Abstract
Fluorescent DNA dyes are broadly used in many biotechnological applications for detecting and imaging DNA in cells and gels. Their binding alters the structural and nanomechanical properties of DNA and affects the biological processes that are associated with it. Although interaction modes like intercalation and minor groove binding already have been identified, associated mechanic effects like local elongation, unwinding, and softening of the DNA often remain in question. We used magnetic tweezers to quantitatively investigate the impact of three DNA-binding dyes (YOYO-1, DAPI, and DRAQ5) in a concentration-dependent manner. By extending and overwinding individual, torsionally constrained, nick-free dsDNA molecules, we measured the contour lengths and molecular forces that allow estimation of thermodynamic and nanomechanical binding parameters. Whereas for YOYO-1 and DAPI the binding mechanisms could be assigned to bis-intercalation and minor groove binding, respectively, DRAQ5 exhibited both binding modes in a concentration-dependent manner.
Introduction
Fluorescent DNA dyes are broadly used in biology and biomedicine to detect and image DNA in separation gels or in fluorescence light microscopy. The specific interaction of these dyes with DNA affects the nanomechanical behavior and the structural properties of DNA, interfering with many of the well-known DNA associated processes like transcription and replication. Although a number of different interaction modes have been identified, the associated molecular mechanics like elongation, untwisting, and softening of the DNA polymer are often poorly understood and debated. In this work we chose three commonly used fluorescent stains (YOYO-1, DAPI, and DRAQ5) and investigated the nanomechanical effects of the DNA-binding dyes using magnetic tweezers (MTs). The green-fluorescent DNA dye YOYO-1 is a tetracationic homodimer of oxazole yellow and belongs to the monomethine cyanine family. It is one of the most commonly used nucleic acid dyes and shows virtually no fluorescence in solution but in contrast a more than 1000-fold increase of fluorescent intensity when bound to DNA (1). YOYO-1 is known to interact with DNA as a bis-intercalator, which leads to a local unwinding the double helix and a considerable lengthening of the DNA strand. The reported elongation varies from 18% to 50% (2, 3, 4, 5, 6, 7). The associated unwinding angle of YOYO-1 has been measured in MT experiments to 24° by Günther et al. (8), whereas Johansen et al. measured an angle of 106° by NMR (9). A further debate was related to how YOYO-1 influences the bending stiffness or softening of the biopolymer. Quake et al. reported a 32% persistence-length increase of DNA with optical tweezers (3), whereas two other groups have measured reduction of 70% with the same methodology (4, 5). Shi et al. have used entropic force microscopy and observed a reduction of 9% of the persistence length (7). By using atomic force microscopy (AFM) in solution, Maaloum et al. reported a reduction of 44% (6), whereas Kundukad et al. suggested a more or less constant persistence length under ambient conditions (10). Recently, Günther et al. (8) employed MTs to perform single-molecule stretching experiments and concluded that the persistence length of dsDNA remained almost constant during intercalation with YOYO-1.
Although YOYO-1 is regarded as an efficient and photobleaching-resistant DNA dye, it is cell-impermeable, which means that it cannot be applied in live- and fixed-cell experiments. Therefore, the classical blue DNA dye DAPI, which belongs to the indol family, is the dye of choice for staining the chromosomes in the nucleus of fixed and living cells. It could be shown that the binding of DAPI to DNA leads to a 20-fold increase of the fluorescence (11). However, since DAPI can only be excited with ultraviolet (UV) light, exposition of this fluorescent dye interferes with the viability of the probed cells and somewhat limits the accessible observation time. DAPI is known as a DNA minor groove binder that binds preferentially to AT-rich regions and theoretically influences the bending rigidity of DNA (12, 13, 14). In a recent AFM study, Japaridze et al. (15) reported that upon DAPI staining the contour length of dsDNA remains unchanged, whereas, the persistence length considerably decreased up to 42%.
In contrast to DAPI, DRAQ5 (deep red fluorescent antraquinone dye Nr. 5) is less harmful for living cells due to the long excitation wavelength in the red spectral range (647 nm) (16). Furthermore it is membrane-permeable and it can be multiplexed with many other fluorophores. Consequently, it is an ideal choice for many live-cell experiments. Yet, the binding mode of DRAQ5 to DNA is still debated. Japardize et al. (15) and Martin et al. (17) suggested DNA intercalation as the dominating binding mechanism whereas Njoh et al. (18) concluded in their molecular docking studies that the binding mode can adequately be described by AT preferred engagement in the DNA minor groove. A more competitive bi-mechanistic interaction was suggested by Islam et al. (19) in their theoretical modeling experiments, however, without experimentally investigating further associated nanomechanical properties.
In this work, we investigated the DNA binding properties of DRAQ5 by means of MT extension and overwinding experiments within a force regime from 5 × 10−3 pN up to 10 pN. The well-known bis-intercalator YOYO-1 and the minor groove binder DAPI served as a reference to interpret and quantify the influence of DRAQ5 association on the structural and nanomechanical properties of dsDNA. Within this mechanistic picture we found that DRAQ5 exposes a bimodal binding behavior. Our results consistently indicate that at concentrations below 0.5 μM, DRAQ5 binds as a minor groove binder whereas at higher dye concentrations intercalation dominates.
Materials and Methods
DNA sample preparation
We prepared λ-DNA fragments (New England BioLabs, Frankfurt, Germany) that were functionalized at one end with several biotins (Biotin-14-dCTP, Metabion, Steinkirchen, Germany) and on the other side with several digoxigenins (Dig-11-dUTP, Hoffmann-LaRoche, Penzberg, Germany) according to a recently published protocol (20). The fragments were repaired with PreCR Repair Mix (New England BioLabs, Frankfurt, Germany) and stored in 10 mM PBS buffer (137 mM NaCl + 2.7 mM KCl (pH 7.4) at 25°C) at 4°C. The DNA fragments were attached via several antidigoxigenins to the surface of a glass cover slip of a custom-made flow cell. As a handle, streptavidin-coated magnetic beads with a diameter of 1 μm (Dynabeads MyOne Streptavidin C1, Life Technologies, Carlsbad, CA) were attached to the other DNA side. For that, the magnetic beads (5 mg/ml) were gently mixed with DNA fragments (60 pM) into the MT buffer and incubated for 15 min. Before every experiment we verified the structural integrity of each probed DNA-molecule (nick-free) and its torsionally constrained immobilization, and we acquired a reference hat curve by MT overwinding experiments. Moreover we verified the contour length by means of stretching experiments (21).
Fluorescent dyes
In the experiments we used three fluorescent dyes, namely YOYO-1, DAPI, and DRAQ5. YOYO-1 iodide was purchased from Life Technologies (stock solution 1 mM). DAPI was obtained from Sigma-Aldrich (Seelze, Germany; stock solution 1 mg/500 μl). DRAQ5 was acquired from Thermo Fisher Scientific (Ulm, Germany; stock solution 5 mM). Before the experiments all three stock solutions were diluted with the MT buffer to concentrations from 1 nM to 10 μM.
Magnetic tweezers analysis
The used MT setup is a commercial system (PicoTwist, Lyon, France) and has been described previously (21, 22, 23) (Fig. 1 a). The DNA-functionalized beads were filled into the chamber of the flow cell, whose surface was functionalized with antidigoxigenins (0.2 mg/ml, at 37°C for 2 h) and allowed to rest for 20–30 min. Then, the flow cell was flushed with the MT buffer so that unbound DNA-beads were removed. All force-extension experiments have been taken from 5 × 10−3 pN up to an upper force limit of 10 pN. The overwinding experiments were performed with a preset force of 0.2 pN. The DNA was stained within the fluid cell by means of flushing the fluorescent dyes into the chamber with stepwise increasing concentrations. To exchange the buffer completely a multitude of the fluid cell volume was infused. Furthermore, the DNA was incubated for another 20 min to achieve thermodynamical equilibrium. To avoid crosstalk of different dyes, we replaced the complete flow cell after every experimental series. All experiments were performed in the MT buffer at 25°C. Of note, we took special attention that DRAQ5 that was incubated in total darkness according to the instructions of the manufacturer.
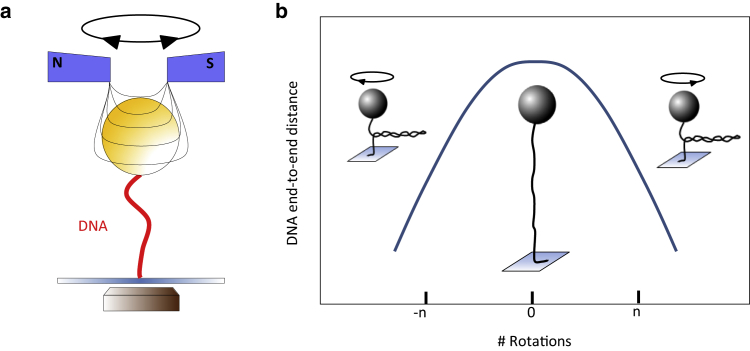
Figure 1.
Schematic of magnetic tweezers. (a) A single dsDNA molecule is attached between a surface and a micrometer-sized magnetic bead that is trapped and manipulated in a variable, external magnetic field. (b) Supercoiling curve (hat curve) of dsDNA is shown. By rotating a magnetic bead, mechanical torque is applied to the DNA molecule and the end-to-end distance is reduced. Hat curves can be divided into two regimes: for a number of twists, R, smaller than the buckling number, N, the torque is released along the double strands (torsional elastic regime). At higher rotation numbers, R > N, the torsional stress is released by the formation of plectonemes that leads to a substantial topological shortening of the DNA end-to-end distance (plectonemic regime). To see this figure in color, go online.
MT buffer
All the experiments were performed with the MT buffer that contained 10 mM phosphate buffered saline (137 mM NaCl + 2.7 mM KCl (pH 7.4) at 25°C) with 0.1 mg/ml additional bovine serum albumin (Sigma-Aldrich) and 0.1% Tween 20 (Sigma-Aldrich).
Analysis of the data
The molecular stretching curves were fitted with the following worm-like chain (WLC) model:
| (1) |
for each dye concentration (24, 25). Here, F, P, L(c), , and d denote the pulling force, the persistence length as a function of the dye concentration, the contour length, the thermal energy, and the DNA molecular extension (end-to-end distance), respectively. Furthermore, the correlation between the fractional DNA elongation and dye concentrations c were analyzed by fitting it with the following McGhee–von Hippel model (26):
| (2) |
where Ka is the equilibrium constant of association for intercalation, n is the binding site size per dye molecule, is the reference distance between two base pairs ( nm), Δx is the DNA elongation due to one binding site of an intercalated dye, and a is a dimensionless geometrical factor for intercalators like DRAQ5 (a = 1) and bis-intercalators like YOYO-1 (a = 2).
Results and Discussions
To investigate the impact of dye association on the nanomechanic properties of DNA we used a joined approach of MT-based extension and overwinding experiments. MTs allow the exact determination of the DNA end-to-end distance, contour, and persistence length at full control of the DNA helical twist. First, we performed extension experiments while the twist was adjusted in such a way that the DNA remained in the torsionally relaxed state in which it exposes the maximum end-to-end length. The DNA was incubated within the MT fluid cell at various dye concentrations for 20 min. We started with concentrations as low as 1 nM to inhibit artifacts due to unspecific adhesion. The stained DNA molecules were extended at zero twist to acquire force-extension curves with a maximum force up to 10 pN (Fig. 2, a, c, and e). After each extension step we allowed the molecular system to settle down for ∼5 s before the force and end-to-end distance were acquired with an integration time of ∼1 min. The force-extension data was approximated to the WLC model to estimate the DNA contour and persistence length and also to the McGhee–von Hippel model to evaluate the equilibrium constant of association, the induced length increase per associated dye, and the binding site size in base pairs per dye molecule, respectively.
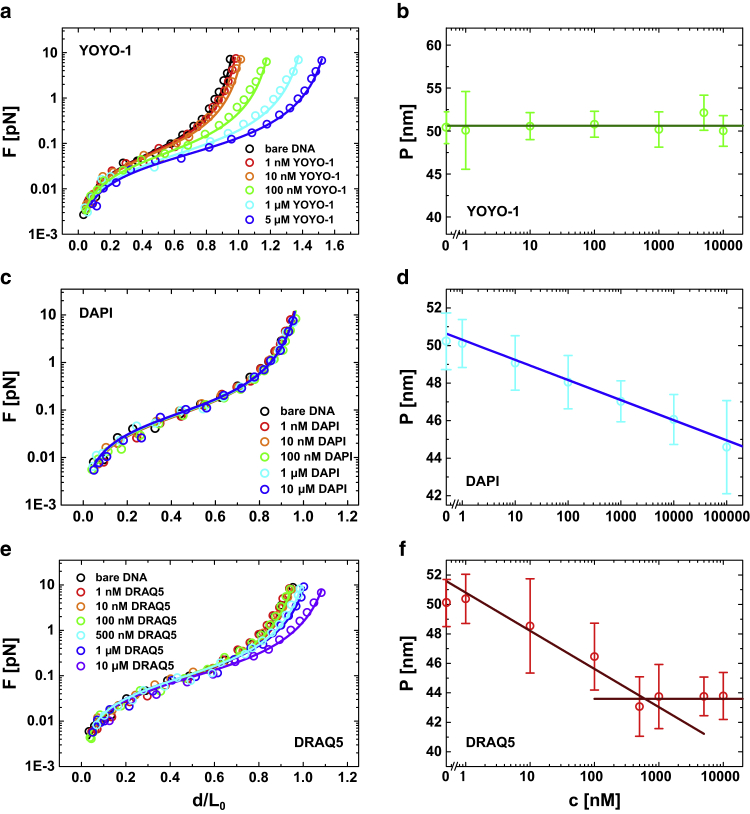
Figure 2.
Force-extension curves of bare and dye-complexed dsDNA (left column) for (a) YOYO-1, (c) DAPI, and (e) DRAQ5. Open circles represent experimental data whereas solid lines stand for WLC fits. Persistence length in dependence of dye concentration is shown (b, d, and f). To see this figure in color, go online.
Contour and persistence length
By analyzing the YOYO-1 bis-intercalator we found a successive shift of the force-extension curves toward larger contour lengths ranging from elongations of 2.3% to 58% at dye concentrations from 1 nM to 10 μM (Fig. 2 a). This finding is in full accordance with recently published results (5). In contrast, the DNA persistence length remained unchanged at ∼50 nm independent of the dye concentration (Fig. 2 b). Interestingly, reports on the impact of YOYO-1 association on the persistence length are not very consistent. Kundukad and co-workers outlined the key data of various publications that were acquired with different experimental techniques such as optical tweezers or MTs, AFM, and entropic force microscopy (10). There, minute or no changes of the persistence length nicely matching our findings stand in contrast to publications reporting on a significant decrease (up to 70%). Though, when regarding the experimental conditions under which these data are obtained, a consistent scheme can be found: whenever the DNA is incubated with YOYO-1 below the saturation concentration (i.e., less than one YOYO-1 per four base pairs), no or only a small change of the persistence length was observed (8, 10). In contrast, when measurements have been performed with a large excess of dye molecules the DNA becomes more flexible, which corresponds to a decrease of the persistence length (3, 4, 5, 6). We ascribe this to an additional unspecific adhesion like mono-intercalated YOYO-1 and electrostatic interaction between the negatively charged DNA backbone and the positively charged dye molecule that generates an electrostatic shielding and thus reduces the DNA stiffness (27).
Furthermore, we also tested the minor groove binder DAPI in a concentration-dependent manner. According to our data, DAPI binding to DNA has virtually no influence on the contour length that can be deduced from the evolution of the force-extension curves (Fig. 2 c). We furthermore fitted our data to the WLC molecular elasticity model (Eq. 1) and found no obvious elongation. However, we observed a progressive but moderate reduction of the persistence length from 48 to 44 nm for increasing dye concentrations (Fig. 2 d), which is consistent with recent results (15). As a consequence, we can conclude that minor groove binding leads to a moderate softening of the dsDNA.
By knowing the influence of intercalation and minor groove binding on DNA nanomechanics, we now analyzed the binding mode of the DNA dye DRAQ5. Interestingly, DRAQ5 exhibits characteristics of both, minor groove binding and intercalation (Fig. 2 e). At low dye concentrations (<0.5 μM), we merely observed a decrease of the persistence length from ∼50 to 43 nm while the contour length remained constant (Fig. 2 f). In accordance with our previous findings this clearly indicates a minor-groove-like binding mechanism. By further increasing the dye concentration we found an incremental shift of the force-extension curves toward a higher contour length (Fig. 2 e). Similar to the results obtained for YOYO-1, this indicates an intercalator-like binding mode. Noteworthy, the increase of the contour length is rather moderate. We estimated the maximum increase to be 14% at a dye concentration of 10 μM. Moreover, the persistence length did not vary anymore at high DRAQ5 concentrations (Fig. 2 f).
Fractional elongation and association constant
As shown before, intercalation induces an increase of the DNA contour length. This elongation can be used to further validate and quantify the intercalating character of a bond. We plotted the fractional elongation as a function of the dye concentration c. Here denotes the contour length of bare DNA whereas is the DNA contour length as a function of the dye concentration. We approximated to the McGhee–von Hippel model (Eq. 2) to determine the equilibrium constant of association Ka, the binding site size per dye molecule n in base pair, and the DNA elongation per intercalated dye molecule a × Δx, respectively (Fig. 3, a and c). Because DAPI as a typical minor groove binder does not affect the contour length, this analytical approach is not applicable (Fig. 3 b).
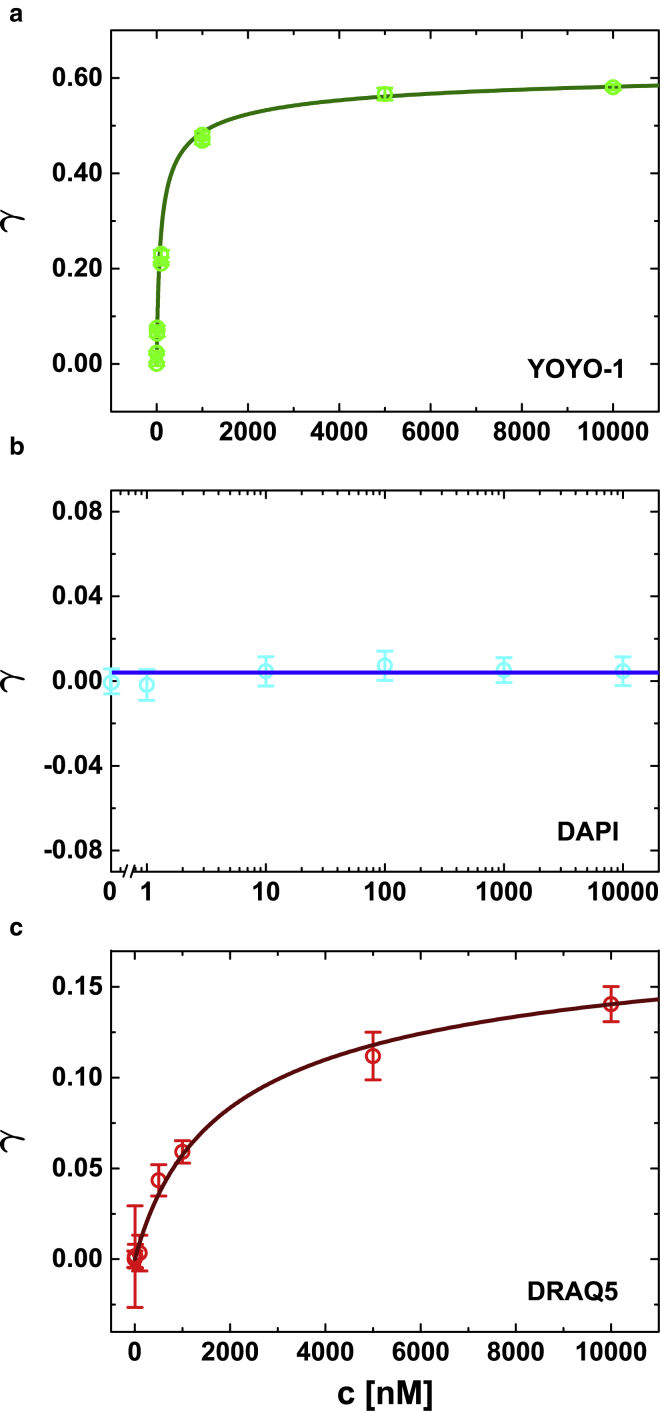
Figure 3.
Plots of the fractional elongation of (a) YOYO-1, (b) DAPI, and (c) DRAQ5. Experimental data (open circles) are approximated to the McGhee–von Hippel model (solid line in a and c). To see this figure in color, go online.
For YOYO-1 the equilibrium constant Ka was calculated to be M−1, which is nicely supported by previously published data (5). The apparently reduced affinity can be attributed to elution effects caused by the high salt concentration of the PBS buffer (137 mM NaCl + 2.7 mM KCl) (28). The binding site size was estimated as bp/dye. This comparably large value reflects the fact that bis-intercalation inhibits the occupation of adjacent base pair voids (29). This negative cooperativity is referred to as “nearest neighbor exclusion principle” (30). The DNA elongation per intercalated molecule was nm/dye.
Analogously, we analyzed the DNA elongation by DRAQ5 binding within the intercalating regime (μM). Here we estimated an equilibrium constant of association of M−1 which, compared with YOYO-1, is more than one magnitude smaller than for YOYO-1. Furthermore, we estimated a binding site size of 3.13 ± 0.286 bp/dye. Compared with a mono-intercalator such as ethidium bromide (∼2 bp/dye), one would expect a smaller binding site size (31, 32). However, as DRAQ5 exposes nonuniform binding characteristics, the McGhee–von Hippel model might be of limited applicability. Probably, minor groove and intercalative binding interfere with each other: apparently, minor groove binding is favored in the first place until a certain degree of binding sites is occupied. Then, intercalative binding dominates. Hence, the amount of free intercalative binding sites is reduced due to previous minor groove association that in turn inhibits a high intercalation density along the dsDNA. Therefore, the estimated binding-site size appears larger than expected.
The intercalation of DRAQ5 to DNA seems to be a less-pronounced secondary binding mode. In line with this finding is also a comparably small elongation per (intercalative) bond. We found a lengthening per dye molecule of nm/dye, which is nearly identical to data obtained for ethidium bromide ( nm/dye) (31).
Dye-induced unwinding
A crucial feature of MTs is the ability to apply torque to single molecules. When rotating magnetic beads, attached DNA molecules are twisted in a controlled manner and plectonemic strands can be produced. These induced conformational changes are often displayed as DNA supercoil curves that expose the end-to-end distance d versus the number of applied turns R (Fig. 1 b). Commonly, these are also referred to as “hat curves.” In the low-force range (∼ <0.5 pN), bare dsDNA exposes a symmetric torsional behavior (33). There, the peak of the hat curve denotes the rotational relaxed state where the DNA exposes the maximum end-to-end distance. Starting from here, the hat curve can be divided into two regimes (Fig. 1 b). At low rotation numbers the torque that is built up by the intrinsic torsional elasticity (torsional elastic regime) of the DNA double strand goes along with a slight decrease of the end-to-end distance. In contrast, at higher rotation numbers the torque induces buckling of the DNA double helix that leads to the formation of topological plectonemes and a significant reduction of the end-to-end length. The rotation number at the transition of these regimes is often referred to as buckling number N (Fig. 1 b) (34).
To strengthen the results of our extension measurements we acquired overwinding curves at pulling forces of 0.2 pN for all three ligands (Fig. 4, a, c, and e). First, YOYO-1 was applied to torsional fixed but relaxed (R = 0) dsDNA with a stepwise increasing concentration. The acquired hat curves expose a successive shift to negative rotation numbers and an increasing end-to-end distance that indicates a gradual unwinding of the dsDNA (Fig. 4 a). Evidently, YOYO-1 intercalation induces a supercoiling that can be removed by applying rotations in the (negative) counterdirection (6, 8, 10). We estimated the untwisting angle per dye molecule as follows:
| (3) |
where ΔR quantifies the unwinding at different dye concentrations c, and # is the corresponding number of intercalated dye molecules. The latter is calculated by the difference of the DNA contour length ΔL divided by lengthening per dye molecule a × Δx. Both quantities have been estimated in the experiments before. We plotted ΔR versus ΔL and estimated by approximating the slope of the plot (Fig. 4, b, d, and f). As a result, we obtained an angle of 0.088 ± 0.0411 turns per dye corresponding to per dye for YOYO-1.
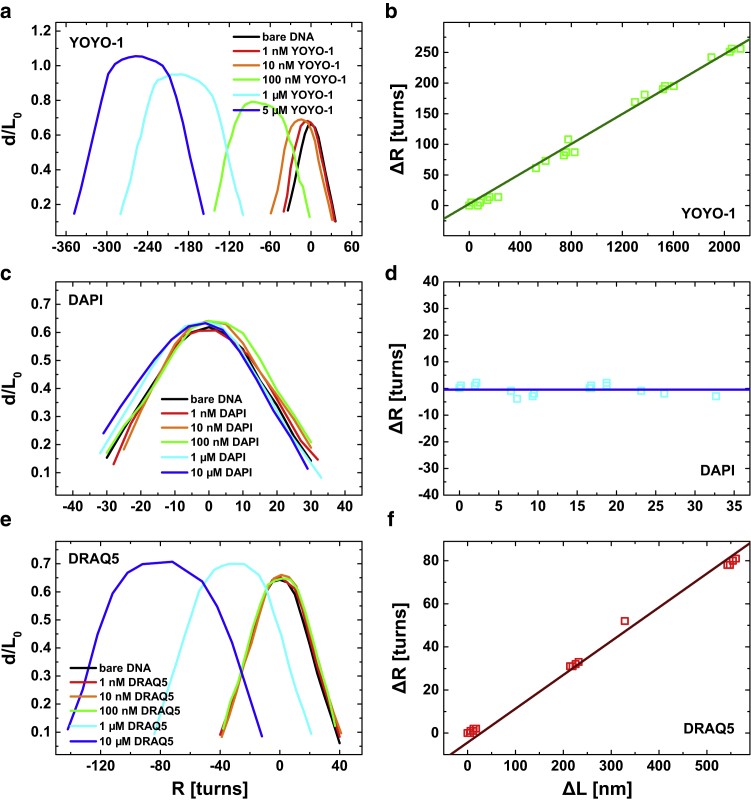
Figure 4.
Supercoiling curves (hat curves) of dsDNA complexed with (a) YOYO-1, (c) DAPI, and (e) DRAQ5. The data were acquired at a load of 0.2 pN. (b, d, and f) Linear approximation of the rotation-elongation plots allows to estimate the unwinding angle per intercalated dye. The error of ΔL was found in the order of 10 nm where ΔR occupied an error of about ± 1 turn. To see this figure in color, go online.
In contrast, supercoil curves that have been acquired upon association of the minor groove binder DAPI hardly differ from those acquired with bare DNA (Fig. 4 d). We neither observed a rotational shift nor a significant increase of the end-to-end distance. This finding verifies that DAPI minor groove binding does not induce supercoiling to dsDNA and therefore elongation due to unwinding cannot be observed.
The results of the rotation experiments of DRAQ5, which are presented in Fig. 4 e, reveal features of both, minor groove binding and intercalation. At dye concentrations below 0.5 μM, DRAQ5 associates to DNA like a minor groove binder that can be seen from the overlaying hat curves (Fig. 4 e). At higher dye concentrations, the hat curves yield a substantial shift to negative rotation numbers and a slight increase of the end-to-end distance indicating an intercalation-like binding mechanism. Notably, as intercalation apparently is a less-distinct secondary binding mode, the increase of the molecule end-to-end distance as compared with YOYO-1 is rather small (Fig. 4 e). We calculated the unwinding angle within the intercalation regime as described before and estimated an unwinding of 0.036 ± 0.0127 turns per dye that is equivalent to per dye (Fig. 4 f). However, the exact quantity of might be affected by potential crosstalk between minor groove binding and intercalation as the lengthening per dye molecule Δx (Eq. 3) was estimated by the McGhee–von Hippel model (Eq. 2).
Effects on torsional elasticity
Disregarding the shift to negative rotation numbers and an increase of the end-to-end distance that has been discussed above, we furthermore observed a substantial broadening of the hat curves within the intercalation regime that can be interpreted alongside within the picture of torsional elasticity and the formation of topological plectonemes. There, the buckling number N defines the crossover regime where the ability of dsDNA to settle torsional stress along the strand is exhausted and topological plectonemes are formed instead. For YOYO-1 the buckling number shifted from N = 6 for bare dsDNA to N = 40 at a concentration of 5 μM. Notably, N was determined with respect to the new shifted center of the hat curve. Furthermore, the hat curves continuously flatten within the torsional elastic regime for increasing dye concentrations (Fig. 4 a). In contrast, the plectonemic regime (R > N) is virtually unchanged throughout the whole experiment. The shift of the buckling transition to higher rotation numbers together with the flattening of the hat curves indicates that YOYO-1 binding softens dsDNA, i.e., the ability to release torque along the double strands is enhanced. Hence, more twists have to be applied until buckling and the formation of plectonemes are induced. To quantify the effect of intercalation we analyzed our data within the following simplified torsional elasticity model (23, 32, 34):
| (4) |
where the torsional elasticity of dsDNA in the presence and absence of dye molecules are denoted by and , respectively. Similarly, L, P, and N represent the contour length, the persistence length, and the buckling number, respectively. At a YOYO-1 concentration of c = 5 μM we observed a relative lengthening of and a virtually unchanged persistence length . Moreover a significantly reduced relative torsional elasticity of was discerned. These results are consistent with recently published data on the (mono-) intercalator ethidium bromide (32).
In contrast, DAPI minor groove binding has only a very small impact on the shape of the hat curves. At a concentration of c = 10 μM, the rotation-extension curve yields only a minimal shift of the buckling number (N = 6 -> N = 8) and a slight broadening of the torsional elastic regime (Fig. 4 c). As DAPI does not unwind the dsDNA, we did not observe any lengthening of the DNA contour . Nevertheless, the reduction of the bending stiffness (persistence length) that we have seen in the extension experiments has a considerable effect on the torsional elasticity. Accordingly, we estimated a torsional softening by a factor of 1.42 .
On the basis of these findings we can interpret the results of DRAQ5 association on the torsional elasticity of dsDNA. In the primary, minor groove binding mode ( μM) the DNA softens due to the decrease of the persistence length , whereas the shift of the buckling number and the elongation of the contour length can be neglected. Evidently, the torsional softening due to minor groove binding is rather moderate (by a factor of 1.61), which is consistent with the results found for DAPI.
At dye concentrations above 0.5 μM, where DRAQ5 intercalation dominates, the persistence length stays constant and the buckling number increases significantly (N = 6 -> N = 28). Furthermore, we observed an increase of the contour length . Yet, this effect is unincisive, which probably is due to the fact that intercalation itself is a less-pronounced binding mode. Finally, minor groove binding and intercalation of DRAQ5 at the concentration of 10 μM lead to a decrease of the torsional stiffness by a factor of 4.35 .
Conclusions
In summary, we investigated the nanomechanical effects on dsDNA due to dye association. Therefore, we used MTs to perform various force-extension and rotation-extension experiments in a concentration-dependent manner.
In case of the bis-intercalator YOYO-1 we found a significant increase of the contour length that we could attribute to a gradual unwinding of the DNA double helix. In contrast the persistence length remained virtually unchanged independent of the YOYO-1 concentration. Presuming mono-intercalation and unspecific adhesion of YOYO-1 to the dsDNA backbone, we could provide a consistent explanation for disparate reports concerning the impact of intercalation on the persistence length. We furthermore approximated our data according to the McGhee–von Hippel model and estimated the binding site size per dye molecule, the induced elongation per dye, and the equilibrium constant of association that are well supported by recently published works.
The minor groove binder DAPI caused no significant elongation of the DNA contour length that is also supported by our rotation extension experiments, where we could not find evidence of DNA unwinding. In contrast, the persistence length decreased substantially upon increasing the DAPI concentration that is in agreement with recently published data.
On the basis of these results we categorized the characteristic phenomena of intercalation and minor groove binding, respectively. Within this scheme we analyzed the DRAQ5 binding data and consistently identified a bimodal association behavior: at low dye concentrations DRAQ5 associates to DNA as a minor groove binder. We observed a decrease of the persistence length but neither an increase of the contour length nor an unwinding of the DNA double helix.
At high DRAQ5 concentrations, we found an intercalator-like behavior, i.e., the lengthening of the DNA due to unwinding at a constant persistence length. In the rotation extension experiments dsDNA decorated with DRAQ5 significant broadening of the hat curves that also clearly indicate intercalation. Furthermore, we estimated a threshold concentration at which the primary minor groove binding declines and intercalation becomes the dominant binding mode.
Finally, we estimated the effect of dye association on the torsional elasticity within the framework of a simplified model. Interestingly, both minor groove binding and intercalation induce a reduction of the dsDNA torsional stiffness. Yet, softening due to intercalation is the major contribution whereas minor groove binding plays only a minor role. In full accordance with our previous results the reduction of the dsDNA elasticity due to DRAQ5 binding can be divided into two “softening regimes”: at low concentrations of DRAQ5 (in the minor groove binding regime) torsional softening is less pronounced whereas in the intercalation regime the torsional elasticity drops by a factor of four.
Author Contributions
D.A., K.T., and A.S. designed the research. K.T. contributed substantially to the sample preparation. Y.W. performed all measurements. Y.W., A.S., and V.W. analyzed the data. Y.W., V.W., and D.A. wrote the manuscript.
Editor: Keir Neuman
Footnotes
Two figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30765-2.
Supporting Material
References
- 1.Rye H.S., Yue S., Glazer A.N. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and applications. Nucleic Acids Res. 1992;20:2803–2812. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins T.T., Smith D.E., Chu S. Single polymer dynamics in an elongational flow. Science. 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 3.Quake S.R., Babcock H., Chu S. The dynamics of partially extended single molecules of DNA. Nature. 1997;388:151–154. doi: 10.1038/40588. [DOI] [PubMed] [Google Scholar]
- 4.Sischka A., Toensing K., Anselmetti D. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophys. J. 2005;88:404–411. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murade C.U., Subramaniam V., Bennink M.L. Interaction of oxazole yellow dyes with DNA studied with hybrid optical tweezers and fluorescence microscopy. Biophys. J. 2009;97:835–843. doi: 10.1016/j.bpj.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maaloum M., Muller P., Harlepp S. DNA-intercalator interactions. structural and physical analysis using atomic force microscopy in solution. Soft Matter. 2013;9:11233–11240. [Google Scholar]
- 7.Shi N., Ugaz V.M. An entropic force microscope enables nano-scale conformational probing of biomolecules. Small. 2014;10:2553–2557. doi: 10.1002/smll.201303046. [DOI] [PubMed] [Google Scholar]
- 8.Günther K., Mertig M., Seidel R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010;38:6526–6532. doi: 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansen F., Jacobsen J.P. 1H NMR studies of the bis-intercalation of a homodimeric oxazole yellow dye in DNA oligonucleotides. J. Biomol. Struct. Dyn. 1998;16:205–222. doi: 10.1080/07391102.1998.10508240. [DOI] [PubMed] [Google Scholar]
- 10.Kundukad B., Yan J., Doyle P.S. Effect of YOYO-1 on the mechanical properties of DNA. Soft Matter. 2014;10:9721–9728. doi: 10.1039/c4sm02025a. [DOI] [PubMed] [Google Scholar]
- 11.Barcellona M.L., Cardiel G., Gratton E. Time-resolved fluorescence of DAPI in solution and bound to polydeoxynucleotides. Biochem. Biophys. Res. Commun. 1990;170:270–280. doi: 10.1016/0006-291x(90)91270-3. [DOI] [PubMed] [Google Scholar]
- 12.Parolin C., Montecucco A., Palu G. The effect of the minor groove binding agent DAPI (2-amidino-diphenyl-indole) on DNA-directed enzymes: an attempt to explain inhibition of plasmid expression in Escherichia coli [corrected] FEMS Microbiol. Lett. 1990;68:341–346. doi: 10.1111/j.1574-6968.1990.tb13962.x. [DOI] [PubMed] [Google Scholar]
- 13.Tanious F.A., Veal J.M., Wilson W.D. DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry. 1992;31:3103–3112. doi: 10.1021/bi00127a010. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson S., Kim S.K., Nordén B. Binding of 4′,6-diamidino-2-phenylindole (DAPI) to AT regions of DNA: evidence for an allosteric conformational change. Biochemistry. 1993;32:2987–2998. doi: 10.1021/bi00063a009. [DOI] [PubMed] [Google Scholar]
- 15.Japaridze A., Benke A., Dietler G. Influence of DNA binding dyes on bare DNA structure studied with atomic force microscopy. Macromolecules. 2015;48:1860–1865. [Google Scholar]
- 16.Thomas J.A. Optical imaging probes for biomolecules: an introductory perspective. Chem. Soc. Rev. 2015;44:4494–4500. doi: 10.1039/c5cs00070j. [DOI] [PubMed] [Google Scholar]
- 17.Martin R.M., Leonhardt H., Cardoso M.C. DNA labeling in living cells. Cytometry A. 2005;67:45–52. doi: 10.1002/cyto.a.20172. [DOI] [PubMed] [Google Scholar]
- 18.Njoh K.L., Patterson L.H., Smith P.J. Spectral analysis of the DNA targeting bisalkylaminoanthraquinone DRAQ5 in intact living cells. Cytometry A. 2006;69:805–814. doi: 10.1002/cyto.a.20308. [DOI] [PubMed] [Google Scholar]
- 19.Islam S.A., Neidle S., Brown J.R. Comparative computer graphics and solution studies of the DNA interaction of substituted anthraquinones based on doxorubicin and mitoxantrone. J. Med. Chem. 1985;28:857–864. doi: 10.1021/jm00145a003. [DOI] [PubMed] [Google Scholar]
- 20.Cheng, W. Protocol to generate half LAMBDA DNA for optical/magnetic tweezer.http://tweezerslab.unipr.it/cgi-bin/mt/documents.pl/Show?_id=ab03&sort=DEFAULT&search=&hits=23. Accessed November 4, 2015.
- 21.Strick T.R., Allemand J.-F., Croquette V. Behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strick T.R., Allemand J.-F., Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 23.Vilfan, I. D., J. Lipfert, …, N. H. Dekker. 2009. Handbook of Single-Molecule Biophysics. P. Hinterdorfer and A. van Oijen, editors. Springer, New York.
- 24.Smith S.B., Finzi L., Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 25.Bouchiat C., Wang M.D., Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 27.Paik D.H., Perkins T.T. Dynamics and multiple stable binding modes of DNA intercalators revealed by single-molecule force spectroscopy. Angew. Chem. Int. Ed. Engl. 2012;51:1811–1815. doi: 10.1002/anie.201105540. [DOI] [PubMed] [Google Scholar]
- 28.Petty J.T., Bordelon J.A., Robertson M.E. Thermodynamic characterization of the association of cyanine dyes with DNA. J. Phys. Chem. B. 2000;104:7221–7227. [Google Scholar]
- 29.Kleimann C., Sischka A., Anselmetti D. Binding kinetics of bisintercalator Triostin a with optical tweezers force mechanics. Biophys. J. 2009;97:2780–2784. doi: 10.1016/j.bpj.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams L.D., Egli M., Rich A. Vol. 1. Adenine Press; New York: 1992. pp. 107–125. (Structure & Function). [Google Scholar]
- 31.Vladescu I.D., McCauley M.J., Williams M.C. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat. Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- 32.Lipfert J., Klijnhout S., Dekker N.H. Torsional sensing of small-molecule binding using magnetic tweezers. Nucleic Acids Res. 2010;38:7122–7132. doi: 10.1093/nar/gkq598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salerno D., Brogioli D., Mantegazza F. Magnetic tweezers measurements of the nanomechanical properties of DNA in the presence of drugs. Nucleic Acids Res. 2010;38:7089–7099. doi: 10.1093/nar/gkq597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strick T.R., Dessinges M.-N., Croquette V. Stretching of macromolecules and proteins. Rep. Prog. Phys. 2003;66:1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.