinant LvTIM followed Michaelis–Menten kinetics for the substrate glyceraldehyde-3-phosphate (Fig. 3A). The Km of LvTIM was 0.7 mM, higher than the Km reported for human TIM (0.49 mM) [5] and TIMs from protozoan parasites such as L. donovani (0.328 mM) [18], L. mexicana (0.30 mM) [37], G. lamblia (0.53 mM) [38], T. brucei (0.25 mM) [39], but lower than the Km from TIMs from Vibrio marinus (1.9 mM) [9] or S. cerevisiae (1.27 mM) [40]. The LvTIM kcat was 1.2 × 105 min−1, similar to the kcat reported for other TIMs such L. mexicana (2.5 ×105 min−1) [37], Helicobacter pylori (8.8 ×104 min−1) [41], T. brucei (3.7 ×105 min−1) [39] and rabbit muscle TIM (5.1 ×105 min−1) [42]. The calculated catalytic efficiency (kcat/Km) was 1.71 × 105 min−1 mM−1, similar to the observed for most TIMs and higher than the observed in T. brucei (1.4 ×106 min−1 mM−1) [39].
Figure 3.
Characterization of LvTIM. Panel (A) Michaelis-Menten kinetics with a Lineweaver-Burk plot at the inset. Panel (B) Optimal temperature and pH.
The LvTIM optimal temperature was 45 °C and the optimal pH was 8.5, with good activity in the range from 7 to 9 pH units (Fig. 3B). The optimal pH is in agreement with reports by Kumar et al. where the L. donovani is more stable at pH 8.8, ranging from 6.5-8.5 [18]. Although enzyme optimal temperature may or not have a correlation with the environment of the organisms, as shrimp does not regulate its body temperature, the structural stability to unfolding will provide more insights into the structure and function relationship.
3.2. Thermal unfolding transitions.
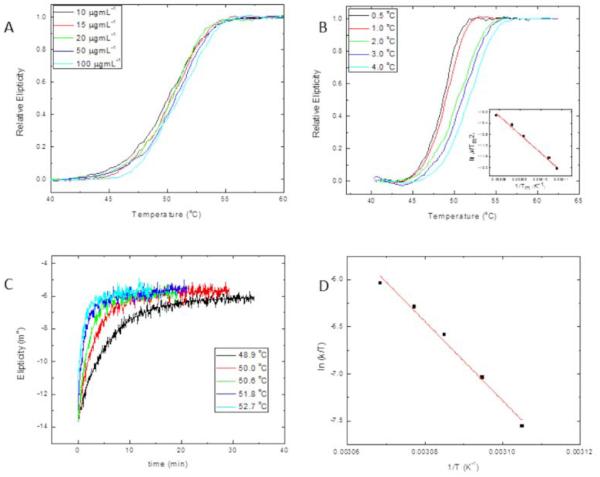
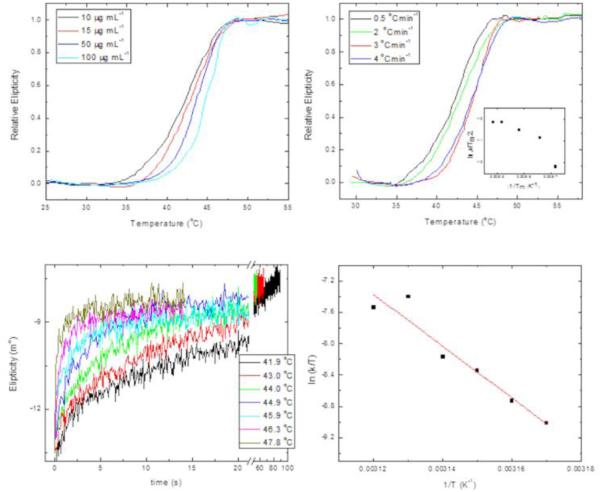
Figure 4A shows the thermal-denaturation profiles of LvTIM, at protein concentrations ranging from 10 to 100 μgmL−1, obtained by continuous monitoring de CD signal at 200 nm, at a constant heating rate. We observe that transitions appear as single sigmoid curves, without showing stable intermediates, over the range of protein concentrations studied. Thermal denaturation profiles shown in figure 4A are very close to each other, but still, a slight shift towards higher temperatures when protein concentration is increased could be observed, as it is expected for dimeric proteins, due to the coupled denaturation and dissociation reactions that occur during unfolding. The denaturation profile corresponding to the lowest protein concentration (10 μg mL−1) shows a midpoint of 50.1 °C, while the pattern obtained at 100 μg mL-1, shows a melting temperature (Tm) of 51 °C. Melting temperature varies with experimental conditions, like heating rate, protein concentration, among others, even though it still can be used for comparison between orthologs, particularly when differences are quite significant. The range of melting temperatures, Tm, observed for LvTIM are substantially lower than the measured for TIMs from other organisms. For comparison we will refer to the TIMs from TvTIM1 and TvTIM2 [16] that were measured under the same experimental conditions as LvTIM (Table 1 and Fig. 4A). Data indicates that crustacean TIM shows lower melting temperature compared to TvTIMs. LvTIM is also unfolded at lower temperatures than TIMs from Homo sapiens, Fasciola hepatica, S. cerevisiae and Leishmania mexicana that present melting temperatures of 66.2 °C, 67 °C, 63 °C and 57 °C respectively [43-47]. In contrast, the melting temperature for T. brucei TIM is 53.1 °C, very similar to the estimated melting temperature for LvTIM [48].
Figure 4.
Temperature-induced denaturation of LvTIM. Panel (A) Thermal denaturation transitions of LvTIM obtained at different protein concentrations as indicated on the legend. All transitions on this panel were obtained at a heating rate of 2 °Cmin−1. Panel (B) Thermal denaturation profiles of LvTIM obtained at different heating rates. All profiles of this panel were measured using a protein concentration of 15 μg mL−1. The inset shows the plot ln versus 1/Tm made with data from transitions of this panel. Panel (C) unfolding kinetic traces of LvTIM obtained at different temperatures. Smooth lines correspond to fitting of experimental data to single exponential decay equation. Protein concentration was 15 μg mL−1. Panel (D) Eyring’s plot for the rate constants of unfolding calculated from kinetic traces shown on panel C. In panels A and B, the data have been normalized for ease of comparison. All the experiments on this figure were performed using Tris 20 mM buffer at pH 7.4.
Table 1.
Comparison of melting temperatures (°C) for LvTIM, LvTIM-Cys47Ala, TvTIM1 and TvTIM2.
| Protein source | μg mL−1 | |||
|---|---|---|---|---|
|
| ||||
| 10 | 15 | 30 | 50 | |
| LvTIMa | 50.1 | 50.3 | 50.5 | 50.7 |
| LvTIM-Cys47Alaa | 42.1 | 42.7 | 43.1 | 43.5 |
| TvTIM1b | 61.5 | 63.2 | 63.8 | 67.1 |
| TvTIM2b | 62.9 | 63.2 | 63.2 | 63.1 |
Denaturation profiles were obtained at a heating rate of 2 °Cmin−1.
Data from Lara-Gonzalez et al. 2014
After thermal denaturation transitions had gone to completion, protein solutions were cooled down to 25 °C either at a scan speed of 4 °C min−1 or by quickly reducing the temperature of the sample. We observed that thermal denaturation reaction of LvTIM was entirely irreversible as judged from the CD signal (data not-shown). Also, the thermal denaturation profiles of LvTIM were obtained at different heating rates. Figure 4B shows the denaturation profiles obtained at 0.5, 1.0, 2.0, 3.0 and 4.0 °C min−1. It can be seen that the unfolding transition is strongly dependent on the heating rate, as expected for an irreversible process. [[46, 49-52]. Given the irreversibility of the thermal denaturation transitions of LvTIM, and since they do not show any noticeable intermediates, the profiles were analyzed regarding a two-state irreversible process represented as:
| (2) |
where N2 is the native dimeric state, D the irreversibly unfolded monomeric state, and k a first-order kinetic constant. The denaturation curves are assumed to start at temperatures low enough to make the unfolding reaction rate negligible, and hence the concentration of the native state is equal to the total protein concentration. The dependence of the melting temperature (Tm) on the heating rate (v) can be analyzed by the relationship
| (3) |
where A is the pre-exponential factor in the Arrhenius equation, and Ea is the activation energy of the process [51]. The data profiles from figure 4B were used to construct the plot of ln versus l/Tm shown in the inset of the same figure. The linearity of this plot confirms the absence of intermediates in the thermal denaturation pathway of LvTIM.
3.3 Denaturation kinetics
Kinetic unfolding studies were performed on LvTIM, by registering changes on far UV-CD at 220 nm after temperature jumps at different temperatures. Figure 4C shows some of the kinetic curves obtained at 48.9, 50.0, 50.6, 51.8 and 52.7 °C. In all cases, single exponential decay curves fitted well to the experimental data. This confirms the lack of intermediates under these experimental conditions. The rate constants associated with the unfolding reaction were calculated from the fitting of the data.
The effect of temperature on the rate constants of the reaction, observed by changes in the secondary for the thermal denaturation of LvTIM (k,) is plotted in figure 4D. The ln (k/T) vs. 1/T plot corresponds to Eyring’s equation:
| (4) |
where, k is the rate constant of the reaction; kB and ħ are the Boltzman’s and Planck’s constants, respectively; ΔH# and ΔS# are the changes on the activation enthalpy and entropy, correspondingly. The data describe linear ln(k/T) vs. 1/T plot. Linearity of Eyring’s plot implies that the dependence of ΔH# on temperature is mostly negligible, indicating a change in the heat capacity between the native and transition state species (ΔCp#) is close to cero, and in consequence that there are small changes in solvent accessibility to reach the transition state [27, 29]. The unfolding ΔH# value obtained for LvTIM is 350 ±20 kJ mol−1. This is much smaller than the enthalpic barrier values of 450-480 kJ mol−1 or 430 ± 25 kJ mol−1calculated for ScTIM or TcTIM, respectively, under similar experimental conditions [50]. The destabilization of the native state of a protein could be attributed to either a high equilibrium constant, or low rates of renaturation, or low enthalpic barriers of denaturation. Since, thermal denaturation of LvTIM is under kinetic control, and reversibility of the reaction could not be observed under our experimental conditions, neither the equilibrium constant, nor the renaturation rate constants could be evaluated: Nevertheless, the low value of ΔH# could be the energetic reason for the low stability of LvTIM compared to other orthologs.
3.4 Crystal structure
The crystal of LvTIM in complex with 2-PG diffracted at a resolution of 1.7 Å. Coordinates and structural data were deposited with the Protein Databank with the accession code 5EYW. The high-resolution structure helped to have a good statistic in the integration and scaling of the data set (Table 2).
Table 2.
Data reduction and refinement statistics of LvTIM structure.
| DATA SET | LvTIM |
|---|---|
| Space group | P1 |
| Unit-cell parameters (Å) |
a = 38.14, b= 45.98, c =71.85 a= 74.567 b=80.240 g=75.179 |
| Data collection | |
| Resolution range (Å) | 27.1–1.7 (1.8–1.7) |
| Unique reflections | 94294 (6728) |
| Rmeas§ | 0.081 (0.335) |
| Completeness (%) | 94.7 (91.2) |
| I/s(I) | 15.67 (4.35) |
| Redundan cy |
2.71 (2.69) |
| Refinement statistics | |
| Rwork/Rfree(
5%) |
0.1920/0.2366 |
| R.M.S.D. form ideal | |
| Bond length (Å) | 0.007 |
| Bond angles (°) | 1.09 |
| Ramachandran plot, residues in Most favored regions |
728(97.5%) |
| Outliers | 0 (0%) |
| PDB ID | 5EYW |
Values in parenthesis represent the statistics at the highest resolution bin.
Rmeas is a redundancy-independent version of Rsymm, , where .
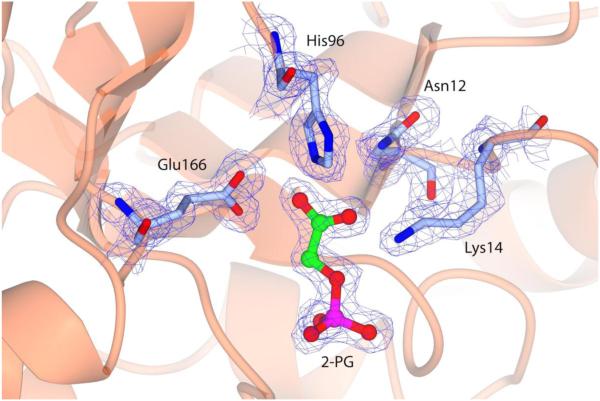
A well-defined electronic density allowed fitting each atom in a proper position and confirming the residue sequence. After several cycles of refinement and manual building, the Rfree and Rwork dropped to 0.23 and 0.19 respectively. As the protein crystallized in a P1 space group, the asymmetric unit contains a dimer. No pseudosymmetric restrictions between monomers were made during crystallographic refinement. The active site was well defined and clear electron density, 2Fo-Fc, was found for the substrate analog 2-PG and catalytic residues Asn 12, Lys 14, His 96 and Glu 166 (Fig. 5)
Figure 5.
LvTIM active site in complex with 2-PG.
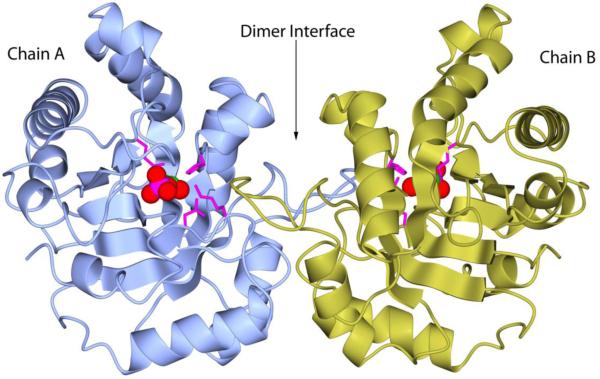
The crystal structure of LvTIM assembles as a homodimer (Fig. 6). The catalytic site is in a closed conformation with a molecule of 2-PG bound to each monomer, and loop 6 (a.a. 166-176) covering the substrate as a lid. In this closed conformation, residues 168-170 (Val, Trp and Ala in LvTIM) form a semi-helix as observed in other substrate-complex TIM structures. At the end of loop 6 a threonine 176 (Thr 176) is found in most of the TIM sequences. However, some TIM present a serine residue in this position. Examples of this substitution are TvTIM 1 and 2, the bacteria Allistipes shahii, Bacteroides fragilis, Flavobacterium psychrophilum and LvTIM. A close analysis of the LvTIM crystal structure indicates that Ser 178 (in LvTIM amino acid sequence) has the same orientation of the −OH group and as a consequence forming the same polar contacts as Thr 176. This suggests that this change, in one of the most conserved residues, does not alter the catalytic function of the enzyme as noted in catalytic parameters of LvTIM.
Figure 6.
Quaternary structure of LvTIM. Dimer interface is formed by interconnected loops. Each monomers shows one 2-PG molecule bound to active site. 2-PG is shown as spheres colored by atom type. While, catalytic residue Glu166 and accessory residues Asn12, Lys14 and His96 are depicted as magenta cylinders.
Using the DALI server [53] to look for similar structures, the root mean square deviation (RMSD) for backbone carbons between LvTIM and chicken TIM (1TPH) was 0.5 Å, human (4POD) or rabbit (1R2R) 0.7 Å, C. elegans (1MO0) 0.9 Å, S. cerevisiae (7TIM) 0.9 Å, T. brucei (6TIM) 1.1 Å, P. falciparum (1WOB) 1.5 Å, Coccidioides immitis (3S6D) 2.1 Å, Methanocaldococcus jannaschii (2H6R) 2.3 Å [53]. As described earlier, some marine shrimp enzymes appear to be more similar to their vertebrate homologs compared to invertebrate organisms [54].
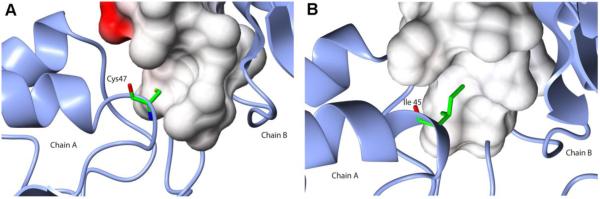
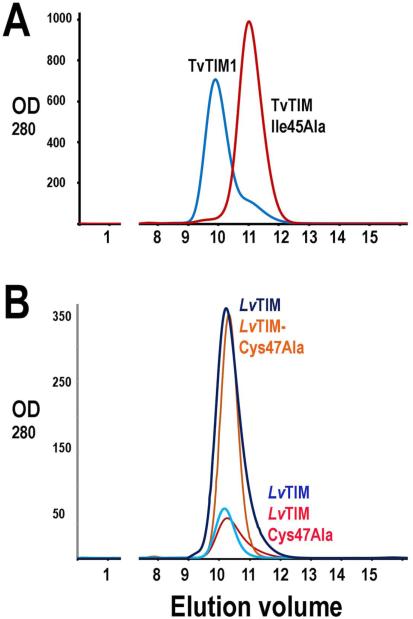
In LvTIM the “ball and socket” interplay contains a cysteine residue (Cys 47) as a “ball” and an unusual cysteine (Met-Val-Lys-Asp-Cys-Gly-Cys-Glu motif) is present at the “socket” (Fig. 7 panel A). The amino acid character of the "ball" is usually an Ala, Val, Ile or Thr and the socket is not conserved, although Phe, Ile or Met are generally present (Fig. 7 panel B) [26]. Considering the properties of cysteine (similar side chain length as an Ala), the prediction of Lara et al [26] is that Cys 47 favors the monomeric state in TIMs The observed structural stability between LvTIM and TvTIMs correlates with the hydrophobicity of the ball: TvTIM1-Ile45 > TvTIM2-Val45 > LvTIM-Cys47.
Figure 7.
Ball and socket dimer interface of LvTIM (panel A) and TvTIM (panel B). Cys47 in the ball motif is presented as cylinder (colored by atom type) at the dimer interface of LvTIM; for comparison also is showing Ile 45 of TvTIM1. In both panels residues in the socket motif as represented as surface colored by electrostatic potential. Net negative charges are in red color and neutral charges as white.
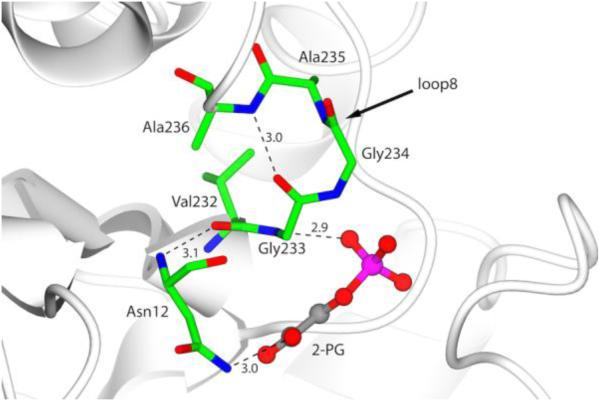
Dimer stability in TIMs has also been related to the presence of a serine residue at positon 236, which is located at loop 8. Few sequences, like LvTIM, contain an amino acid that is different at this position (Fig. 1). LvTIM harbors Ala 236, in the psychrophilic bacterium Vibrio marinus, an alanine residue is also found at this position [9]. The Ala 236 Ser in TIM from substitution V. marinus (VmTIM) increased its thermal stability and catalytic efficiency, and the crystal structure of the mutant showed a network of hydrogen bonds that stabilize the Asn 11 residue in the active site (PDB 1AW1). The crystal structure of LvTIM reveals several features conserved with VmTIM [9]: Ala 236 makes a hydrogen bond with the carbonyl group of Gly 233 and the nitrogen-amide contacts the substrate analog 2PG keeping the hydrogen network found in V. marinus TIM (Fig. 8), however by modeling the mutant Ala 236 Ser the hydroxyl group seems to be capable of making additional hydrogen bonds with the backbone carbonyl group Og Val 232 and 2-PG, as is the case for VmTIM [9].
Figure 8.
Hydrogen network in loop 8 of LvTIM associated to psychrophilic TIM. Residues that comprise the loop 8 and catalytic Asn12 are represented as cylinders colored by atom type. Hydrogen bonds are show as black lines and 2-PG is depicted as ball and stick with carbon atom in grey color.
3.5 Is the character of the ball-socket necessary for dimer stability?
The structural localization of Cys47 at the “ball” in LvTIM lead us to hypothesize that this amino acid is essential for dimer stability. Cysteine harbors an ionizable thiol. However this residue is often found in the hydrophobic core of proteins and has a considerable hydrophobic character [55, 56]. To understand if the “ball and socket” interplay is necessary for dimer formation in LvTIM, we constructed a mutant in which cysteine 47 was replaced by an alanine (LvTIM-Cys47Ala), this mutant eliminated the thiol group and reduced the accessible surface area (ASA) by 37 Å2. In the case of TvTIM2 the substitution of Valine 45 to Alanine decrease the ASA by 50 Å2 and completely alters the equilibrium from dimer to monomer [16, 26]. A gel filtration experiment of purified LvTIM-Cys47Ala with wild-type LvTIM shows that both proteins are dimers at concentrations of 2 and 0.2 mg ml−1. The latter is in contrast with TvTIM1-Ile45Ala mutant that behaves a monomeric protein in comparison to the wild-type TvTIM1 (Fig. 9)[26]. An enzymatic characterization of LvTIM-Cys47Ala indicates that its calculated catalytic efficiency (kcat/Km) was 1.4 × 105 min−1 mM−1, this catalytic efficiency is similar to the one observed for the wild-type LvTIM. As monomeric TIMs exhibit a decrease in catalytic efficiency from 25 to 1000-fold, the wild-type-like enzymatic activity of LvTIM-Cys47Ala implicates that this mutant is a dimer in solution. Thus, gel filtration experiments and biochemical characterization indicate that residue Cys47 is not indispensable for dimer formation. However, when we carried out an analysis of the thermal denaturation of LvTIM-Cys47Ala, we observed that LvTIM-Cys47Ala is even more unstable than wild-type LvTIM. Thermal denaturation transitions of LvTIM-Cys47Ala are single sigmoidal curves which depend on both, protein concentration and heating rate (Fig. 10A and 10B). The range of melting temperatures was reduced to 42-45 °C (Table 1). In this case, the plot of ln versus l/Tm (Inset of Fig. 10B) shows a marked curvature, indicating that the reaction does not proceed through a single reaction step with a single transition state. Kinetic denaturation curves (Fig. 10C) show biphasic behavior and could be fitted to double exponential decay equations; (θt = θf + A1 exp [−k1t] + A2 exp [−k2t]). Where θt is the signal measured at time t, θf is the final signal value, A1 and A2 represent amplitudes of each phase, and k1 and k2 are the unfolding rate constants for each reaction. Thus the unfolding pathway of LvTIM-Cys47Ala involves a kinetic intermediate. Using Eyring’s equation the main enthalpic barrier, associated with the unfolding reaction could be calculated (280 ± 20 kJmol−1). This indicates that apart from the hydrophobic interactions within the ball-socket moiety described for other TIMs [16] there should be other amino acid residues responsible for the conformational stability of LvTIM. The full description of interactions responsible for the low stability of LvTIM remains unclear. Therefore, the thermodynamics and chemical induced denaturation of LvTIM-Cys47Ala and some other mutants should be subject to future investigation
Figure 9.
Size-exclusion chromatography of wt-LvTIM and Cys 47Ala mutant showing the elution of those proteins in comparison to TvTIM and TvTIM-Ile45Ala. A) Elution profile of TvTIM and TvTIM-Ile45A. Dimeric TvTIM elutes at 9.5 ml whereas monomeric TvTIM-Ile45Ala elutes at 11.7 ml. The monomeric and dimeric peaks can be separated in a Superdex 75 10/300 GL column as previously described [26]. B) Elution profile of wt-LvTIM and LvTIM-Cys47Ala mutant at 2 mg ml−1 and 0.2 mg ml−1. wt-LvTIM elutes at 10.2 ml at both protein concentrations, whereas LvTIM-Cys47Ala mutant elutes at 10.4 ml.
Figure 10.
Temperature-induced denaturation of LvTIM-Cys47Ala. Panel (A) Thermal denaturation transitions of LvTIM-Cys47Ala obtained at different protein concentrations as indicated on the legend. All transitions at this panel were obtained at a heating rate of 2 °C min−1. Panel (B) Thermal denaturation profiles of LvTIM-Cys47Ala obtained at different heating rates. All profiles at this panel were measured using a protein concentration of 15 μg mL−1. The inset shows the plot ln versus 1/Tm made with data from transitions of this panel. Panel (C) unfolding kinetic traces of LvTIM-Cys47Ala obtained at different temperatures. Protein concentration was 15 μg mL−1. Panel (D) Eyring’s plot for the rate constants of unfolding calculated from kinetic traces shown on panel C. In panels A and B, the data have been normalized for ease of comparison. All the experiments on this figure were performed using Tris 20 mM buffer at pH 7.4.
4. CONCLUSIONS AND SIGNIFICANCE
Marine organisms rely on glycolysis during stages of hypoxia [57] and TIM is at the core of that metabolic pathway. The biochemical characterization of LvTIM indicates that this enzyme is more similar to TIMs from higher eukaryotes compared to invertebrates, protozoans or parasites. The LvTIM active site residues are conserved as well the interactions with the substrate analog. We calculated a small value of enthalpic barrier of denaturation (350 ± 20 kJmol−1), as well as a low range of melting temperatures compared to other orthologs.
White shrimp is a thermo conformer organism (which mean that is not able to regulate its temperature) that lives in environments with moderate temperature changes (20-30 °C). Therefore, should have metabolic pathways (as glycolysis) with adapted-enzymes that cope with these natural changes. Also, LvTIM shows an uncommon Ala in the position 236, which could explain the lower thermal stability compared with other mesophilic species, but further studies are recommended. Functional studies indicate that TIMs are moonlighting proteins involved in cell-adhesion [58, 59] and that these new functions imply dimer-monomer equilibrium [58]. We propose that the low stability of LvTIM may be part of a regulatory mechanism that regulates glycolysis and that this mechanism may also involve dimer-monomer transitions. Our structural data confirms that LvTIM is an obligated dimer molecule that is structurally more similar to higher organisms than protozoan. Also, this novel described TIM is a cornerstone for further studies to reveal the relation with psychrophilic species.
Highlights.
- Triosephosphate isomerase from Litopenaeus vannamei (LvTIM) is the first structural characterized TIM from of a crustacean.
- LvTIM is the least stable TIM characterized to date.
- The lower stability of LvTIM may play a role in glycolysis regulation
ACKNOWLEDGMENTS
Dr. Lopez-Zavala thanks CONACYT for support by Fondo Consolidación Institucional (RET-I0007-2015-01, #250973). Drs. Lopez-Zavala and Carrasco-Miranda were supported as doctoral fellows from CONACYT. Drs. Sotelo-Mundo and Brieba acknowledge financial support from CONACYT grant CB-2014-237963 and Fronteras de la Ciencia #11 respectively. We thank the staff at BNL-NSLS beam lines X6A and X4C (Drs. Vivian Stojanoff and Dr. John Schwanoff respectively) for data-collection facilities. Beamline X6A was funded by NIGMS (GM-0080) and the US Department of Energy (No. DE-AC02-98CH10886).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author’s contributions
AALZ collected crystallographic data and solved the structure, designed and prepared figures and wrote the paper. JSCM set up crystallization experiments and size exclusion chromatography, solved the crystal structure and wrote the paper. CDRA and CDQ did enzyme kinetic experiments, analyzed data, performed site-directed mutagenesis and enzyme purification, MLH realized circular dichroism stability experiments and analyzed data. CGBC designed, performed and analyzed circular dichroism data of thermal stability, and wrote the paper. AOL analyzed data and wrote the paper. CSCF designed and realized crystallization experiments. ERP collected crystallographic data, solved the structure and wrote the paper, RRSM conceptualized and planned the project and experiments, analyzed data, designed figures and wrote the paper. LGB designed and supervised experiments, analyzed the data, designed figures and wrote the paper.
References
- [1].Tellez LA, Blancas-Mejia LM, Carrillo-Nava E, Mendoza-Hernandez G, Cisneros DA, Fernandez-Velasco DA. Thermal unfolding of triosephosphate isomerase from Entamoeba histolytica: dimer dissociation leads to extensive unfolding. Biochemistry. 2008;47:11665–11673. doi: 10.1021/bi801360k. [DOI] [PubMed] [Google Scholar]
- [2].Mainfroid V, Terpstra P, Beauregard M, Frere JM, Mande SC, Hol WG, Martial JA, Goraj K. Three hTIM mutants that provide new insights on why TIM is a dimer. Journal of molecular biology. 1996;257:441–456. doi: 10.1006/jmbi.1996.0174. [DOI] [PubMed] [Google Scholar]
- [3].Kohlhoff M, Dahm A, Hensel R. Tetrameric triosephosphate isomerase from hyperthermophilic Archaea. FEBS Lett. 1996;383:245–250. doi: 10.1016/0014-5793(96)00249-9. [DOI] [PubMed] [Google Scholar]
- [4].Sharma P, Guptasarma P. 'Super-perfect' enzymes: Structural stabilities and activities of recombinant triose phosphate isomerases from Pyrococcus furiosus and Thermococcus onnurineus produced in Escherichia coli. Biochem Biophys Res Commun. 2015;460:753–758. doi: 10.1016/j.bbrc.2015.03.102. [DOI] [PubMed] [Google Scholar]
- [5].Mande SC, Hol WGJ, Mainfroid V, Goraj K, Martial JA, Kalk KH. Crystal structure of recombinant human triosephosphate isomerase at 2.8 A resolution. Triosephosphate isomerase-related human genetic disorders and comparison with the trypanosomal enzyme. Protein Science. 1994;3:810–821. doi: 10.1002/pro.5560030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wierenga RK, Kapetaniou EG, Venkatesan R. Triosephosphate isomerase: a highly evolved biocatalyst. Cellular and Molecular Life Sciences. 2010;67:3961–3982. doi: 10.1007/s00018-010-0473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Knowles JR. Enzyme catalysis: not different, just better. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- [8].Neria E, Karplus M. Molecular dynamics of an enzyme reaction: proton transfer in TIM. Chemical Physics Letters. 1997;267:23–30. [Google Scholar]
- [9].Alvarez M, Zeelen JP, Mainfroid V, Rentier-Delrue F, Martial JA, Wyns L, Wierenga RK, Maes D. Triose-phosphate Isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties. Journal of Biological Chemistry. 1998;273:2199–2206. doi: 10.1074/jbc.273.4.2199. [DOI] [PubMed] [Google Scholar]
- [10].Banner DW, Bloomer AC, Petsko GA, Phillips DC, Pogson CI, Wilson IA, Corran PH, Furth AJ, Milman JD, Offord RE, Priddle JD, Waley SG. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975;255:609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- [11].Lolis E, Alber T, Davenport RC, Rose D, Hartman FC, Petsko GA. Structure of yeast triosephosphate isomerase at 1.9-A resolution. Biochemistry. 1990;29:6609–6618. doi: 10.1021/bi00480a009. [DOI] [PubMed] [Google Scholar]
- [12].Wierenga RK, Noble MEM, Vriend G, Nauche S, Hol WGJ. Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4-ammonium sulphate: A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex. Journal of molecular biology. 1991;220:995–1015. doi: 10.1016/0022-2836(91)90368-g. [DOI] [PubMed] [Google Scholar]
- [13].Noble MEM, Zeelen JP, Wierenga RK, Mainfroid V, Goraj K, Gohimont AC, Martial JA. Structure of triosephosphate isomerase from Escherichia coli determined at 2.6 A resolution. Acta Crystallographica Section D: Biological Crystallography. 1993;49:403–417. doi: 10.1107/S0907444993002628. [DOI] [PubMed] [Google Scholar]
- [14].Delboni LF, Mande SC, Rentier-Delrue F, Mainfroid V, Turley S, Vellieux FM, Martial JA, Hol WG. Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions. Protein Science. 1995;4:2594–2604. doi: 10.1002/pro.5560041217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Velanker SS, Ray SS, Gokhale RS, Balaram H, Balaram P, Murthy MRN. Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design. Structure. 1997;5:751–761. doi: 10.1016/s0969-2126(97)00230-x. [DOI] [PubMed] [Google Scholar]
- [16].Lara-Gonzalez S, Estrella-Hernandez P, Ochoa-Leyva A, Del Carmen Portillo-Tellez M, Caro-Gomez LA, Figueroa-Angulo EE, Salgado-Lugo H, Miranda Ozuna JF, Ortega-Lopez J, Arroyo R, Brieba LG, Benitez-Cardoza CG. Structural and thermodynamic folding characterization of triosephosphate isomerases from Trichomonas vaginalis reveals the role of destabilizing mutations following gene duplication. Proteins. 2014;82:22–33. doi: 10.1002/prot.24333. [DOI] [PubMed] [Google Scholar]
- [17].Perez-Montfort R, de Gomez-Puyou M, Gomez-Puyou A. The interfaces of oligomeric proteins as targets for drug design against enzymes from parasites. Current Topics in Medicinal Chemistry. 2002;2:457–470. doi: 10.2174/1568026024607454. [DOI] [PubMed] [Google Scholar]
- [18].Kumar K, Bhargava P, Roy U. Cloning, overexpression and characterization of Leishmania donovani triosephosphate isomerase. Experimental Parasitology. 2012;130:430–436. doi: 10.1016/j.exppara.2012.01.016. [DOI] [PubMed] [Google Scholar]
- [19].Tellez-Valencia A, Avila-Rios S, Perez-Montfort R, Rodriguez-Romero A, Tuena de Gomez-Puyou M, Lopez-Calahorra F, Gomez-Puyou A. Highly specific inactivation of triosephosphate isomerase from Trypanosoma cruzi. Biochem Biophys Res Commun. 2002;295:958–963. doi: 10.1016/s0006-291x(02)00796-9. [DOI] [PubMed] [Google Scholar]
- [20].Ostoa-Saloma P, Garza-Ramos G, Ramirez J, Becker I, Berzunza M, Landa A, Gomez-Puyou A, Tuena de Gomez-Puyou M, Perez-Montfort R. Cloning, expression, purification and characterization of triosephosphate isomerase from Trypanosoma cruzi. European Journal of Biochemistry. 1997;244:700–705. doi: 10.1111/j.1432-1033.1997.00700.x. [DOI] [PubMed] [Google Scholar]
- [21].Enriquez-Flores S, Rodriguez-Romero A, Hernandez-Alcantara G, De la Mora-De la Mora I, Gutierrez-Castrellon P, Carvajal K, Lopez-Velazquez G, Reyes-Vivas H. Species-specific inhibition of Giardia lamblia triosephosphate isomerase by localized perturbation of the homodimer. Molecular and Biochemical Parasitology. 2008;157:179–186. doi: 10.1016/j.molbiopara.2007.10.013. [DOI] [PubMed] [Google Scholar]
- [22].Gonzalez-Mondragon E, Zubillaga RA, Saavedra E, Chanez-Cardenas ME, Peez-Montfort R, Hernandez-Arana A. Conserved cysteine 126 in triosephosphate isomerase is required not for enzymatic activity but for proper folding and stability. Biochemistry. 2004;43:3255–3263. doi: 10.1021/bi036077s. [DOI] [PubMed] [Google Scholar]
- [23].Chanez-Cardenas ME, Perez-Hernandez G, Sanchez-Rebollar BG, Costas M, Vazquez-Contreras E. Reversible equilibrium unfolding of triosephosphate isomerase from Trypanosoma cruzi in guanidinium hydrochloride involves stable dimeric and monomeric intermediates. Biochemistry. 2005;44:10883–10892. doi: 10.1021/bi047687a. [DOI] [PubMed] [Google Scholar]
- [24].Najera H, Costas M, Fernandez-Velasco DA. Thermodynamic characterization of yeast triosephosphate isomerase refolding: insights into the interplay between function and stability as reasons for the oligomeric nature of the enzyme. Biochem J. 2003;370:785–792. doi: 10.1042/BJ20021439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Figueroa-Angulo EE, Estrella-Hernandez P, Salgado-Lugo H, Ochoa-Leyva A, Gomez Puyou A, Campos SS, Montero-Moran G, Ortega-Lopez J, Saab-Rincon G, Arroyo R, Benitez-Cardoza CG, Brieba LG. Cellular and biochemical characterization of two closely related triosephosphate isomerases from Trichomonas vaginalis. Parasitology. 2012;139:1729–1738. doi: 10.1017/S003118201200114X. [DOI] [PubMed] [Google Scholar]
- [26].Lara-Gonzalez S, Estrella P, Portillo C, Cruces ME, Jimenez-Sandoval P, Fattori J, Migliorini-Figueira AC, Lopez-Hidalgo M, Diaz-Quezada C, Lopez-Castillo M, Trasvina-Arenas CH, Sanchez-Sandoval E, Gomez-Puyou A, Ortega-Lopez J, Arroyo R, Benitez-Cardoza CG, Brieba LG. Substrate-Induced Dimerization of Engineered Monomeric Variants of Triosephosphate Isomerase from Trichomonas vaginalis. PloS one. 2015;10:e0141747. doi: 10.1371/journal.pone.0141747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ghaffari N, Sanchez-Flores A, Doan R, Garcia-Orozco KD, Chen PL, Ochoa-Leyva A, Lopez-Zavala AA, Carrasco JS, Hong C, Brieba LG, Rudino-Pinera E, Blood PD, Sawyer JE, Johnson CD, Dindot SV, Sotelo-Mundo RR, Criscitiello MF. Novel transcriptome assembly and improved annotation of the whiteleg shrimp (Litopenaeus vannamei), a dominant crustacean in global seafood mariculture. Sci Rep. 2014;4:7081. doi: 10.1038/srep07081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Science. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rozacky EE, Sawyer TH, Barton RA, Gracy RW. Studies on human triosephosphate isomerase. I. Isolation and properties of the enzyme from erythrocytes. Arch Biochem Biophys. 1971;146:312–320. doi: 10.1016/s0003-9861(71)80069-3. [DOI] [PubMed] [Google Scholar]
- [30].Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- [32].Knobeloch D, Schmidt A, Scheerer P, Krauss N, Wessner H, Scholz C, Kuttner G, von Rintelen T, Wessel A, Hohne W. A coleopteran triosephosphate isomerase: X-ray structure and phylogenetic impact of insect sequences. Insect Molecular Biology. 2010;19:35–48. doi: 10.1111/j.1365-2583.2009.00928.x. [DOI] [PubMed] [Google Scholar]
- [33].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].DeLano WL. The PyMOL Molecular Graphics System. DLano Scientific.; San Carlos, CA, USA: 2002. [Google Scholar]
- [37].Kohl L, Callens M, Wierenga RK, Opperdoes FR, Michels PA. Triose-phosphate isomerase of Leishmania mexicana mexicana. Cloning and characterization of the gene, overexpression in Escherichia coli and analysis of the protein. European Journal of Biochemistry. 1994;220:331–338. doi: 10.1111/j.1432-1033.1994.tb18629.x. [DOI] [PubMed] [Google Scholar]
- [38].Reyes-Vivas H, Diaz A, Peon J, Mendoza-Hernandez G, Hernandez-Alcantara G, De la Mora-De la Mora I, Enriquez-Flores S, Dominguez-Ramirez L, Lopez-Velazquez G. Disulfide bridges in the mesophilic triosephosphate isomerase from Giardia lamblia are related to oligomerization and activity. Journal of molecular biology. 2007;365:752–763. doi: 10.1016/j.jmb.2006.10.053. [DOI] [PubMed] [Google Scholar]
- [39].Lambeir AM, Opperdoes FR, Wierenga RK. Kinetic properties of triose-phosphate isomerase from Trypanosoma brucei brucei. A comparison with the rabbit muscle and yeast enzymes. European journal of biochemistry / FEBS. 1987;168:69–74. doi: 10.1111/j.1432-1033.1987.tb13388.x. [DOI] [PubMed] [Google Scholar]
- [40].Krietsch WKG, Pentchev PG, Klingenburg H, Hofstatter T, Bucher T. The isolation and crystallization of yeast and rabbit liver triose phosphate isomerase and a comparative characterization with the rabbit muscle enzyme. European Journal of Biochemistry. 1970;14:289–300. doi: 10.1111/j.1432-1033.1970.tb00289.x. [DOI] [PubMed] [Google Scholar]
- [41].Chu CH, Lai YJ, Huang H, Sun YJ. Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori. Proteins. 2008;71:396–406. doi: 10.1002/prot.21709. [DOI] [PubMed] [Google Scholar]
- [42].Krietsch WK. Triosephosphate isomerase from rabbit liver. Methods in enzymology. 1975;41:438–442. doi: 10.1016/s0076-6879(75)41095-3. [DOI] [PubMed] [Google Scholar]
- [43].Olah J, Orosz F, Keseru GM, Kovari Z, Kovacs J, Hollan S, Ovadi J. Triosephosphate isomerase deficiency: a neurodegenerative misfolding disease. Biochemical Society transactions. 2002;30:30–38. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- [44].Zinsser VL, Hoey EM, Trudgett A, Timson DJ. Biochemical characterisation of triose phosphate isomerase from the liver fluke Fasciola hepatica. Biochimie. 2013;95:2182–2189. doi: 10.1016/j.biochi.2013.08.014. [DOI] [PubMed] [Google Scholar]
- [45].Benitez-Cardoza CG, Rojo-Dominguez A, Hernandez-Arana A. Temperature-induced denaturation and renaturation of triosephosphate isomerase from Saccharomyces cerevisiae: evidence of dimerization coupled to refolding of the thermally unfolded protein. Biochemistry. 2001;40:9049–9058. doi: 10.1021/bi010528w. [DOI] [PubMed] [Google Scholar]
- [46].Labastida-Polito A, Garza-Ramos G, Camarillo-Cadena M, Zubillaga RA, Hernandez-Arana A. Complex kinetics and residual structure in the thermal unfolding of yeast triosephosphate isomerase. BMC biochemistry. 2015;16:20. doi: 10.1186/s12858-015-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Williams JC, Zeelen JP, Neubauer G, Vriend G, Backmann J, Michels PAM, Lambeir AM, Wierenga RK. Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power. Protein engineering. 1999;12:243–250. doi: 10.1093/protein/12.3.243. [DOI] [PubMed] [Google Scholar]
- [48].Costas M, Rodriguez-Larrea D, De Maria L, Borchert TV, Gomez-Puyou A, Sanchez-Ruiz JM. Between-species variation in the kinetic stability of TIM proteins linked to solvation-barrier free energies. Journal of molecular biology. 2009;385:924–937. doi: 10.1016/j.jmb.2008.10.056. [DOI] [PubMed] [Google Scholar]
- [49].Freire E, van Osdol WW, Mayorga OL, Sanchez-Ruiz JM. Calorimetrically determined dynamics of complex unfolding transitions in proteins. Annu Rev Biophys Biophys Chem. 1990;19:159–188. doi: 10.1146/annurev.bb.19.060190.001111. [DOI] [PubMed] [Google Scholar]
- [50].Mixcoha-Hernandez E, Moreno-Vargas LM, Rojo-Dominguez A, Benitez-Cardoza CG. Thermal-unfolding reaction of triosephosphate isomerase from Trypanosoma cruzi. Protein J. 2007;26:491–498. doi: 10.1007/s10930-007-9090-3. [DOI] [PubMed] [Google Scholar]
- [51].Sanchez-Ruiz JM. Theoretical analysis of Lumry-Eyring models in differential scanning calorimetry. Biophys J. 1992;61:921–935. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tello-Solis SR, Hernandez-Arana A. Effect of irreversibility on the thermodynamic characterization of the thermal denaturation of Aspergillus saitoi acid proteinase. Biochem J. 1995;311:969–974. doi: 10.1042/bj3110969. Pt 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Arvizu-Flores AA, Aispuro-Hernandez E, Garcia-Orozco KD, Varela-Romero A, Valenzuela-Soto E, Velazquez-Contreras EF, Rojo-Dominguez A, Yepiz-Plascencia G, Maley F, Sotelo-Mundo RR. Functional identity of the active sites of crustacean and viral thymidylate synthases. Comp Biochem Physiol C Toxicol Pharmacol. 2009;150:406–413. doi: 10.1016/j.cbpc.2009.06.008. [DOI] [PubMed] [Google Scholar]
- [55].Nagano N, Ota M, Nishikawa K. Strong hydrophobic nature of cysteine residues in proteins. FEBS Lett. 1999;458:69–71. doi: 10.1016/s0014-5793(99)01122-9. [DOI] [PubMed] [Google Scholar]
- [56].Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- [57].Wu RSS. Hypoxia: from molecular responses to ecosystem responses. Marine Pollution Bulletin. 2002;45:35–45. doi: 10.1016/s0025-326x(02)00061-9. [DOI] [PubMed] [Google Scholar]
- [58].Miranda-Ozuna JF, Hernandez-Garcia MS, Brieba LG, Benitez-Cardoza CG, Ortega-Lopez J, Gonzalez-Robles A, Arroyo R. The Glycolytic Enzyme Triosephosphate Isomerase of Trichomonas vaginalis (TvTIM) is a Surface-Associated Protein Induced by Glucose that Functions as a Laminin-and Fibronectin-Binding Protein. Infect Immun. 2016 doi: 10.1128/IAI.00538-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Furuya H, Ikeda R. Interaction of triosephosphate isomerase from Staphylococcus aureus with plasminogen. Microbiol Immunol. 2011;55:855–862. doi: 10.1111/j.1348-0421.2011.00392.x. [DOI] [PubMed] [Google Scholar]