Abstract
Autophagy is emerging as a key regulatory process during skeletal muscle development, regeneration and homeostasis, and deregulated autophagy has been implicated in muscular disorders and age-related muscle decline. We have monitored autophagy in muscles of mdx mice and human Duchenne muscular dystrophy (DMD) patients at different stages of disease. Our data show that autophagy is activated during the early, compensatory regenerative stages of DMD. A progressive reduction was observed during mdx disease progression, in coincidence with the functional exhaustion of satellite cell-mediated regeneration and accumulation of fibrosis. Moreover, pharmacological manipulation of autophagy can influence disease progression in mdx mice. Of note, studies performed in regenerating muscles of wild-type mice revealed an essential role of autophagy in the activation of satellite cells upon muscle injury. These results support the notion that regeneration-associated autophagy contributes to the early compensatory stage of DMD progression, and interventions that extend activation of autophagy might be beneficial in the treatment of DMD. Thus, autophagy could be a ‘disease modifier' targeted by interventions aimed to promote regeneration and delay disease progression in DMD.
Duchenne muscular dystrophy (DMD) is a lethal muscle degenerative disease caused by mutations in the dystrophin gene.1 Dystrophin is a scaffold protein that binds several structural and signaling proteins to the sarcolemma, thus having an essential role in membrane integrity2 and in regulating signaling pathways such as those activating nitric oxide (NO) and reactive oxygen species (ROS) production.3 Accordingly, loss of dystrophin results in increased myofiber fragility4 and deregulated signaling pathways. Recent evidence demonstrates that defects in dystrophin lead to intrinsic satellite cells dysfunction, including loss of cell polarity and reduced number of self-renewing cells, resulting in compromised muscle regeneration.5 Early in life, DMD patients can temporarily counter the continuous degeneration imposed by contractile activity of dystrophin-deficient myofibers through a compensatory regeneration mediated by adult muscle stem (satellite) cells (MuSCs).6, 7 However, the regenerative potential of MuSCs declines at later stages of DMD progression and muscle tissues are supplanted by fibrosis, calcium deposits and fat infiltration leading to the clinical manifestations in DMD patients.8, 9
Muscle regeneration is orchestrated by the coordinate interplay between MuSCs and different cell types that compose the regenerative environment. Alterations of these networks have been associated to muscular chronic diseases and aging.10, 11, 12, 13
Different biological processes evolved to preserve tissue homeostasis in eukaryotes. Among them, autophagy is emerging as a key protective mechanism in many tissues.14 So far, three types of autophagy have been described: macroautophagy; microautophagy; and chaperone-mediated autophagy. The most studied mechanism is macroautophagy, referred hereafter as autophagy, consisting of trafficking machinery, in which portions of cytoplasm, organelles and long-lived proteins are engulfed into double-membrane vesicles (named autophagosomes) and fused with the lysosome for content degradation. This process allows recycling of macromolecules to provide energy-rich compounds and to eliminate toxic molecules, thereby enabling cells and tissues adaptation under stress conditions15, 16 or permitting structural remodeling (that is, during development).17 However, in specific circumstances, the self-cannibalistic or the same pro-survival functions of autophagy may be deleterious and contribute to the pathogenesis of different diseases.18
Growing evidence indicates that autophagy is required to maintain muscle mass and myofiber integrity.19 Indeed, muscle-specific deletion of autophagy genes, such as atg7 and atg5 results in muscle atrophy and age-dependent decrease in force.20 Therefore, it is possible to outline a dual role of autophagy in muscle homeostasis; on one side, defective autophagy compromises tissue clearance of damaged proteins, toxic compounds and organelles; conversely, excessive autophagy leads to muscle loss and atrophy, pointing to autophagy as a highly sensitive process for fine-tuning muscle mass and functions.
In muscular dystrophies, autophagy has been reported to be impaired in collagen VI-deficient muscles, which accumulate dysfunctional organelles and undergo muscle wasting.21 Reactivation of the autophagic flux by nutritional, pharmacological or genetic tools ameliorates the dystrophic phenotype by removing dysfunctional mitochondria.21, 22, 23 Recent investigations have begun to elucidate the role of autophagy in other muscular disorders, including DMD.24, 25, 26, 27 Moreover, it has been shown that autophagy is implicated in loss of muscle mass in the elderly, referred as sarcopenia,28, 29 as well as in myofiber survival.30 Multiple lines of evidence indicate that autophagy diminishes with aging,31 premature aging correlates with autophagy inhibition,29 while increased autophagy (as by mean of calorie restriction) delays aging and extends longevity.32, 33 Of note, the evidence that calorie restriction enhances MuSCs availability and activity34 indicates a close relationship between autophagy, the regenerative potential of skeletal muscles, MuSCs activation35 and self-renewal.36 A progressive reduction of basal autophagy from young to old and geriatric MuSCs triggering numerical and functional MuSCs decline during ageing has been recently described.37 However, whether autophagy is implicated in MuSC-mediated regeneration of DMD muscles remains elusive.
Here we show that autophagy is activated during muscle regeneration at early compensatory stages of DMD progression both in a mouse model of disease (the mdx mice) and in human biopsies isolated from DMD boys. Impaired activation of autophagy at late stages of disease progression correlates with the decline in regeneration and fibrotic tissue deposition in dystrophic muscles. We also show that autophagy is activated in MuSCs from regenerating muscles of wild-type (WT) mice. Finally, we demonstrated that modulating autophagy impacts the regenerative response in WT and mdx mouse model.
Results
Autophagy is modulated during disease progression in mdx mice
We set a systematic analysis of autophagy during DMD by monitoring the autophagic flux during the sequential stages of disease progression in the mdx mouse model. LC3 lipidation (LC3II form) was monitored by western blot from extracts of tibialis anterior (TA) and diaphragm (D) muscles isolated from WT and mdx mice at early (1.5 months), intermediate (5 months) and late (8 and 12 months) stages of disease progression (Supplementary Figure S1a). We observed increased LC3II/GAPDH ratio in mdx mice at early-middle stages, as compared with the age-matched WT mice (Supplementary Figure S1b–g). Although the LC3II/GAPDH ratio from WT mice decreases at 12 months in TA and remains unchanged in D muscles, in mdx mice the LC3II/GAPDH ratio gradually declines from the fifth through the eighth and twelfth month in both TA and D (Supplementary Figure S1b–g). We also monitored the accumulation of the sequestosome1 (SQSTM1), also called p62 that links ubiquitinated proteins to the autophagic machinery for degradation. To assess whether autophagy is active or inactive, we analyzed p62 at protein and RNA levels, being the transcriptionally activation or reduction associated to active or inactive autophagy, respectively.38 We show p62 protein levels increased at later stages of the disease in TA and D from mdx while remaining rather stable in TA and accumulating at 12 months in D from WT mice (Supplementary Figure S1b–g). By monitoring lc3 and p62 expression levels, as well as other genes involved in the autophagic process, such as beclin1 and vps34 we found again an increase of lc3 and p62 transcripts in TA muscles of mdx mice at early stages (1.5 month), as compared with the WT counterpart. At later stages, we observed a reduction of all the autophagic markers included p62, indicating a block of autophagy (Supplementary Figure S1h).
Dynamic changes in autophagy distinguish mdx mice from the WT counterpart
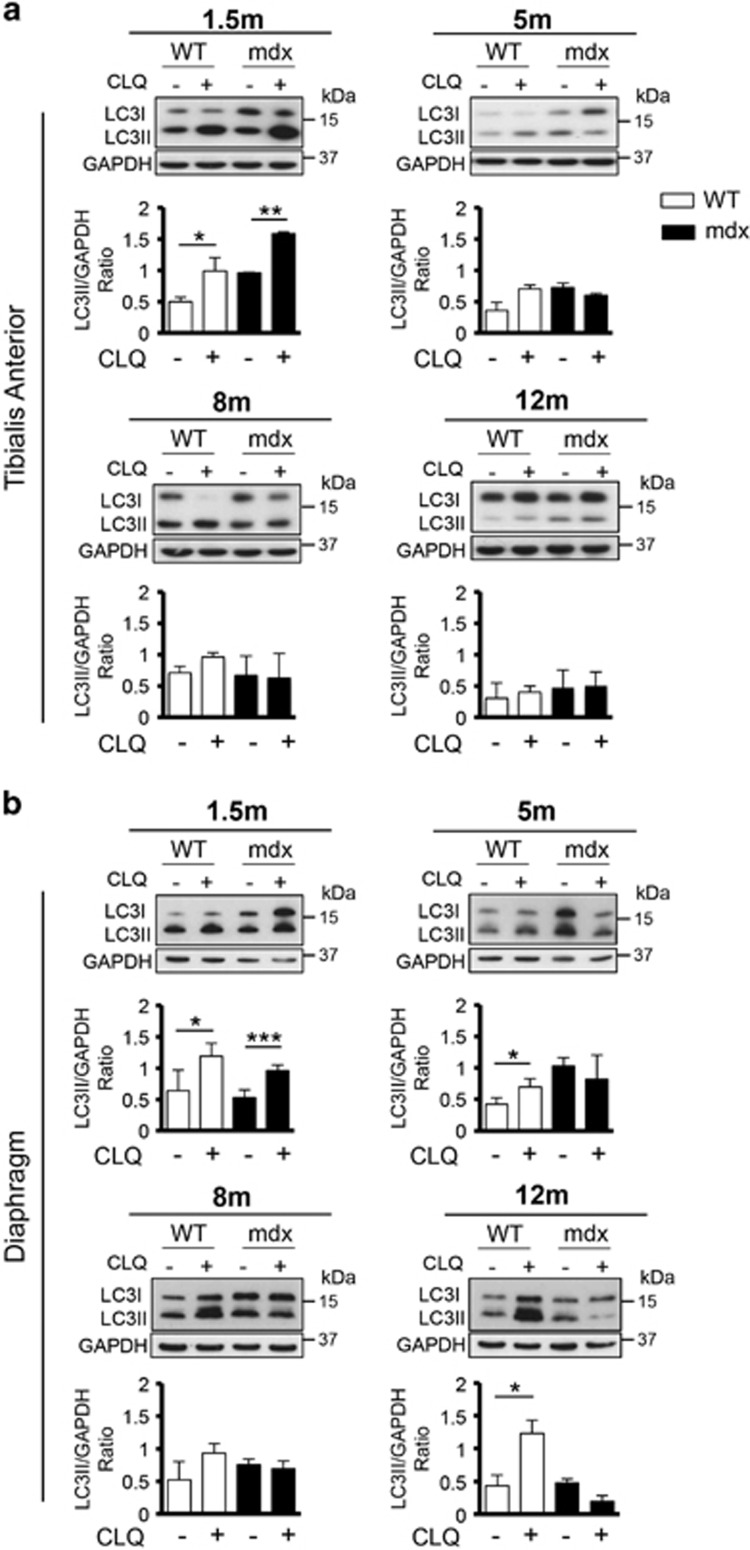
Given the highly dynamic nature of autophagy and the fact that increased LC3II levels may be a consequence of both activation or block of the autophagic flux, we treated WT and mdx mice at different ages with chloroquine (CLQ), an agent that causes a block of the autophagic flux39 at the dosage of 50 mg/kg by intra-peritoneal (i.p.) injections executed daily for 4 days. Under these experimental conditions, accumulation of LC3II is indicative of active autophagy. CLQ was administered to 1.5, 5, 8 and 12-month-old mdx mice and their WT counterpart, and accumulation of the LC3-lipidated form was analyzed in both TA (Figure 1a) and D (Figure 1b) muscles. In unperturbed muscles of WT mice, we observed an active autophagic flux at 1.5 month, a moderate activation of autophagy at 5 and 8 months, and a block of autophagy in 12-month-old mice (Figure 1a), as previously reported in other aged animals.28, 29 In dystrophic mice, we show a significant increase of LC3II in TA at 1.5 month of age indicative of active autophagic flux that precociously decreases at middle and late stages of disease (Figure 1a). As the impairment of the diaphragm in DMD patients is responsible for respiratory failure, we extended our analysis to this muscle in mdx mice.40 We detected again differences in autophagic flux between diaphragms from WT, in which autophagy is active in all age groups, and mdx mice that display an impairment of the autophagic process starting from 5 months of age (Figure 1b). Overall, our data reveal a dynamic regulation of autophagy in mdx mice, characterized by an active autophagic flux during the early stages, and its premature blockade at later stages of disease progression.
Figure 1.
Premature block of autophagic flux in mdx mice compared with WT mice. To assess the autophagic flux in control and pathological conditions, WT and mdx mice at 1.5, 5, 8 and 12 months were treated with CLQ (50 mg/kg every 24 h for 4 days). Protein extracts from TA (a) and D (b) muscles were probed for LC3 and GAPDH. A representative western blot for every time point is shown. The plots represent LC3II/GAPDH ratio relative to WT (white bars) and mdx (black bars) mice; n=4 for each experimental group. Statistical significance assessed by t-test, *P≤0.05, **P≤0.01, ***P≤0.001
Failure to activate autophagy coincides with increased fibrosis and the exhaustion of the regeneration potential of dystrophic muscles
To establish a relationship between autophagy and the pathological transitions during the course of DMD, we monitored the deposition of connective tissue, a feature of advanced stages of disease,41 by performing a Masson's Trichrome staining in tissue sections from TA and D muscles isolated from WT and mdx mice at the same time points analyzed in Figure 1 (Supplementary Figure S2a–d). We observed a moderate accumulation of fibrotic tissue in TA and D from WT mice throughout 12 months of life-span (Supplementary Figure S2a, c), while in mdx mice the fibrotic deposition starts at 5 months of age and increases at later stages both in D (Supplementary Figure S2b) and in TA muscles (Supplementary Figure S2d). To identify the newly formed fibers generated upon productive regeneration, we performed a staining for embryonal myosin heavy chain (eMyHC) on TA muscle sections (Supplementary Figure S2e, f). Although in unperturbed WT mice there is no evidence of regeneration (Supplementary Figure S2e), we detected eMyHC+ fibers in mdx mice at early stages of the disease, followed by a progressive decline already manifest in 5-month old and a complete absence of eMyHC+ fibers in 12-month-old mdx mice (Supplementary Figure S2f). Collectively, these data reveal that the failure to activate autophagy in mdx mice coincides with an incipient fibrosis and impaired regeneration as the disease progresses.
Compromised autophagic response in human biopsies from older DMD boys displaying altered MuSC activation
We next analyzed the activation of autophagy in biopsies isolated from DMD boys at two distinct stages of disease progression—that is, 2- and 8-year-old patients. Masson's trichrome staining showed a progressive increase of fibrosis at advanced stages of the pathology (Figure 2a and b). LC3 staining reveals no detectable levels in biopsies isolated from control samples, while increased LC3 expression was detected in 2-year-old DMD boys. LC3 levels declined at later stages of DMD progression, as shown by biopsies isolated from 8-year-old boys (Figure 2a and c). We next sought to determine the cell type in which autophagy is activated, by performing a double staining of LC3 together with either MyoD (Figure 2d and e) or Pax7 (Figure 2f and g). Our results show an overall reduction of LC3+, MyoD+ and double-positive cells in older patients, while the number of Pax7+ cells that activates LC3 remains unchanged along DMD progression. Interestingly, the increased amounts of LC3 co-segregate with MyoD-committed cells when the compensatory regeneration is sustained in young DMD boys, whereas in older DMD patients the number of LC3/MyoD double-positive cells significantly declines, in coincidence with compromised muscle morphology. Altogether, these data suggest a potential relationship between autophagy and the control of muscle stem cell activity during compensatory regeneration in DMD muscles.
Figure 2.
Altered autophagic response in human biopsies from DMD boys at different ages. Autophagy was monitored in human biopsies isolated from vastus medialis of 2- and 8-year-old dystrophic boys. (a, left) Representative images of Masson's trichrome staining on 10 μm slices of human biopsies from control (CTR), 2- and 8-year-old DMD patients. (right) Representative images of slices of human biopsies from control, 2- and 8-year-old DMD patients were probed for LC3 (green) and laminin (blue); nuclei were detected with DAPI (white). (b) Graph representing the fibrotic index (quantification of collagen deposition) of muscles measured as the percentage of blue fibrotic area on total muscle area (10 fields for each sample). (c) Plot representing the percentage of LC3-positive cells counted in slices of human biopsies from control, 2- and 8-year-old DMD patients based on the average of 12 fields for each experimental group. (d) Representative images of immunostaining for LC3 (green), MyoD (red) and laminin (blue) of 2- and 8-year-old DMD patients muscle biopsies; nuclei were detected with DAPI (white). (e) Schematic representation of the percentage of LC3-/MyoD+, LC3+/MyoD-, LC3+/MyoD+ cells counted in 2- and 8-year-old DMD patients muscle biopsies based on the average of 12 fields for each experimental group. (f) Representative images of immunostaining for LC3 (green), Pax7 (red) and Laminin (blue) of 2- and 8-year-old DMD patients muscle biopsies; nuclei were detected with DAPI (white). (g) Schematic representation of the percentage of LC3-/Pax7+, LC3+/Pax7- and LC3+/Pax7+ cells counted in 2- and 8-year-old DMD patients muscle biopsies based on the average of 12 fields for each experimental group. n.s., not significative. Scale bar, 50 μm. Statistical significance assessed by t-test, *P≤0.05, **P≤0.01, ***P≤0.001
Autophagy contributes to activate compensatory regeneration in dystrophic muscles
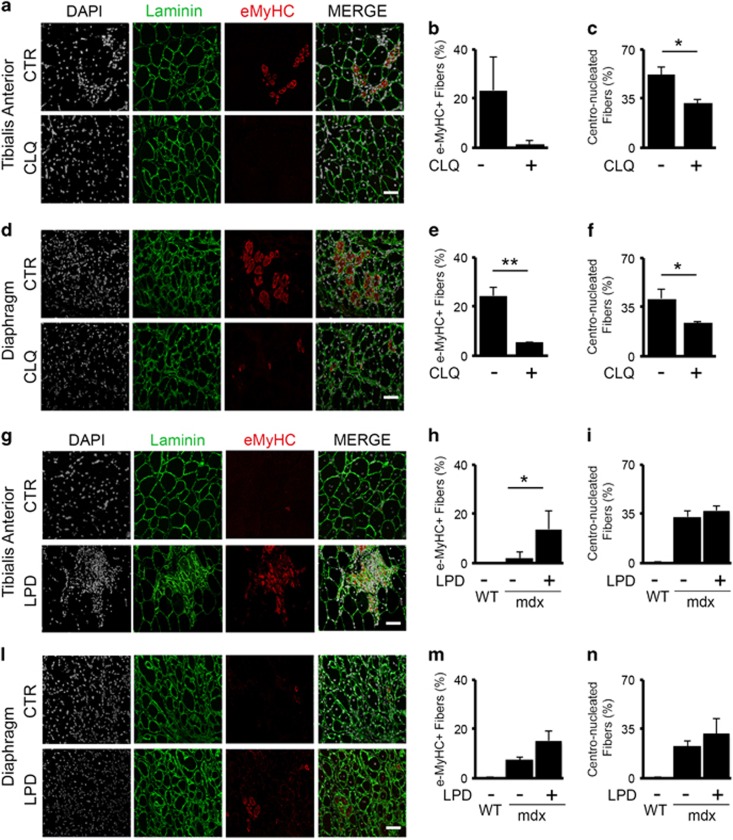
To directly investigate the impact of autophagy on disease progression, we interfered with the autophagic process in 2-month-old mdx mice, when autophagy is active, by in vivo treatments with CLQ administered at the dosage of 50 mg/kg by i.p. injections executed daily for 14 days (Figure 3a–f and Supplementary Figure S3a–b). Conversely, we stimulated autophagy in 5-month-old mdx mice – a stage in which autophagy begins to decline – by feeding animals with a low-protein diet (LPD) for 60 days24 (Figure 3g–n and Supplementary Figure S3c–d). CLQ administration (Supplementary Figure S3a–b) led to reduced number of eMyHC+ fibers in both TA (Figure 3a and b) and D (Figure 3d and e) muscles. Accordingly, upon CLQ treatment we measured a lower percentage of centro-nucleated fibers in both TA (Figure 3c) and D (Figure 3f), suggesting an impairment of the regenerative process. On the other hand, reactivating autophagy in 5-month-old mdx mice by LPD (Supplementary Figure S3c, d) counteracts the decline of muscle regeneration, as measured by the increased percentage of eMyHC+ fibers in both TA (Figure 3g and h) and D (Figure 3l and m). LPD does not affect the total percentage of centro-nucleated fibers counted in TA (Figure 3i) and D (Figure 3n) from mdx mice possibly as a result of the switch of former centro-nucleated to mature fibers. Altogether, this evidence supports the conclusion that autophagy promotes compensatory regeneration in DMD.
Figure 3.
Autophagic modulation impacts muscle regeneration in dystrophic mice. Representative images of TA (a) and D (d) isolated from 2-month-old mdx mice untreated (CTR) or treated with CLQ, administered at the dosage of 50 mg/kg by i.p. injections executed daily for 14 days, immunostained with laminin (green), eMyHC (red); nuclei were detected with DAPI (white) (n=5 for each experimental group). Plot representing the percentage of eMyHC-positive fibers in untreated and CLQ-treated mdx mice isolated from TA (b) and D (e) based on the average of eight fields for mice. Graph representing the percentage of centro-nucleated fibers counted in untreated and CLQ-treated mdx mice isolated from TA (c) and D (f) based on the average of eight fields for mice. Representative images of TA (g) and D (l) isolated from 5-month-old mdx mice fed with standard diet (CTR) or fed with LPD for 60 days, immunostained with laminin (green), eMyHC (red); nuclei were detected with DAPI (white) (n=3 for each experimental group). Plot representing the percentage of eMyHC-positive fibers in 5-month-old WT mice, in age-matched mdx mice fed with standard diet and LPD-fed isolated from TA (h) and D (m) based on the average of eight fields for mice. Graph representing the percentage of centro-nucleated fibers counted in WT, mdx standard-fed and LPD-fed mdx mice isolated from TA (i) and D (n) based on the average of eight fields for mice. Statistical significance assessed by t-test, *P≤0.05, **P≤0.01
Autophagy regulates MuSC cell activity
The correlation observed between autophagy and the compensatory regeneration at early stages of DMD progression prompted our interest to further investigate whether autophagy could directly regulate MuSCs activity, in context of the conventional model of muscle damage induced by cardiotoxin injection to elicit a regenerative response in WT muscles. We injured muscles of 2-month-old WT mice analyzing two different time points, 5 and 15 days after injury, to evaluate the peak of the regenerative response and the resolution of the lesions, respectively.
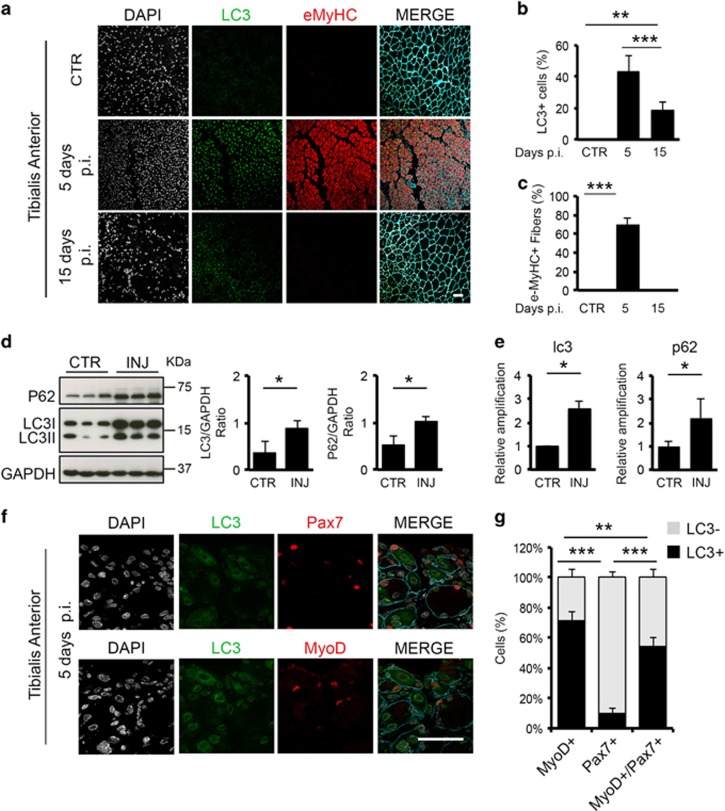
Albeit we detected almost no cells expressing LC3 under basal conditions (Figure 4a and b), we observed a marked increase of LC3 5 days post injury (p.i.) (Figure 4a and b) – a timing consistent with the presence of differentiation-committed MuSCs. Indeed, the appearance of LC3+ cells coincided with a robust regenerative response identified by the formation of new myofibers marked by eMyHC (Figure 4a and c). A biochemical analysis of protein extracts obtained from muscles harvested 5 days p.i. demonstrated that upon muscle damage p62 is highly expressed, LC3 becomes lipidated (Figure 4d), and both are transcriptionally activated (Figure 4e), indicating the induction of autophagy during regeneration of injured WT muscles.
Figure 4.
The autophagic process contributes to muscle regeneration. Muscle damage was induced in the right leg of WT mice by injection of 20 μl of cardiotoxin (10 μM), while contro-lateral leg was left uninjured (CTR) (n=3 for each experimental group). (a) Representative images of control (CTR), 5 and 15 days p.i. of TA sections immunostained for eMyHC (red), LC3 (green) and laminin (blue); nuclei were detected with DAPI (white). (b) Graphs showing the percentage of LC3-positive cells in uninjured muscles (CTR) and after 5 and 15 days p.i. (c) Graphs showing the percentage of eMyHC-positive fibers in uninjured muscles (CTR) and after 5 and 15 days p.i. based on the average of eight fields for mice. (d) Protein extracts from whole muscles isolated from uninjured (CTR) and injured (INJ) TA 5 days p.i. were probed for p62, LC3 and GAPDH as loading control. Plots represent LC3II/GAPDH ratio and p62/GAPDH ratio based on the average for each experimental point (n=3). (e) qRT-PCR analysis of lc3 and p62 gene expression assessed in control (CTR) and after 5 days p.i. (INJ) TA muscles. (f) Representative images of serial cryosections from injured TA 5 days p.i. immunostained with LC3 (green), Pax7 (red/top panel), MyoD (red/bottom panel) and Laminin (blue); nuclei were detected with DAPI (white). (g) Schematic representation of the percentage of LC3 negative (grey bars) and LC3 positive (black bars) cells within MyoD-positive, Pax7-positive and MyoD/Pax7-double-positive cells, based on the average of eight fields for each mice. Scale bar, 50 μm. Statistical significance assessed by t-test, *P≤0.05, **P≤0.01, ***P≤0.001
We therefore extended our studies and analyzed the co-localization of LC3 with markers of MuSCs quiescence/self renewal (Pax7+/MyoD-), transient amplification (Pax7+/MyoD+) or commitment to differentiation (Pax7-/MyoD+) in TA sections from WT mice 5 days p.i. (Figure 4f). The percentage of LC3+/MyoD-/Pax7- cells was ~82%, indicating that cells other than MuSCs display active autophagy (data not shown); however, focusing on MuSCs, we where able to detect that among MyoD+ cells the 71.8% displayed LC3 positivity, while only the 9.6% of the Pax7+ cells are LC3+. The percentage of MyoD/Pax7 double positive is almost equally distributed among the LC3-positive and LC3-negative cells, suggesting that autophagy is selectively active in MyoD+ MuSCs (Figure 4g). This evidence supports the hypothesis that autophagy might have a part in cellular determination during muscle regeneration affecting the myogenic differentiation but not the self-renewing properties of adult stem cells.
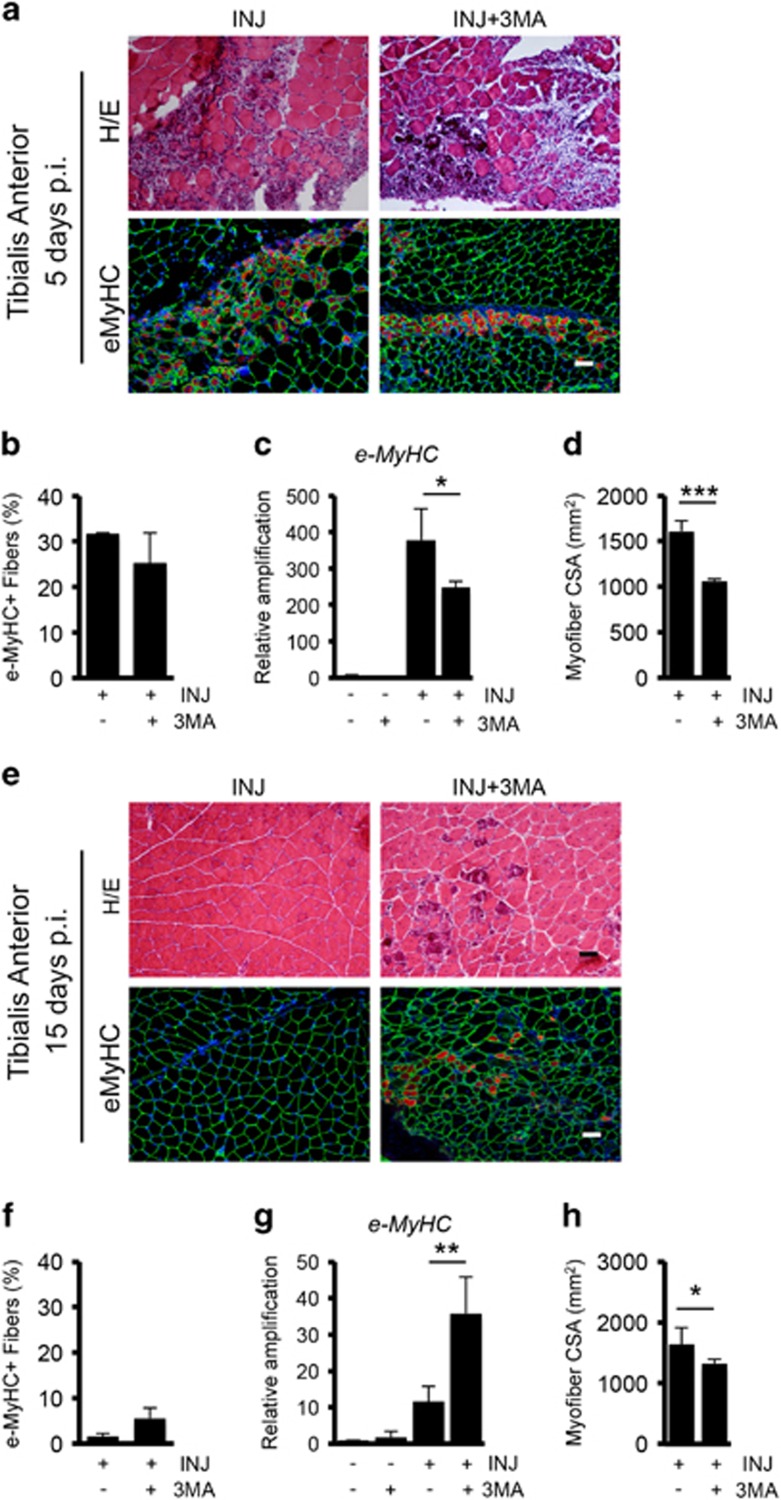
To determine the functional contribution of autophagy to muscle repair activity, we treated WT mice with the autophagy inhibitor 3-methyladenine (3-MA), administered daily at the dosage of 10 mg/kg by i.p. injections after muscle damage as described in Supplementary Figure S4a. We first validated that upon 3-MA in vivo treatment, the autophagic process was reduced, evaluating the percentage of LC3+ cells in TA muscle tissue sections (Supplementary Figure S4b), by western blot analysis for LC3 and p62 (Supplementary Figure S4c), and by real-time quantitative PCR (qRT-PCR) for autophagic markers (Supplementary Figure S4d). Next, we analyzed the effect of reduced autophagy on the regenerative process. At 5 days p.i. the formation of eMyHC+ fibers was slightly reduced in injured mice treated with 3-MA (Figure 5a–c). Importantly, we observed a concomitant reduction of myofibers cross-sectional area (CSA) in regenerating muscle of 3-MA-treated mice after 5 days p.i., indicating a less efficient regeneration process when autophagy is blocked (Figure 5d). Indeed, while control injured mice displayed a total recovery of muscle damage after 15 days, in 3-MA-treated mice the regenerative response was not completely resolved, as indicated by the persistence of eMyHC+ fibers (Figure 5e–g). The reduced CSA observed in 3-MA treated respect to the control mice (Figure 5h) suggests a delay in muscle repair upon autophagy blockade.
Figure 5.
The inhibition of autophagy reduces muscle repair. (a, top) Representative images of Hematoxilin/Eosin (H/E) staining on TA sections from injured WT mice treated or not with 3-MA (10 mg/kg daily i.p. injection for 5 days) analyzed 5 days p.i., n=3 for each experimental group. (bottom) Immunostaining for eMyHC (red) and Laminin (green) on TA sections from injured WT mice treated or not with 3-MA for 5 days analyzed at 5 days p.i. Nuclei were counterstained with DAPI (blue). (b) Plot representing the percentage of eMyHC based on the average of six fields for each sample. (c) qRT-PCR analysis of eMyHC gene expression analyzed in uninjured and injured WT mice treated or not with 3-MA and harvested 5 days p.i. (d) Quantification of average myofiber CSA 5 days after injury based on the average of six fields for each sample. (e, top) Representative images of H/E staining on TA sections from injured WT mice treated or not with 3-MA (10 mg/kg daily i.p. injection for 15 days) analyzed 15 days p.i. n=3 for each experimental group. (bottom) Immunostaining for eMyHC (red) and laminin (green) on TA sections from injured WT mice treated or not with 3-MA for 15 days analyzed 15 days p.i. (f) Plot representing the percentage of eMyHC based on the average of six fields for each sample. (g) qRT-PCR analysis of eMyHC gene expression analyzed in uninjured and injured WT mice treated or not with 3-MA and harvested 15 days p.i. (h) Quantification of average myofiber CSA 15 days after injury based on the average of six fields for each sample. Scale bar, 50 μm. Statistical significance assessed by t-test, *P≤0.05, **P≤0.01, ***P≤0.001
The autophagic process is induced in MuSCs during muscle regeneration
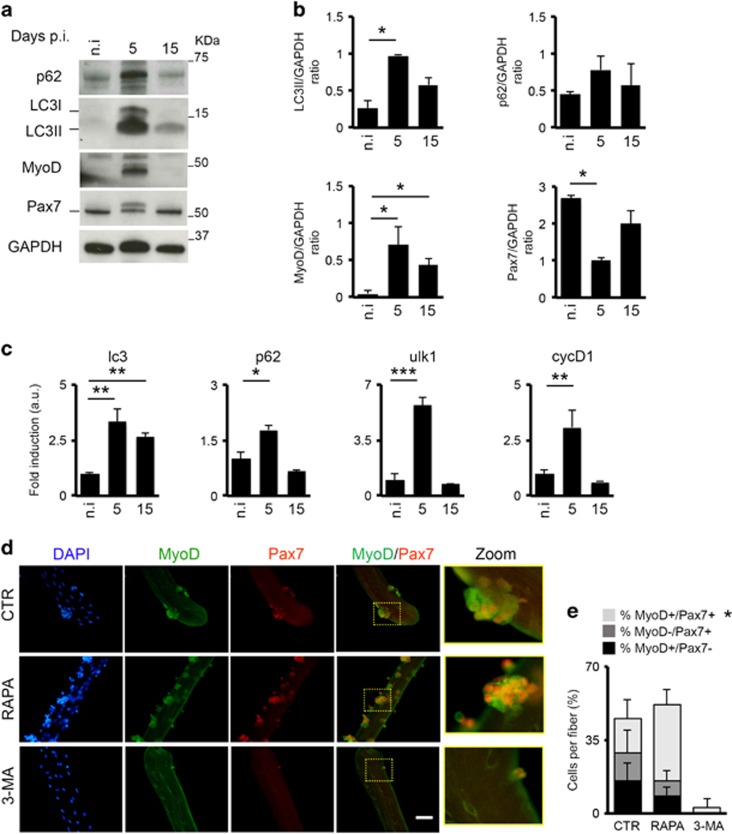
To further dissect the role of autophagy in MuSCs activation, we performed an ex vivo experiment isolating MuSCs by fluorescence-activated cell sorting (FACS) from uninjured and injured mice to monitor the autophagic process during the regenerative response at 5 and 15 days p.i. (Figure 6). MuSCs were isolated by FACS as Ter119-/CD45-/CD31-/α7integrin+/Sca1- and immediately analyzed for the autophagy markers. By western blot analysis, we observed increased level of LC3II during the transition from unperturbed to activated MuSCs at 5 days p.i. and the return to the baseline levels once muscle repair is completed (15 days p.i.) (Figure 6a and b). LC3 lipidation is associated with the activation of MuSCs that down-regulate Pax7 and start to express MyoD at 5 days p.i. (Figure 6a and b). The activation of autophagy coincides with MuSCs activation and decreases once the regenerative process ended. This was confirmed by the transcriptional analysis that shows the activation of autophagic-related transcripts – lc3, p62 and ulk1 (ref. 42), as well as cyclinD1 at MuSC transition from quiescence to the activated state (Figure 6c) and the later downregulation once the regeneration is resolved at 15 days p.i. These data provide direct evidence that autophagy increases in activated MuSCs.
Figure 6.
The autophagic process is induced in MuSCs during muscle regeneration. Ex vivo analysis of the autophagic process in FACS-sorted MuSCs from uninjured (n.i.) and injured mice. (a) MuSCs were isolated from three different mice for each time point as Ter119-/CD45-/CD31-/α7+/integrin+/Sca1- cells, collected and immediately analyzed after isolation by western blot analysis for p62, LC3, MyoD and Pax7 expression; GAPDH were used as loading control. (b) Plots representing the average of the quantization of LC3II/GAPDH, p62/GAPDH, MyoD/GAPDH and Pax7/GAPDH ratio in uninjured mice (n.i.) and after 5 and 15 days after injury based on three different experiments. (c) qRT-PCR analysis of lc3, p62, ulk1 and cyclinD1 expression in uninjured and after 5 and 15 days after injury in freshly isolated MuSCs. (d) Representative images of freshly isolated fibers untreated or treated with rapamycin (100 nM) and 3-MA (10 mM) for 96 h immunostained with MyoD (green) and Pax7 (red); nuclei were detected with DAPI (blue). The area within the drawn square is enlarged in the zoom panels. (e) Schematic representation of the percentage of MyoD+/Pax7-, MyoD-/Pax7+ and MyoD+/Pax7+ cells counted in fibers untreated (CTR) and treated with rapamycin (RAPA) and 3-MA based on n=3 mice per experimental group. Statistical significance assessed by t-test, *P≤0.05, **P≤0.01, ***P≤0.001
We extended our analysis to freshly isolated fibers from WT mice, in which autophagy was ex vivo activated with rapamycin and inhibited with 3-MA for 96 h. In Figure 6d, we show that stimulating autophagy enhances MuSC activation and proliferation, while its inhibition leads to a complete impairment of both processes. Indeed, upon rapamycin treatment we measured an overall twofold increase of MyoD+/Pax7+ cells as compared with control untreated fibers (Figure 6e). Conversely, the amount of these cells is significantly reduced in 3-MA-treated fibers.
Altogether, these data indicate that autophagy influences MuSC activation and proliferation possibly exhibiting distinct functions at different stages of MuSCs state – proliferative/myogenic – and in muscle fibers. Likely, a combination of autophagy impairment in both compartments cooperates in reduced regeneration and muscle wasting during DMD progression.
Discussion
Collectively our data reveal a central role of autophagy in MuSCs activation during muscle regeneration in normal and dystrophic muscles. Although our work reiterates the potential importance of the relationship between dysfunctional autophagy and DMD progression anticipated by previous works, it also points to autophagy as a key process required for compensatory regeneration, by regulating MuSC activity. Autophagy is induced in MuSCs during muscle regeneration of healthy muscles, and inhibition of autophagy with 3-MA delays the regenerative response. This evidence indicates that autophagy is a general regulator of MuSC activity. It is important to note that in vivo drug delivery or nutritional schemes such as 3-MA, CLQ and LPD may affect all cell types leading to the resulting phenotype throughout a multifactorial mechanism on MuSCs, myofibers and other resident cell types. As MuSCs are the cellular effectors of the compensatory regeneration at early stages of DMD, and their long-term activity is compromised by dystrophin deficiency,5 we speculate that autophagy could be a potential ‘disease modifier' exploitable by new interventions aimed at extending the compensatory stage in DMD progression.
To date, steroid treatment is the only treatment for DMD leading to a moderate increase in muscle strength, albeit with significant side-effects.43 Yet, glucocorticoids induce atrogin-1 and MuRF-1 expression44 increasing the rate of protein degradation and muscle atrophy. Despite these data are apparently in contrast with the beneficial effects of autophagy stimulation in dystrophies, our work rather emphasizes the necessity to develop cell-specific strategies to modulate autophagy in the satellite compartment to improve muscle regeneration. Pharmacological approaches that restore the autophagic flux as LPD24 and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) – an AMP-activated protein kinase (AMPK) agonist –26 have been proven to be effective in ameliorating muscular functions in mdx mice. Other studies demonstrates that the E3 ubiquitin ligase TNF receptor-associated factor 6 (TRAF6), which regulates autophagy, displays a stage-dependent effect in mdx mice,25 further supporting the notion that unbalanced autophagy occurs in DMD. Defective autophagy of mdx animals is restored by rapamycin delivery through nanoparticles45 and Simvastin treatment that reduces ROS by enhancing the autophagic process27, 46 leading to improved physical performance.
Defective neuronal NO synthase signaling has been associated to DMD and other myopathies and affects muscle functions, performance, mitochondrial biogenesis and autophagy,47 suggesting use of NO-donating drugs as candidate treatment for DMD.48 Impaired nitrosylation is associated with deregulated activation of histone deacetylase2 (HDAC2) in mdx mice.49 Accordingly, pharmacological treatment with HDAC inhibitors (HDACi) leads to morphological and functional recovery of the dystrophic phenotype in mdx mice.50, 51, 52, 53 Still a stage-dependent beneficial effect of HDACi treatment has been reported being HDACi efficacy restricted to early stages of the disease.52, 54 Interestingly, the refractoriness to HDACi treatment coincides with the premature block of autophagy reported in this work. Recent evidence underscores a role of HDACi in inducing autophagy55, 56 by directly regulating LC3 levels. The HDACi-mediated induction of autophagy has been associated with ROS and damaged mitochondria clearance that imparts a pro-survival effect.
The new perspective is to combine different therapeutic approaches to improve the efficacy of the single treatment. Here we show that autophagy promotes regeneration and is associated to the early stage of disease in mdx mice and DMD boys thus it appears of particular interest the possibility to activate autophagy by dietary interventions, thereby paving the way for a new rationale in the control of calorie intake in DMD patients to extend the compensatory regeneration.
Materials and Methods
Mouse model and in vivo treatments
Normal WT C57/BL6 (WT) and C57/BLl0/mdx (mdx) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). WT and mdx mice were analyzed in this study at different ages that are 1.5, 5, 8 and 12 months. When indicated, WT and mdx mice were treated daily with intraperitoneal (i.p.) injections of autophagic lisosomotrophic agent CLQ (Sigma-Aldrich, St Louis, MO, USA, 50 mg/kg in PBS every 24 h for 4 days). CLQ was administered for 4 days to assess the autophagic flux and, at the same dosage, for 14 days to inhibit the autophagic process in vivo. In order to activate autophagy, mdx mice were fed with LPD for 60 days as described in ref. 24. 3-methyladenine (3-MA; Sigma) or vehicle (PBS) administration was performed under physiological conditions by daily i.p. injections (10 mg/kg) for 5 and 15 days. Muscle injury was performed by intramuscular injection of 20 μl of cardiotoxin (10 μM) into TA muscles. At indicated times, TA and D muscles were collected from the mice immediately after killing. Killing of all animal was executed at 0900 h in order to avoid fluctuation of the autophagic process due to nutritional and feeding habits. Mice were bred and maintained according to the standard animal facility procedures, and all experimental protocols were approved by the internal Animal Research Ethical Committee according to the Italian Ministry of Health and complied with the NIH Guide for the Care and Use of Laboratory Animals.
Muscles and histology
Vastus medialis from DMD boy's biopsies, D and TA from WT and mdx muscles were snap-frozen in liquid nitrogen-cooled isopentane to obtain 10 μm transversal cryosections. Eosin/hematoxylin staining was performed as described in ref. 50. Fibrotic tissue was detected by Masson's trichrome staining. The CSA was calculated using the ImageJ software downloaded from http://rsb.info.nih.gov/ij. Fibrotic areas were measured by selecting at least six representative fields at × 10 magnification. The total fibrosis was calculated from sections evaluating image analysis algorithms for color deconvolution. ImageJ was used for image processing, the original images were segmented with three clusters and the plugin assumes images generated by color subtraction (white representing background, blue collagen and magenta non-collagen regions).57 The fibrotic area was normalized to the entire muscle area of each image field.
Isolated fibers
Freshly isolated fibers from extensor digitorum longus and TA of WT mice were obtained by digesting muscles with 0.2% collagenase type I (SIGMA) in DMEM incubated in shaking water bath for 1 h then kept in culture in DMEM containing 20% FBS (Gibco, Waltham, MA, USA), 10% horse serum (Gibco) and 1% chicken embryo extract for 96 h to allow MuSCs activation and division. When indicated, fibers were treated with rapamycin (100 nM) and 3-MA (10 mM) for 96 h.
Immunofluorescence
Cells and cryosections were fixed in PFA 4% (Sigma) and permeabilized with 100% methanol at −20 °C. The following antibodies were used: embryonic MyHC (DSHB); laminin (Sigma); p62 (Sigma); MyoD (Santa Cruz); LC3 (Cell Signaling, Technology, Danvers, MA, USA), Pax7 (Hybridoma Bank). Alexa Fluor secondary antibodies (Molecular Probes, Eugene, OR, USA) were used and nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole). Images were acquired with fluorescent microscope and by four-laser Leica confocal microscopy (Microsystems, Concord, ON, Canada) integrated with image capture system and analytical software.
RT-PCR and qPCR
Total RNA was extracted with Trizol and retro-transcribed with Taqman reverse transcription kit (Applied Biosystems) following the manufacturer's indication. qRT-PCR was performed using SYBR Green Master mix (Applied Biosystems, Foster City, CA, USA) following manufacturer indications. Relative expression values were normalized to the housekeeping gene gapdh. Primers sequences are listed in Table 1.
Table 1. Primers List.
| Gene | Forward | Reverse |
|---|---|---|
| gapdh | 5′-CACCATCTTCCAGGAGCGAG-3′ | 5′-CCTTCTCCATGGTGGTGAAGAC-3′ |
| eMyHC | 5′-CAATAAACTGCGGGCAAAGAC-3′ | 5′-CTTGCTCACTCCTCGCTTTCA-3′ |
| lc3 | 5′-CACTGCTCTGTCTTGTGTAGGTTG-3′ | 5′-TCGTTGTGCCTTTATTAGTGCATC-3′ |
| p62 | 5′-CCCAGTGTCTTGGCATTCTT-3′ | 5′-AGGGAAAGCAGACCAAGCTC-3′ |
| beclin1 | 5′-AGCCTCTGAAACTGGACACG-3′ | 5′-GCTGTGGTAAGTAATGGAGCTGT-3′ |
| ulk | 5′-TTACCAGCGCATCGAGCA-3′ | 5′-TGGGGAGAAGGTGTGTAGGG-3′ |
| vps34 | 5′-CAAGAACTCCCGGCAGAACA-3′ | 5′-ATGAGCCTTGGTGAGCTTGG-3′ |
| cycD1 | 5′-ACAAGCAGACCATCCGCAAG-3′ | 5′-GCTCCCAGCAGCTACCATG-3′ |
Proteins extraction and western blotting
Protein extracts from muscles and FACS-sorted MuSCs were obtained by tissue homogenization with TissueRuptor in lysis buffer (320 mM sucrose; 50 mM NaCl; 50 mM Tris pH7.5, 10% glycerol, 1% Triton) plus protease and phosphatase inhibitors. After being sonicated, samples were loaded in SDS gel and transferred to PVDF membranes. The following antibodies were used: LC3B (Cell Signaling); p62 (Sigma); MyoD (Santa Cruz); Pax7 (Hybridoma Bank); and GAPDH (Santa Cruz). HRP-conjugated secondary antibodies were revealed with the ECL chemiluminescence kit (Amersham, GE Healthcare, Little Chalfont, UK).
Cell preparation and FACS sorting
For FACS, hind-limb muscles were homogenized and digested in HBSS (Gibco) with 2 μg/ml CollagenaseA (Roche, Mannheim, Germany), 2.4 U/ml Dispase II (Roche), 10 ng/ml DNase I (Roche), 0.4 mM CaCl2 and 5 mM MgCl2 for 90 min at 37 °C. Samples are then resuspended in HBBS with 0.1% BSA to stop the digestion. After serial filtering (100, 70 and 40 μm strainers), cells were stained with primary antibodies (10 ng/ml) CD31-PacificBlue (Invitrogen, Waltham, MA, USA), CD45-eFluor450 (eBioscience, San Diego, CA, USA), Ter119-eFluor450 (eBioscience), Sca1-FITC (BD Pharmingen, Franklin Lakes, NJ, USA) and α7integrin-APC for 30 min on ice. Flow cytometry analysis and cell sorting were performed on a DAKO-Cytomation MoFlo High Speed Sorter (Glostrup, Denmark). MuSCs were isolated as Ter119−/CD45−/CD31−/α7-integrin+/Sca1− cells. Right after isolation, MuSCs were processed to obtain protein extract and RNA.
Biopsies
Human biopsies from 2- and 8-year-old DMD boys, performed for diagnostic purposes after informed consent, were collected from vastus medialis at Bambino Gesù Children's Hospital, Rome, Italy. Human biopsies were assayed for Masson's trichrome staining and immunofluorescence for LC3, MyoD, Pax7 and laminin antibodies.
Statistical methods and analyses
Data are presented as mean±S.E.M. Comparisons between two groups were made using the Student's t-test assuming a two-tailed distribution, with significance being defined as *P<0.05, **P<0.01, ***P<0.001. Comparisons between three or more groups were made using one-way ANOVA test, with Tukey's multiple comparisons post test and with significance being defined as *P<0.05; **P<0.01; ***P<0.001.
Acknowledgments
We thank all laboratory members for critical discussion and daily support. We thank Dr. Alessandra Sacco for careful reading of the manuscript. We thank Giovanna Borsellino and Luca Battistini (Fondazione Santa Lucia, Roma, Italy) for isolation of MuSCs by cell sorter. We thank the head of the administrative office Dr. Pamela Papa and the directorate secretary Dr. Matilde Paggiolu (Istituto di Farmacologia Traslazionale, Roma, Italy) for their technical support. This work was supported by the following grants (to PLP): R01AR056712; R01AR052779; and P30 AR061303 from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases, French Association Against Myopathies (AFM). This work has promoted from research funding to LL from the National Institutes of Health NIH R01 AR064873 and Epigen Project PB.P01.001.019/Progetto Bandiera Epigenomica IFT.
Glossary
- DMD
Duchenne muscular dystrophy
- ROS
reactive oxygen species
- MuSCs
muscle satellite cells
- WT
Wild-type
- TA
Tibialis anterior
- D
diaphragms
- LC3
microtubule-associated protein light chain 3
- DAPI
4',6-diamidino-2-phenylindole
- CLQ
chloroquine
- p62
Sequestosome 1-SQSTM1
- 3-MA
3-methyladenine
- eMyHC
embryonal myosin heavy chain
- CSA
fibers cross-sectional area
- LPD
low-protein diet
- p.i.
post injury
- H/E
hematoxilin/eosin
- i.p.
intra peritoneal
- n.i.
not injured
- HDACi
histone deacetylase inhibitor
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Hoffman EP, Brown RH Jr., Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987; 51: 919–928. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 1990; 345: 315–319. [DOI] [PubMed] [Google Scholar]
- Allen DG, Whitehead NP, Froehner SC. Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol Rev 2016; 96: 253–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Campbell KP. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve 1994; 17: 2–15. [DOI] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med 2015; 21: 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Partridge TA. Self-renewal of the adult skeletal muscle satellite cell. Cell Cycle 2005; 4: 1338–1341. [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science 2005; 309: 2064–2067. [DOI] [PubMed] [Google Scholar]
- Heslop L, Morgan JE, Partridge TA. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J Sci 2000; 113: 2299–2308. [DOI] [PubMed] [Google Scholar]
- Serrano AL, Mann CJ, Vidal B, Ardite E, Perdiguero E, Munoz-Canoves P. Cellular and molecular mechanisms regulating fibrosis in skeletal muscle repair and disease. Curr Top Dev Biol 2011; 96: 167–201. [DOI] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature 2012; 490: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, Dumont NA, Rudnicki MA. Cellular dynamics in the muscle satellite cell niche. EMBO Rep 2013; 14: 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaro L, Latella L. Forever young: rejuvenating muscle satellite cells. Front Aging Neurosci 2015; 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney MT, Aydogdu T, Sala D, Malecova B, Gatto S, Puri PL et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med 2014; 20: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol 2011; 23: 198–206. [DOI] [PubMed] [Google Scholar]
- Jiang P, Mizushima N. Autophagy and human diseases. Cell Res 2014; 24: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752. [DOI] [PubMed] [Google Scholar]
- Sandri M. Autophagy in skeletal muscle. FEBS Lett 2010; 584: 1411–1416. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M et al. Autophagy is required to maintain muscle mass. Cell Metab 2009; 10: 507–515. [DOI] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med 2010; 16: 1313–1320. [DOI] [PubMed] [Google Scholar]
- Chrisam M, Pirozzi M, Castagnaro S, Blaauw B, Polishchuck R, Cecconi F et al. Reactivation of autophagy by spermidine ameliorates the myopathic defects of collagen VI-null mice. Autophagy 2015; 11: 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Sandri M, Bonaldo P. Autophagy induction rescues muscular dystrophy. Autophagy 2011; 7: 426–428. [DOI] [PubMed] [Google Scholar]
- De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis 2012; 3: e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindi SM, Sato S, Choi Y, Kumar A. Distinct roles of TRAF6 at early and late stages of muscle pathology in the mdx model of Duchenne muscular dystrophy. Hum Mol Genet 2014; 23: 1492–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Daussin F, Burelle Y, Li T, Godin R, Fauconnier J et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol 2012; 181: 583–592. [DOI] [PubMed] [Google Scholar]
- Whitehead NP, Kim MJ, Bible KL, Adams ME, Froehner SC. A new therapeutic effect of simvastatin revealed by functional improvement in muscular dystrophy. Proc Natl Acad Sci USA 2015; 112: 12864–12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnio S, LoVerso F, Baraibar MA, Longa E, Khan MM, Maffei M et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep 2014; 8: 1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell 2011; 146: 682–695. [DOI] [PubMed] [Google Scholar]
- Sandri M, Coletto L, Grumati P, Bonaldo P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J Cell Sci 2013; 126: 5325–5333. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 2008; 105: 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 2009; 325: 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Licastro D, Cava E, Veronese N, Spelta F, Rizza W et al. Long-term calorie restriction enhances cellular quality-control processes in human skeletal muscle. Cell Rep 2016; 14: 422–428. [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell 2012; 10: 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J 2014; 33: 2782–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath SD, Webb AE, Brunet A, Rando TA. FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Rep 2014; 2: 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E et al. Autophagy maintains stemness by preventing senescence. Nature 2016; 529: 37–42. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- Perry CN, Kyoi S, Hariharan N, Takagi H, Sadoshima J, Gottlieb RA. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol 2009; 453: 325–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosqueira M, Zeiger U, Forderer M, Brinkmeier H, Fink RH. Cardiac and respiratory dysfunction in Duchenne muscular dystrophy and the role of second messengers. Med Res Rev 2013; 33: 1174–1213. [DOI] [PubMed] [Google Scholar]
- Kharraz Y, Guerra J, Pessina P, Serrano AL, Munoz-Canoves P. Understanding the process of fibrosis in Duchenne muscular dystrophy. Biomed Res Int 2014; 2014: 965631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15: 406–416. [DOI] [PubMed] [Google Scholar]
- Markham LW, Spicer RL, Khoury PR, Wong BL, Mathews KD, Cripe LH. Steroid therapy and cardiac function in Duchenne muscular dystrophy. Pediatr Cardiol 2005; 26: 768–771. [DOI] [PubMed] [Google Scholar]
- Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology 2008; 23: 160–170. [DOI] [PubMed] [Google Scholar]
- Bibee KP, Cheng YJ, Ching JK, Marsh JN, Li AJ, Keeling RM et al. Rapamycin nanoparticles target defective autophagy in muscular dystrophy to enhance both strength and cardiac function. FASEB J 2014; 28: 2047–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Palmieri M, Loehr JA, Li S, Abo-Zahrah R, Monroe TO et al. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nat Commun 2014; 5: 4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma C, Morisi F, Pambianco S, Assi E, Touvier T, Russo S et al. Deficient nitric oxide signalling impairs skeletal muscle growth and performance: involvement of mitochondrial dysregulation. Skelet Muscle 2014; 4: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli S, Sciorati C, D'Antona G, Innocenzi A, Covarello D, Galvez BG et al. Nitric oxide release combined with nonsteroidal antiinflammatory activity prevents muscular dystrophy pathology and enhances stem cell therapy. Proc Natl Acad Sci USA 2007; 104: 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi C, Mozzetta C, Gurtner A, Illi B, Rosati J, Straino S et al. HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc Natl Acad Sci USA 2008; 105: 19183–19187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti GC, Colussi C, Adami R, Serra C, Mozzetta C, Parente V et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med 2006; 12: 1147–1150. [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Consalvi S, Saccone V, Forcales SV, Puri PL, Palacios D. Selective control of Pax7 expression by TNF-activated p38alpha/polycomb repressive complex 2 (PRC2) signaling during muscle satellite cell differentiation. Cell Cycle 2011; 10: 191–198. [DOI] [PubMed] [Google Scholar]
- Saccone V, Consalvi S, Giordani L, Mozzetta C, Barozzi I, Sandona M et al. HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Genes Dev 2014; 28: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consalvi S, Mozzetta C, Bettica P, Germani M, Fiorentini F, Del Bene F et al. Preclinical studies in the mdx mouse model of duchenne muscular dystrophy with the histone deacetylase inhibitor givinostat. Mol Med 2013; 19: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C, Consalvi S, Saccone V, Tierney M, Diamantini A, Mitchell KJ et al. Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol Med 2013; 5: 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011; 471: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ng S, Wang J, Zhou J, Tan SH, Yang N et al. Histone deacetylase inhibitors induce autophagy through FOXO1-dependent pathways. Autophagy 2015; 11: 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, Smith LH, Rong J, Huang X, Hyer ML, Zeps N et al. Image analysis algorithms for immunohistochemical assessment of cell death events and fibrosis in tissue sections. J Histochem Cytochem 2009; 57: 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.