Abstract
In vitro polymerized type I collagen hydrogels have been used extensively as a model system for three-dimensional (3D) cell and tissue culture, studies of fibrillogenesis, and investigation of multiscale force transmission within connective tissues. The nanoscale organization of collagen fibrils plays an essential role in the mechanics of these gels and emergent cellular behavior in culture, yet quantifying 3D structure with nanoscale resolution to fully characterize fibril organization remains a significant technical challenge. In this study, we demonstrate that a new imaging modality, focused ion beam scanning electron microscopy (FIB-SEM), can be used to generate 3D image datasets for visualizing and quantifying complex nanoscale organization and morphometry in collagen gels. We polymerized gels at a number of concentrations and conditions commonly used for in vitro models, stained and embedded the samples, and performed FIB-SEM imaging. The resulting image data had a voxel size of 25 nm, which is the highest resolution 3D data of a collagen fibril network ever obtained for collagen gels. This resolution was essential for discerning individual fibrils, fibril paths, and their branching and grouping. The resulting volumetric images revealed that polymerization conditions have a significant impact on the complex fibril morphology of the gels. We segmented the fibril network and demonstrated that individual collagen fibrils can be tracked in 3D space, providing quantitative analysis of network descriptors such as fibril diameter distribution, length, branch points, and fibril aggregations. FIB-SEM 3D reconstructions showed considerably less lateral grouping and overlap of fibrils than standard 2D SEM images, likely due to artifacts in SEM introduced by dehydration. This study demonstrates the utility of FIB-SEM for 3D imaging of collagen gels and quantitative analysis of 3D fibril networks. We anticipate that the method will see application in future studies of structure-function relationships in collagen gels as well as native collagenous tissues.
Introduction
Fibril-forming collagens (e.g., type I, II, III) are ubiquitous proteins that bear loads and maintain shape within the extracellular matrix (ECM) of vertebrate organisms (1, 2, 3). Type I collagen is essential for bone formation and it is the primary organic constituent of bone (2, 3). Type I collagen is also the predominant load-bearing protein within connective tissues such as ligament, tendon, and the dermis (4, 5, 6). Type II collagen is the primary constituent of articular cartilage, while type III collagen forms granular tissue during wound healing and colocalizes with other collagens (7, 8, 9, 10). Fibrillar collagens undergo an entropy-driven process known as fibrillogenesis, in which single tropocollagen monomers self-assemble into nanoscale collagen fibrils with a high aspect ratio. In vivo, fibrillogenesis is a cell-mediated process whereby procollagen monomers are secreted (e.g., via fibroblasts) by vesicles and modified by membrane bound enzymes into tropocollagen monomers, which then assemble into fibrils within the ECM (11, 12). Fibrillogenesis can also be induced in vitro, whereby acid-solubilized collagen is neutralized in the presence of phosphate buffered saline (PBS), which causes tropocollagen monomers to precipitate from solution and self-assemble into a fibrillar network. Once a critical concentration (known as the gelation point) of collagen is reached, fibrils begin to interconnect into a physically cross-linked network to form a collagen hydrogel (13).
In vitro polymerization of type I collagen has played a crucial role as both a cell culture scaffold and as a model system to study the process of fibrillogenesis (14, 15, 16). The fibril network of a collagen gel is tightly linked to the polymerization conditions, including collagen source, collagen concentration, pH, temperature, and salt content, presence of other proteins and molecules, and other physical and chemical factors (15, 16, 17). Altering these variables changes the rate of polymerization and the resulting diameter distributions and organization of fibrils within the collagen network. Such results are highly relevant, as numerous experimental and computational studies have demonstrated that the microstructural organization of collagen fibrils plays a central role in the resulting macroscopic properties of gels in vitro and tissues in vivo (15, 18, 19, 20, 21), as well as the emergent cellular behavior observed in culture.
For in vitro polymerized collagen gels, a number of methods are utilized to study the structure-function relationships between collagen fibril microstructure and resulting macroscopic properties. Macroscopic properties are assessed using turbidity assays to study the kinetics of fibrillogenesis, mechanical testing to study the elastic, plastic, and viscoelastic properties of gels and a number of other assays to examine the permeability and diffusion coefficients of collagen gels (15, 16, 22, 23, 24). The microscopic organization of collagen gels has been studied using a number of different microscopy techniques, including optical methods such as confocal reflectance microscopy, confocal fluorescence microscopy, second harmonic generation, and electron microscopy methods such as scanning electron microscopy (SEM) and transmission electron microscopy (TEM) (Fig. 1) (15, 25, 26, 27, 28, 29). The major advantage of the aforementioned optical methods is that they are inherently three dimensional (3D). However, the diffraction limit of light, which is ∼λ/2 (where λ is the imaging wavelength), has proven to be a significant limitation. Individual collagen fibrils within in vitro polymerized gels typically have diameters of <100 nm, which is below the diffraction limit for the wavelengths used in optical microscopy (15). Therefore, optical methods cannot resolve structural organization at the fibril level. Electron microscopy methods overcome this limitation and can provide nanometer resolution for observing fibril diameters and organizations. However, both SEM and TEM are inherently two dimensional (2D), so information regarding the 3D organization of collagen fibrils is not obtained.
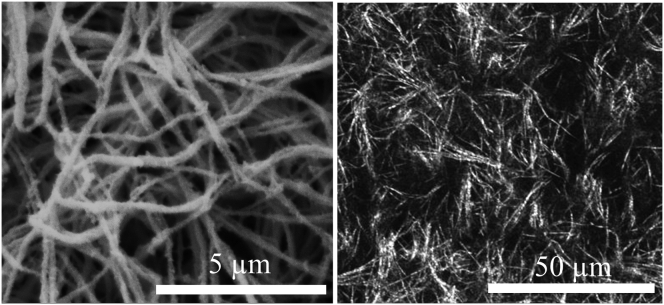
Figure 1.
(Left) 15,000× SEM image of a 2 mg/mL collagen gel demonstrates the ability of electron microscopy to reveal complex organization of individual collagen fibrils and fibril aggregates within a gel. (Right) 60× confocal reflection image of a 2 mg/mL collagen gel reveals the limited ability of optical microscopy to distinguish individual collagen fibrils and their complex nanoscale interactions.
In the absence of a 3D imaging modality with nanoscale resolution, significant questions remain concerning the 3D organization of collagen fibrils, and fibril branching, merging, and grouping within collagen gels. Since collagen gels garner their mechanical stability (at least initially) from physical cross links of fibrils, this information is highly relevant in understanding force transmission within collagen fibril networks and the effects of polymerization conditions on the resulting networks and damage propagation within the networks, to name a few. An important phenomenon seen in vitro is the tendency for fibrils to laterally aggregate into microscale fibers. We have found that studying this grouping of fibrils is challenging using traditional 2D EM methods due to the intrinsic 3D nature of the groupings. There exists a clear need for new methods and modalities capable of providing 3D data sets of collagen gels with nanoscale resolution.
Previous studies have utilized serial block face scanning microscopy (SBF-SEM), whereby a microtome is mounted inside a vacuum chamber and serial images are taken of a sample after successive layers of material are removed (30, 31, 32, 33). These methods have been used to reconstruct 3D images and track collagen fibrils within a mouse embryo tendon (34). Such methods have not been applied to the study of collagen fibril networks in collagen gels. Although SBF-SEM can provide 3D reconstructions, obtaining very thin sections (<50 nm) in hard substrates is challenging, thus limiting the resolution in the slicing direction.
This research examined the use of focused ion beam scanning electron microscopy (FIB-SEM) as a means to image the 3D organization of collagen fibrils within a collagen gel and establishes a pipeline for preparing, nanoscale imaging, and processing data sets for the extraction of 3D fibril network parameters. In FIB-SEM, a focused ion beam (positively charged gallium ions in this case) is used to mill serial sections of the sample, with an SEM image being acquired after each face is exposed (35, 36). This method can yield 3D reconstructions with nanometer resolution, overcomes some of the shortcoming of SBF-SEM such as slice thickness, and provides an automated means of obtaining these reconstructions. This study demonstrates that complex fibrillar organization can be observed in 3D with adequate resolution to distinguish individual collagen fibrils in collagen gels polymerized under varied conditions. Furthermore, a validated 3D tracing protocol is demonstrated that yields network parameters of the fibril organization, and these parameters vary as a result of polymerization conditions. By analyzing the extracted 3D network, fibril parameters, such as segment length and the frequency of branching, have been measured in a collagen gel 3D fibril network for the first time to our knowledge. Furthermore, an algorithm was developed for measuring fibril aggregation into fibers, and it was shown that such aggregation appears to be concentration dependent. By facilitating the 3D imaging of collagen fibril networks at the nanoscale, these methods provide a novel tool for the study of structure-function relationships within collagen gels as well as native collagenous tissues.
Materials and Methods
Collagen gel polymerization
Type I collagen was purified from rat tail tendons as previously described and dissolved in 0.02 M acetic acid at a concentration of 10 mg/mL (15, 16, 25). This solution was diluted to either 2 or 4 mg/mL using 10× PBS containing phenol red and water such that the final concentration of PBS was 1×. While on ice, the collagen solution was neutralized with 1 N sodium hydroxide to obtain a pH of ∼7.5. The solution was poured into silicone elastomer molds, which were pressed onto glass slides to keep them flat and seal the mold while providing a method to gently remove the gels from the mold after polymerization. The gels were allowed to polymerize in the molds overnight at either room temperature (RT) or 37°C. The mold cavities were cylindrical in cross section and had a thickness of 3 mm and diameter of 10 mm (Fig. 2). Additional samples were polymerized either in the presence of the glycosaminoglycan dermatan sulfate (DS) or proteoglycan decorin, as these have previously been shown to alter the microstructural organization of collagen gels (15, 37). A total of six groups of gels (six gels in each group) were polymerized (Table 1).
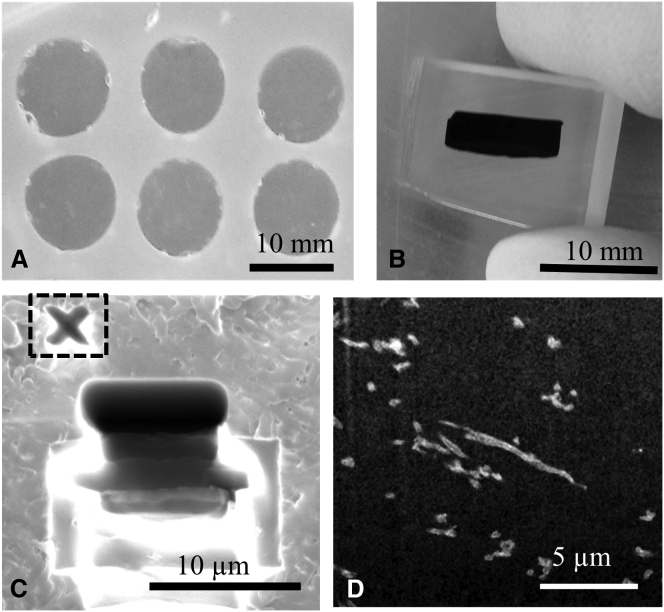
Figure 2.
(A) Collagen gels were polymerized in silicone elastomer molds that were pressed onto glass slides. (B) Stained gels were embedded in PMMA resin and sectioned. (C) A typical FIB-SEM excavation site with the milled fiducial marker identified by a dashed box. (D) Representative SEM image from an ion beam milled face, with collagen fibrils shown in white.
Table 1.
Six Different Groups of Gels Were Polymerized with Varied Collagen Concentration, Polymerization Temperature, and the Presence of Additional Molecules
| Sample Description | Collagen Concentration (mg/mL) | Polymerization Temperature | Additional Molecules |
|---|---|---|---|
| 4 mg/mL RT | 4 | RT | N/A |
| 4 mg/mL 37°C | 4 | 37°C | N/A |
| 2 mg/mL RT | 2 | RT | N/A |
| 2 mg/mL 37°C | 2 | 37°C | N/A |
| 2 mg/mL DS | 2 | RT | DS |
| 2 mg/mL decorin | 2 | RT | decorin |
Staining
Immediately after polymerization, gels were fixed, stained, and embedded. Gels were first placed in a primary fixative (2.5% glutaraldehyde, 2% formaldehyde in 1× PBS) overnight on a shaker plate. Gels were subsequently stained twice with a 2% osmium tetroxide solution overnight and once with a 2% Uranyl acetate solution for two days (34, 35). This created a positive stain in which fibril material generates a signal that can be measured using SEM. After staining, gels to be imaged by FIB/SEM were subjected to a solvent exchange whereby the water phase was replaced with ethanol using an ethanol series (50, 75, 95, and 100%) and were subsequently embedded in PMMA using previously described methods (38). These gels were then cut in half using a diamond point saw and the face was polished using a fine-grit lapidary polishing disk. This exposed the center of the gel, where the fibril geometry was free from edge effects (Fig. 2 B). Gels that were prepared for SEM imaging were dried with hexamethyldisilazane (15, 25).
SEM imaging
Samples dried with hexamethyldisilazane were mounted onto metal posts with carbon tape and imaged at 15,000× in high vacuum using a Quanta 600 FEG SEM (FEI, Hillsboro, OR). Six images were acquired in six separate locations in a sample from each polymerization group. High-resolution images (3775 × 4096 pixels) were saved.
FIB-SEM imaging
A Helios Nanolab 650 (FEI) was used to perform the FIB-SEM imaging of the embedded gels. First, the site was prepared by using the ion beam to excavate a pit around the volume to be imaged. Then the ion beam was used to etch fiducial markers into the surrounding material (Fig. 2 C). Platinum was deposited on the periphery to provide additional strength during the milling process. The focused ion beam was used to sequentially remove either 20 or 25 nm layers of material while the SEM obtained a 10,000× image from the milled face after removal of each layer. An autoslicing program automated this process. This yielded a series of sequential images that were used to reconstruct the 3D volume of the collagen fibril network. The dimensions of the milled face were ∼20 μm in width and 15 μm in height. The total number of slices acquired varied from 250 to 650. During milling, debris tended to accumulate on and around the fiducial markers. If this debris obscured the fiducial marker, the autoslicing program would fail, resulting in a decreased number of slices, thus explaining the variance in the number of slices. A FIB-SEM scan was obtained for each of the six sample types. Additionally, we obtained three scans from three separate regions of a single 4 mg/mL sample to determine site-to-site variability.
Volume rendering and 3D tracing
Image stacks were imported into ImageJ (National Institutes of Health, Bethesda, MD) and were cropped so that unusable portions of the image stack were removed. The usable portion of the stack varied between scans, with milled material debris often obscuring the bottom and edges of the image. After cropping all images within a stack, the resulting images were enhanced using auto level correction, background subtraction using a rolling ball algorithm, a despeckle algorithm, and a 3D median filter (3 × 3). Image stacks were then binarized using the autothreshold function with the Otsu method. Processed image stacks were saved as 3D TIFF images and were imported into Amira (FEI), where the 3D volume was resampled to obtain an isotropic voxel size that had an edge length equal to the slice thickness.
Fibril diameter distributions and volume fraction
Resampled binary datasets were imported into the Farsight Open Snake Tracing System (OSTS) for quantitative analysis (39, 40). This system, which was originally developed to track neuronal axons, proved well suited for tracking collagen fibrils in the 3D data sets. Using an active contouring algorithm, this program extracts the fibril network and related information (Fig. 3). Additional values including fibril diameter, fibril volume, fibril segment length, and the number of branchpoints were obtained using another Farsight executable, Trace Edit (41). The volume of the traced networks was used to compute the volume fraction of collagen in each sample. Histograms were generated and normalized of the fibril segment diameters for each sample so that the integral of the histogram had a value of unity. The mean fibril diameter, fibril segment length, and the number of branchpoints per unit of fibril length were computed for each sample. The samples were grouped by collagen concentration and the mean and standard deviations for these network parameters were computed and compared.
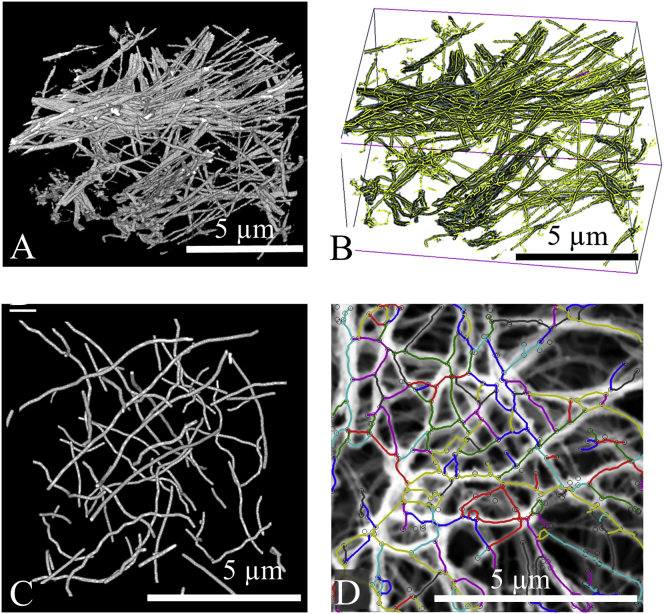
Figure 3.
(A) A typical 3D reconstruction of a collagen fibril network from the FIB-SEM data. (B) The traced fibril network obtained using the Farsight OSTS system, where traces are shown in yellow and the fibril volume is shown in gray. (C) A typical synthetic fibril network (2 mg/mL, 6 pixel diameter) generated using the MATLAB algorithm. (D) An SEM image of a 2 mg/mL sample with the traced network overlaid, with individual traces alternating in color. To see this figure in color, go online.
Fibril grouping measurement
The tendency for collagen fibrils to form into groups of parallel fibrils was quantified by using a custom MATLAB program (The MathWorks, Natick, MA). The coordinates of the fibril networks from the OSTS were analyzed to determine the number of fibril groupings (fibers) relative to the number of detected fibrils, and the diameters of those fibers. The algorithm recognized fibers within the network by checking for fibrils that are oriented within 5° of each other with respect to each fibril axis within a specified distance of each other. A 3D best fit line was determined for each fibril. The dot product of a unit vector along the line with each coordinate axis was used to approximate the orientation of the fibril in 3D space. Fibrils whose orientations were within 5° of each other were to be grouped as possible fibers. Subsequently within each grouping, the distance between all possible two-fiber combinations was taken to determine which fibrils were within a specified input distance of each other. This was done by looking at the shorter fibril of the two fibrils being compared and taking the distance between each of its points and all points on the longer fibril, and saving the minimum measured distances. If 70% of these minimum distance values were within the specified distance criteria, the two fibrils were classified as a fiber. This created an initial set of fibers that were then further grouped with other fibers if they had matching index values. Grouping was continually repeated until no two fibers contained the same fibrils. Fiber diameter was measured by taking the largest measured average distance between any two fibrils within each fiber and adding the respective average radii of each fibril involved with that maximum distance.
Validation of tracing algorithm
To verify the accuracy of the Farsight OSTS for use on collagen fibrils, synthetic images of fibril networks with known parameters were generated and analyzed. Synthetic fibril networks were created using a previously developed MATLAB algorithm (26). The algorithm creates a random fibril network resembling 3D image data of collagen gels (Fig. 3). Synthetic network image data were created with different collagen concentrations (2 and 4 mg/mL) and diameters (2, 4, and 6 pixels). These image datasets were analyzed with OSTS to obtain the traced network along with measured diameters. These recreated images were compared to the original synthetic image data to determine the accuracy of the traced network in OSTS. To check accuracy of diameter measurements, the average measured diameters along with their standard deviations were compared to the known input diameter. To verify that the MATLAB algorithm was able to effectively identify fibers, artificial networks were created in which the addition of a known number of fibril groupings and known diameters were generated. The input values of the synthetic fibrils groupings were then compared to the values obtained from the fiber tracking algorithm in MATLAB.
Results
3D volume rendering of fibril networks
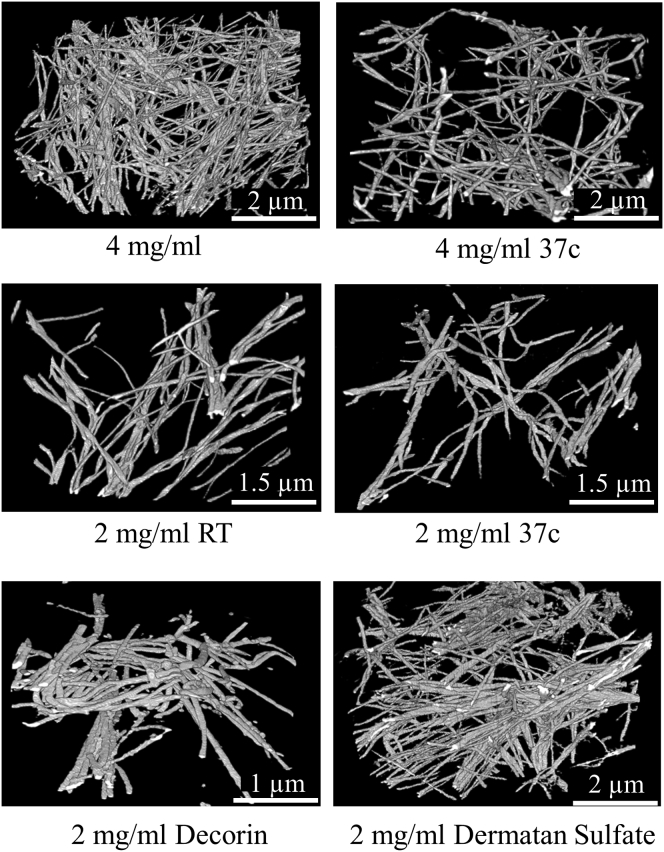
3D reconstructions of FIB-SEM data revealed a complex network of collagen fibrils that display splitting, merging, groupings, and interweaving (Fig. 4). The 20 and 25 nm voxel size provided adequate resolution to reconstruct these complicated features. We attempted to acquire a similar volume of image data across samples. However, controlling the deposition of milled material was problematic, resulting in varied sample dimensions and volumes (Table 2). For all samples, fibril networks were highly connected. Lower-density samples had a correspondingly lower density of fibrils. Qualitative observation of the 3D networks alone was inadequate to distinguish most samples. The addition of the glycosaminoglycan DS resulted in considerably more grouping of fibrils than in the other sample types.
Figure 4.
3D reconstructions for each condition. Within each data set, individual fibrils, grouped fibrils, and complex fibril interactions such as merging, splitting, and interweaving are present. The considerable differences between networks are primarily due to the varied polymerization conditions, although some there are some contributions from varied sample volumes between data sets and spatial variance of collagen density. Scale bars vary across samples due to differences in sample volume inherent in the FIB-SEM technique.
Table 2.
Imaged Sample Dimensions and Volumes
| Sample Description | Width (μm) | Height (μm) | Depth (μm) | Volume (μm3) |
|---|---|---|---|---|
| 4 mg/mL RT | 10.1 | 6.7 | 12.1 | 816 |
| 4 mg/mL 37°C | 9.1 | 6.6 | 6.2 | 375 |
| 2 mg/mL RT | 9.0 | 6.7 | 4.9 | 298 |
| 2 mg/mL 37°C | 10.1 | 6.7 | 9.8 | 664 |
| 2 mg/mL DS | 10.1 | 6.7 | 13.2 | 895 |
| 2 mg/mL decorin | 10.1 | 6.7 | 4.7 | 318 |
Tracing results and comparison of fibril network parameters
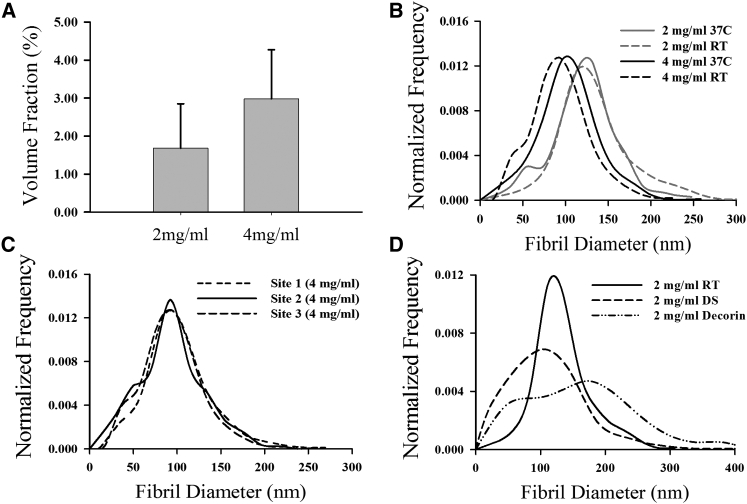
Overlay of the traced networks on the original data set revealed that the OSTS algorithm tracked the fibrils with high fidelity (Fig. 3 B). The volume fraction averaged over all 4 mg/mL samples (2.98 ± 1.29%) was nearly double that of the 2 mg/mL samples (1.67 ± 1.17), but this difference was not statistically significant (Fig. 5 A). The volume fractions for both groups were nearly eight times higher than the theoretical value, which is 0.2 and 0.4% for the 2 and 4 mg/mL samples. The mean fibril diameters for the 2 mg/mL gels were 128 ± 41 and 99 ± 36 nm for the 4 mg/mL gels, with this trend consistent throughout the histograms for samples polymerized at both RT and 37°C (Fig. 5 B). The fibril diameters were similar between the two polymerization temperatures. Although the volume fraction varied significantly from site to site within a single 4 mg/mL sample (2.3–4.9%), the mean fibril diameter and distributions were very similar (Fig. 5 C). The 2 mg/mL DS sample and 2 mg/mL decorin sample had fibril distributions that were markedly different from the other 2 mg/mL samples (Fig. 5 D). Although the mean fibril segment length was greater for the 4 mg/mL gels (1020 ± 150 nm) than the 2 mg/mL gels (900 ± 32 nm), this difference was not statistically significant (p = 0.098). The mean distance between branch points was less for the 4 mg/mL gels (970 ± 32 nm) than the 2 mg/mL gels (1019 ± 151 nm), but this was not statistically significant (p = 0.58). There was an increased tendency for 2 mg/ml gels to have more fibril groupings than the 4 mg/mL gels, with 1.67 ± 0.45% of fibrils being grouped into aggregates of two or more for the former and 1.08 ± 0.78%. This difference was not significant (p = 0.26).
Figure 5.
(A) Average volume fraction for all 2 and 4 mg/mL samples, where error bars represent standard deviations. (B) Normalized histograms comparing the fibril diameter distributions of the 2 mg/mL gels (gray) and 4 mg/ml gels (black), where samples polymerized at RT are dashed and at 37°C are solid. These results demonstrate that increasing the collagen concentration results in a decrease in the fibril diameters, while changing the polymerization temperature had a small but measurable effect. (C) Comparing the normalized histograms of the fibril diameter distributions obtained from FIB-SEM imaging at three different sites within the same 4 mg/mL sample revealed that the fibril diameter within the sample was consistent across the sample volume. (D) Normalized histograms comparing the 2 mg/mL RT sample (solid) with the 2 mg/mL DS sample (dashed) and 2 mg/mL Decorin sample (dot-dash) revealed the significant effect that the addition of these molecules has on the resulting fibril network.
Tracing validation
The artificial fibril networks with varied concentration and diameter were traced using the OSTS algorithm and the mean fibril diameters were computed and compared to the value used to generate the network. The computed fibril diameter for networks generated with a 2 pixel diameter was 3.15 ± 0.21 pixels, which represents an error of 57%. The computed fibril diameter for networks generated using a 4 and 6 pixel diameter was 4.05 ± 0.21 and 5.93 ± 0.34 pixels, respectively. This represents an error of <1.5% for networks with fibrils >4 pixels in diameter, which is the case for all of the networks traced from the volumetric FIB-SEM data. 2D projections were obtained of these same networks and were used to validate the 2D tracking algorithm, where the traced diameters were within 7% of the diameter used to generate the network. The synthetic networks with fibril grouping were tracked using the aforementioned MATLAB algorithm and the results for the number of groups and diameter of the group were compared to the true values and an error of <8% was observed. These data demonstrate that both tracing algorithms produce highly accurate results for the sparse fibril networks traced in this study.
Comparison to 2D SEM data
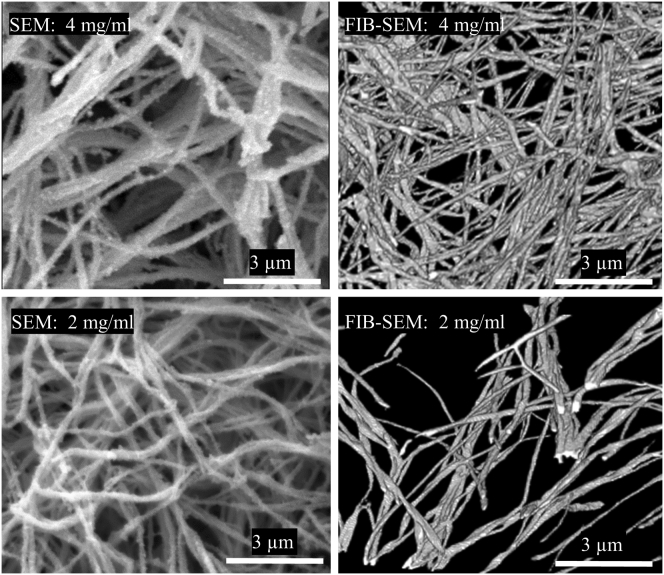
Qualitative side-by-side comparisons of SEM images to the reconstructed FIB-SEM images of the sample within the same group revealed that although SEM images generally corresponded to the FIB-SEM reconstructions, there were distinct differences (Fig. 6). In general, the SEM images showed more aggregation of adjacent fibrils and more overlap of fibrils than the FIB-SEM image, presumably due to the 2D projection of the 3D fibril network.
Figure 6.
Qualitative comparison of the SEM 2D images and FIB-SEM 3D reconstructions (top row, 4 mg/mL; bottom row, 2 mg/mL). The SEM images showed more aggregation and more overlap of adjacent fibrils than the FIB-SEM 3D reconstructions.
Discussion
The results of this study demonstrate that our new approach for characterization of fibril networks in collagen gels via FIB-SEM imaging in combination with 3D tracing provides accurate, high-resolution, automated analysis of fibril network geometry. Raw data sets had a voxel size of 25 nm, which are the highest resolution 3D data sets of a collagen fibril network from a gel ever obtained. This resolution was essential for discerning the individual fibrils, fibril paths, and branching and grouping within the samples. The OSTS algorithm provided a means of obtaining network descriptors such as fibril diameter, length, and number of branch points, as well as number of fibril aggregations.
Validation using synthetic data demonstrated that the traced networks were highly accurate for fibrils with a diameter of 4 pixels or more, which was the case for most fibrils in the 3D FIB-SEM data sets. The resulting fibril diameter histograms demonstrated that the technique can quantify differences in the collagen fibril networks in gels polymerized under different conditions. This technique can be readily applied to other types of hydrogels, including gels made from naturally occurring materials such as fibrin, hyaluronic acid, and alginate, and synthetic scaffolds such as poly(ethylene glycol).
Qualitative comparison of the SEM images to the 3D FIB-SEM reconstructions revealed that there was considerably more lateral grouping of fibrils than in the FIB-SEM data. To image the gels with SEM, they first had to be dehydrated and have all liquid phase material removed, which caused significant shrinking of the gels. We hypothesize that this shrinking resulted in distinct but adjacent fibrils coming into contact, thus appearing to be fused. This result highlights the artifacts that can be introduced by dehydration of collagen gels for SEM, and these artifacts cannot be corrected by postprocessing of the images.
In contrast to SEM, FIB-SEM imaging allows quantification of fibril volume fraction within the gels. In 2D SEM images it is only possible to quantify the percentage of the image area occupied by the fibrils. Because this area is a result of projecting 3D fibrils onto a 2D plane, there is no way to directly correlate these measurements to the 3D volume fraction. It is interesting to note that the volume fractions measured from the FIB-SEM data sets were nearly eight times higher than would be predicted if tropocollagen was densely packed within fibrils. This result suggests that collagen fibrils are highly hydrated, consisting of ∼85% water, a result supported by a previous study (25). FIB-SEM also allows for the computation of fibril segment lengths and the measurement of true branchpoints, something that is not possible using 2D projections of networks, as in SEM.
Although FIB-SEM offers many advantages relative to other imaging modalities for studying collagen gels, there are many factors to be taken into consideration. While the 3D datasets have a resolution that greatly exceeds that of optical methods, the imaging volume is much smaller. However, by measuring fibril network parameters in three different sites within the same sample, we found that even with a small sample volume, representative parameters for the entire sample can be measured. Furthermore, the scan time required to obtain a full data set for FIB-SEM is 6–12 h, while optical methods may only take 1–2 h. One possible solution to decrease imaging time is to reduce the resolution of the image acquired in on each face (we actually had to down-sample our image resolution) and by taking thicker slices. Increasing slice thickness by 50% should have a minimal impact on the accuracy of the reconstructed and traced networks, but should significantly reduce scan time. Additionally, FIB-SEM requires that the sample is stained and embedded within resin, which increases the processing time for the samples. Although this is necessary for obtaining flat faces when milling with the ion beam, by embedding smaller samples, this process could be significantly sped up. Despite the many processing steps for staining and embedding the gels in resin, we believe that the nanoscale network structure was well preserved. The concentrations of gels used in this study result in a physically cross-linked network that is stable under its own weight and can be handled and even tested under tension. The structure of these cross-linked networks should not be affected by the gentle processing steps of staining and solvent exchange during the embedding process, at least at the spatial scale examined in this study. We note that we did not examine the condition of the finer components of the ECM after fixation. Additional considerations include the challenges associated with obtaining a consistent sample volume and the necessity of embedding in a hard resin. PMMA was chosen because it mills well by an ion beam and can fully infiltrate the gel. Many of these issues can likely be addressed by further optimization of the preparation, embedding, and imaging methods used with FIB-SEM.
Although this study is, to our knowledge, the first of its kind for collagen gels, a previous study by Bushby et al. (35) describes a similar method for observing collagen fibrils in a chick embryo. Although this study was used as a reference, unique methods had to be developed to deal with the highly hydrated nature of collagen gels. Furthermore, the aforementioned study did not perform any image processing on the volumetric data sets. In this study, a pipeline is developed that allows for a detailed qualitative analysis of fibril networks with nanoscale resolution, as well as detailed quantitative comparisons using network parameters such as fibril diameter, length, branching, and fibril grouping.
In summary, FIB-SEM is an effective method for imaging the complex morphological organization of fibrils within a collagen gel with nanometer resolution, and the image datasets provide both qualitative and quantitative metrics of comparison for in vitro studies utilizing collagen gels. Due to its high spatial resolution, FIB-SEM represents an ideal tool for examining the nanoscale organization of fibrillar networks in studies of collagen gels in a variety of applications, including their use for in vitro cell culture.
Author Contributions
S.P.R. and J.A.W. conceived and designed the study; S.P.R., N.F., R.P., and G.P. participated in data acquisition; S.P.R., N.F., and J.A.W. devised the methods for image analysis and quantification; all authors participated in data analysis and interpretation; S.P.R. and J.A.W. drafted the article; and all authors edited the article and gave final approval for publication.
Acknowledgments
Financial support from National Institutes of Health grants No. R01EB015133 (to S.P.R. and J.A.W.) and No. R01AR047369 (to S.P.R., G.P., and J.A.W.) is gratefully acknowledged. The facility housing and maintenance of the FIB/SEM microscope was supported by National Science Foundation Materials Research Science and Engineering Center Program No. DMR-1121252.
Editor: Stanislav Shvartsman.
References
- 1.Boot-Handford R.P., Tuckwell D.S. Fibrillar collagen: the key to vertebrate evolution? A tale of molecular incest. BioEssays. 2003;25:142–151. doi: 10.1002/bies.10230. [DOI] [PubMed] [Google Scholar]
- 2.Nair A.K., Gautieri A., Buehler M.J. Molecular mechanics of mineralized collagen fibrils in bone. Nat. Commun. 2013;4:1724. doi: 10.1038/ncomms2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viguet-Carrin S., Garnero P., Delmas P.D. The role of collagen in bone strength. Osteoporosis Int. 2006;17:319–336. doi: 10.1007/s00198-005-2035-9. [DOI] [PubMed] [Google Scholar]
- 4.Kannus P. Structure of the tendon connective tissue. Scand. J. Med. Sci. Sports. 2000;10:312–320. doi: 10.1034/j.1600-0838.2000.010006312.x. [DOI] [PubMed] [Google Scholar]
- 5.Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol. Rev. 2004;84:649–698. doi: 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- 6.Wang J.H. Mechanobiology of tendon. J. Biomech. 2006;39:1563–1582. doi: 10.1016/j.jbiomech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Liu S.H., Yang R.S., Lane J.M. Collagen in tendon, ligament, and bone healing. A current review. Clin. Orthop. Relat. Res. 1995;(318):265–278. [PubMed] [Google Scholar]
- 8.Weiss J.A., Woo S.L., Newton P.O. Evaluation of a new injury model to study medial collateral ligament healing: primary repair versus nonoperative treatment. J. Orthop. Res. 1991;9:516–528. doi: 10.1002/jor.1100090407. [DOI] [PubMed] [Google Scholar]
- 9.Cremer M.A., Rosloniec E.F., Kang A.H. The cartilage collagens: a review of their structure, organization, and role in the pathogenesis of experimental arthritis in animals and in human rheumatic disease. J. Mol. Med. 1998;76:275–288. doi: 10.1007/s001090050217. [DOI] [PubMed] [Google Scholar]
- 10.Eyre D. Collagen of articular cartilage. Arthritis Res. 2002;4:30–35. doi: 10.1186/ar380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banos C.C., Thomas A.H., Kuo C.K. Collagen fibrillogenesis in tendon development: current models and regulation of fibril assembly. Birth Defects Res. C Embryo Today. 2008;84:228–244. doi: 10.1002/bdrc.20130. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G., Ezura Y., Birk D.E. Decorin regulates assembly of collagen fibrils and acquisition of biomechanical properties during tendon development. J. Cell. Biochem. 2006;98:1436–1449. doi: 10.1002/jcb.20776. [DOI] [PubMed] [Google Scholar]
- 13.Na G.C., Butz L.J., Carroll R.J. Mechanism of in vitro collagen fibril assembly. Kinetic and morphological studies. J. Biol. Chem. 1986;261:12290–12299. [PubMed] [Google Scholar]
- 14.Chevallay B., Herbage D. Collagen-based biomaterials as 3D scaffold for cell cultures: applications for tissue engineering and gene therapy. Med. Biol. Eng. Comput. 2000;38:211–218. doi: 10.1007/BF02344779. [DOI] [PubMed] [Google Scholar]
- 15.Reese S.P., Underwood C.J., Weiss J.A. Effects of decorin proteoglycan on fibrillogenesis, ultrastructure, and mechanics of type I collagen gels. Matrix Biol. 2013;32:414–423. doi: 10.1016/j.matbio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roeder B.A., Kokini K., Voytik-Harbin S.L. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J. Biomech. Eng. 2002;124:214–222. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- 17.Achilli M., Mantovani D. Tailoring mechanical properties of collagen-based scaffolds for vascular tissue engineering: the effects of pH, temperature and ionic strength on gelation. Polymers (Basel) 2010;2:664–680. [Google Scholar]
- 18.Chandran P.L., Barocas V.H. Deterministic material-based averaging theory model of collagen gel micromechanics. J. Biomech. Eng. 2007;129:137–147. doi: 10.1115/1.2472369. [DOI] [PubMed] [Google Scholar]
- 19.Roeder B.A., Kokini K., Voytik-Harbin S.L. Fibril microstructure affects strain transmission within collagen extracellular matrices. J. Biomech. Eng. 2009;131:031004. doi: 10.1115/1.3005331. [DOI] [PubMed] [Google Scholar]
- 20.Sander E.A., Stylianopoulos T., Barocas V.H. Image-based multiscale modeling predicts tissue-level and network-level fiber reorganization in stretched cell-compacted collagen gels. Proc. Natl. Acad. Sci. USA. 2009;106:17675–17680. doi: 10.1073/pnas.0903716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurschler C., Provenzano P.P., Vanderby R., Jr. Scanning electron microscopic characterization of healing and normal rat ligament microstructure under slack and loaded conditions. Connect. Tissue Res. 2003;44:59–68. [PubMed] [Google Scholar]
- 22.Ramanujan S., Pluen A., Jain R.K. Diffusion and convection in collagen gels: implications for transport in the tumor interstitium. Biophys. J. 2002;83:1650–1660. doi: 10.1016/S0006-3495(02)73933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori H., Shimizu K., Hara M. Dynamic viscoelastic properties of collagen gels with high mechanical strength. Mater. Sci. Eng. C. 2013;33:3230–3236. doi: 10.1016/j.msec.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan L., Weiss J.A., Hoying J.B. Design and application of a test system for viscoelastic characterization of collagen gels. Tissue Eng. 2004;10:241–252. doi: 10.1089/107632704322791880. [DOI] [PubMed] [Google Scholar]
- 25.Raub C.B., Suresh V., George S.C. Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophys. J. 2007;92:2212–2222. doi: 10.1529/biophysj.106.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein A.M., Vader D.A., Sander L.M. An algorithm for extracting the network geometry of three-dimensional collagen gels. J. Microsc. 2008;232:463–475. doi: 10.1111/j.1365-2818.2008.02141.x. [DOI] [PubMed] [Google Scholar]
- 27.Toki F., Honkura N., Nanba D. Second harmonic generation reveals collagen fibril remodeling in fibroblast-populated collagen gels. Cell Struct. Funct. 2013;38:227–236. doi: 10.1247/csf.13017. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y.L., Leone L.M., Kaufman L.J. Elastic moduli of collagen gels can be predicted from two-dimensional confocal microscopy. Biophys. J. 2009;97:2051–2060. doi: 10.1016/j.bpj.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi S., Chen B., Connon C.J. Plastic compression of a collagen gel forms a much improved scaffold for ocular surface tissue engineering over conventional collagen gels. J. Biomed. Mater. Res. A. 2010;95:447–453. doi: 10.1002/jbm.a.32861. [DOI] [PubMed] [Google Scholar]
- 30.Denk W., Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horstmann H., Körber C., Kuner T. Serial section scanning electron microscopy (S3EM) on silicon wafers for ultra-structural volume imaging of cells and tissues. PLoS One. 2012;7:e35172. doi: 10.1371/journal.pone.0035172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes L., Hawes C., Vaughan S. Serial block face scanning electron microscopy—the future of cell ultrastructure imaging. Protoplasma. 2014;251:395–401. doi: 10.1007/s00709-013-0580-1. [DOI] [PubMed] [Google Scholar]
- 33.Pinali C., Kitmitto A. Serial block face scanning electron microscopy for the study of cardiac muscle ultrastructure at nanoscale resolutions. J. Mol. Cell. Cardiol. 2014;76:1–11. doi: 10.1016/j.yjmcc.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Starborg T., Kalson N.S., Kadler K.E. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat. Protoc. 2013;8:1433–1448. doi: 10.1038/nprot.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bushby A.J., P’ng K.M., Quantock A.J. Imaging three-dimensional tissue architectures by focused ion beam scanning electron microscopy. Nat. Protoc. 2011;6:845–858. doi: 10.1038/nprot.2011.332. [DOI] [PubMed] [Google Scholar]
- 36.Knott G., Marchman H., Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwasaki S., Hosaka Y., Takehana K. The modulation of collagen fibril assembly and its structure by decorin: an electron microscopic study. Arch. Histol. Cytol. 2008;71:37–44. doi: 10.1679/aohc.71.37. [DOI] [PubMed] [Google Scholar]
- 38.Rippstein P., Black M.K., O’Brien E.R. Comparison of processing and sectioning methodologies for arteries containing metallic stents. J. Histochem. Cytochem. 2006;54:673–681. doi: 10.1369/jhc.5A6824.2006. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Narayanaswamy A., Roysam B. A broadly applicable 3-D neuron tracing method based on open-curve snake. Neuroinformatics. 2011;9:193–217. doi: 10.1007/s12021-011-9110-5. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Y., A. Narayanaswamy, …, B. Roysam. 2011. Novel 4-D open-curve active contour and curve completion approach for automated tree structure extraction. In CPVR ‘11 Proceedings of the 2011 IEEE Conference on Computer Vision and Pattern Recognition. 1105–1112.
- 41.Luisi J., Narayanaswamy A., Roysam B. The FARSIGHT Trace Editor: an open source tool for 3-D inspection and efficient pattern analysis aided editing of automated neuronal reconstructions. Neuroinformatics. 2011;9:305–315. doi: 10.1007/s12021-011-9115-0. [DOI] [PubMed] [Google Scholar]