Abstract
Tudor staphylococcal nuclease (TSN, also known as Tudor-SN, SND1 or p100) is an evolutionarily conserved protein with invariant domain composition, represented by tandem repeat of staphylococcal nuclease domains and a tudor domain. Conservation along significant evolutionary distance, from protozoa to plants and animals, suggests important physiological functions for TSN. It is known that TSN is critically involved in virtually all pathways of gene expression, ranging from transcription to RNA silencing. Owing to its high protein–protein binding affinity coexistent with enzymatic activity, TSN can exert its biochemical function by acting as both a scaffolding molecule of large multiprotein complexes and/or as a nuclease. TSN is indispensible for normal development and stress resistance, whereas its increased expression is closely associated with various types of cancer. Thus, TSN is an attractive target for anti-cancer therapy and a potent tumor marker. Considering ever increasing interest to further understand a multitude of TSN-mediated processes and a mechanistic role of TSN in these processes, here we took an attempt to summarize and update the available information about this intriguing multifunctional protein.
Facts:
TSN is an evolutionarily conserved protein having a pivotal role in the regulation of gene expression.
TSN acts both as a scaffolding protein and as a nuclease.
TSN sustains cell viability and its cleavage by proteases facilitates cell death.
TSN is implicated in key cancer-related processes and is a potent marker of various types of tumors.
Open questions:
How is nucleolytic activity of TSN regulated in vivo and how this affects interaction of TSN with downstream targets?
What are the structural bases and functional consequences of distinct intracellular localization patterns of TSN in animals and plants?
What are the molecular mechanisms of the TSN-mediated cancerogenesis?
God has given you one face, and you make yourself another (William Shakespeare)
Being true for all living organisms, accurate spatio-temporal regulation of gene expression is a fundamental mechanism underlying development, homeostasis and adaptation to the environment. Eukaryotic gene expression is a cumulative outcome of a multitude of molecular processes, including transcription, mRNA splicing, stability and translation, chromatin modification, as well as protein stability and modification. Each of these processes is in turn controlled by a highly dedicated repertoire of proteins. However, one protein, Tudor staphylococcal nuclease (TSN, also known as Tudor-SN, SND1 or p100) appears to act in most of the gene expression pathways.
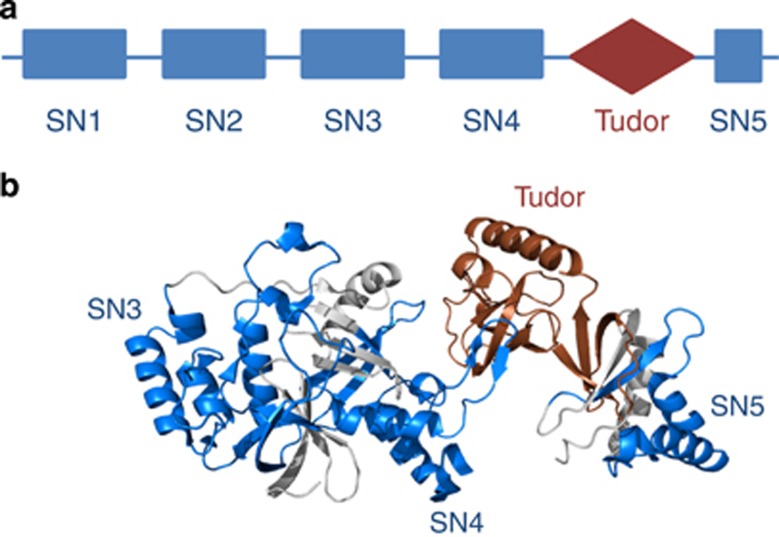
TSN is an evolutionarily conserved protein found in all eukaryotic lineages, except budding yeast, Saccharomyces cerevisiae. Conservation along significant evolutionary distance suggests important physiological functions for TSN. Invariant domain composition of TSN comprises tandem repeat of four staphylococcal nuclease (SN)-like domains (hereafter referred to as SN domains) at the N terminus and a fusion of a Tudor domain with a partial SN domain at the C terminus (Figures 1a and b).1, 2, 3 The simultaneous presence of both Tudor and SN domains in the same protein molecule is intriguing and suggests that during evolution TSN might have acquired novel molecular functions in addition to canonical Tudor- and SN-specific functions. Indeed, owing to the presence of multiple domains TSN protein can interact with nucleic acids, individual proteins and protein complexes in a promiscuous manner.4
Figure 1.
Domain composition and tertiary structure of TSN. (a) TSN has invariant domain architecture including a tandem of four SN-like domains followed by a Tudor domain and a fifth SN domain. (b) The ribbon model of truncated human TSN.17 Tudor domain and three SN domains are colored in brown and blue, respectively
TSN was initially discovered as a transcriptional co-activator interacting with Epstein-Barr nuclear antigen 2 (EBNA2) and promoting EBNA2-dependent transcription.1, 5 Importantly, TSN can also act as a co-activator of several other transcription factors, such as Signal transducer and activator of transcription 5 (STAT5) (ref. 6) and STAT6 (ref. 7). Besides transcriptional role, TSN has been subsequently shown to function in post-transcriptional regulation, including RNA interference (RNAi), splicing and both degradation and stabilization of mRNA. Participation of TSN in several gene regulatory pathways appears to correlate with its ability to shuttle between nucleus and cytoplasm8, 9 and under certain conditions to re-localize to cytoplasmic foci.10, 11, 12
To-date TSN is known to be critically involved in virtually all pathways of gene expression, ranging from transcription to RNA silencing. Moreover, increased expression of TSN has been found to be closely associated with various types of cancer.13, 14, 15 Considering ever increasing interest to further understand a multitude of TSN-mediated processes and a mechanistic role of TSN in these processes, here we took an attempt to summarize and update the available information about this intriguing multifunctional protein.
Structure and intracellular localization of TSN: clues to multiple functions
A combination of the modeled three-dimensional structures and X-ray crystallography revealed that four N-terminally localized SN domains resemble a stick and the C-terminal Tudor fused to a partial SN domain resemble a hook (Figure 1b).16, 17 This indicates that different parts of TSN protein may recruit diverse protein complexes to perform various functions (Figure 2).
Figure 2.
Multi-domain composition of TSN enables interaction with functionally diverse repertoires of proteins. TSN is involved in numerous cellular pathways via interactions with key components of these pathways, through distinct domains. Thus, N-terminal SN domains act as a bridge between several components of the basal transcription machinery such as EBNA2-TFIIE, STAT6-RHA, C-Myb-Pim1, STAT6-PC1 and STAT6-CBP. In addition, TSN interacts through SN domains with the transcription factor PPARγ, several components of the RISC complex, including AEG-1, Ago1 and Ago2, and with different SG-associated proteins, such as ADAR1, G3BP, TIAR, Pabp1 and eIF4E. C-terminal Tudor domain interacts with several components involved in RNA splicing, including the Sm proteins SmB, SmD1/D3, SmD1, SAM68 and Prp8. In addition, C-terminal Tudor domain interacts with the Piwi protein PIWIL1/Miwi
The SN domains belong to the oligonucleotide/oligosaccharide-binding fold (OB-fold) superfamily remarkably conserved throughout evolution and comprising proteins that participate in nucleic acid binding.1, 18 Each SN domain consists of the characteristic five-stranded β-barrel (OB-fold) flanked by three α-helices (Figure 1b). In fact, the presence of SN domains indicates that TSN could share the primary function of bacterial enzyme, that is, nucleic acid binding and processing.1, 9, 10, 19, 20 The C-terminally situated SN domains of TSN specifically interact with several components of RNA-induced silencing complex (RISC) such as the protein astrocyte elevated gene-1 (AEG-1), Argonaute 1 (Ago1) and 2 (Ago2) (Figure 2). In addition, human TSN is directly involved in the degradation of hyper-edited double-stranded RNA (dsRNA) and miRNA precursors produced by adenosine deaminases acting on RNA (ADARs) enzymes.21 Notably, human TSN interacts with ADAR1, RNA-binding Ras-GAP SH3 binding protein (G3BP) and with several core components of stress granules (SGs) under stress conditions, whereupon SN domains are essential for both interaction and SG-specific localization of TSN (Figure 2 and Table 1).3, 11, 22 Apart from binding cytoplasmic partners, SN domains of mammalian TSN bind several components of basal transcription machinery in the nucleus (Figure 2). All these findings suggest that TSN, through its SN domains, may participate in both transcriptional and post-transcriptional regulation of gene expression.
Table 1. Summary of all thus far known functions of TSN, including organisms and interacting proteins.
| Function | Model system/species | Interacting proteins | References |
|---|---|---|---|
| Transcription | Human cells | c-Myb and Pim-1 | 39 |
| STAT6 and CBP | 40 | ||
| STAT6 and RHA | 41 | ||
| STAT6 and PC1 | 42 | ||
| STAT5 | 6 | ||
| EBNA2 and TFIIE | 5 | ||
| PPARγ | 43 | ||
| P. falciparum | 33 | ||
| Splicing | Human cells | Prp8 | 32 |
| SmB, SmD1 and SmD3 | 30 | ||
| SAM68 | 31 | ||
| P. falciparum | SmD1 | 33 | |
| RISC | Various animal models | Ago2, FMRP, VIG and FXR | 9 |
| Human cells | AEG-1 | 70 | |
| Ago1 and Ago2 | 3 | ||
| B. mori | Ago1 and Ago2 | 12 | |
| X. laevis | 21 | ||
| Regulation of noncoding RNAs | Human cells | PIWIL1/Miwi | 25 |
| D. melanogaster | Piwi | 34 | |
| RNA stabilization | Human cells | 3 | |
| Arabidopsis | 36, 38 | ||
| Stress response | Human cells | G3BP | 22 |
| ADAR1 and eIF4E | 11 | ||
| TIAR, Pabp1 and eIF5A | 3 | ||
| B. mori | eIF4E and TIA1 | 12 | |
| Arabidopsis | CML38 | 35 | |
| 10, 27, 38 | |||
| Cell death and cancer | Human cells | NF-kB | 44 |
| 76, 83 | |||
| Human cells and plants (P. abies and Arabidopsis) | Cleaved by caspases/metacaspases | 20 |
Arabidopsis, Arabidopsis thaliana; B. mori, Bombyx mori; D. melanogaster, Drosophila melanogaster; P. abies, Picea abies; P. falciparum, Plasmodium falciparum; X. laevis, Xenopus laevis
The Tudor domain of TSN is a conserved structural motif of ~60 amino acids that is characterized by a strongly bent antiparallel β-sheet composed of five β-strands with a barrel-like fold (Figure 1b).23 This domain has been reported to recognize and bind methylated lysine and arginine of target proteins, which is a key factor for their ability to facilitate the assembly of larger protein complexes at discrete cellular compartments.24, 25, 26, 27, 28, 29 It seems that a common function of proteins containing Tudor domain, including TSN, is to act as an adaptor in order to link methylated arginine or lysine to effector molecules with specific catalytic activities.24 It has recently been shown that human TSN interacts through its Tudor domain with core components of RNA splicing machinery, including Prp8 (human U5-220 kD Protein), two Sm proteins, SmB and SmD1/D3 and the splicing factor SAM68 (Src-associated in mitosis of 68 kDa).30, 31, 32 The interaction of Tudor domain with SmD1, a Sm protein from Plasmodium falciparum suggests an evolutionary conserved role of TSN in splicing.33 The interaction with PIWIL1/Miwi, a specific member of the Argonaute family, suggests a role for TSN in the biogenesis of noncoding RNAs in mammals.25, 34
Because of the cross-kingdom conservation of TSN sequence and its molecular structure, this protein might perform similar functions in different organisms. Indeed, both animal and plant TSN proteins interact with several SG-associated proteins11, 12, 35, 36 and control the fate (stabilization or degradation) of specific mRNAs during stress.10, 36, 37, 38 However, lineage-specific intracellular localization of TSN raises doubts about cross-kingdom conservation of all TSN functions. For example, unlike nuclear-cytoplasmic localization of TSN in animal cells, plant TSN is exclusively cytoplasmic, indicating that the probability of its involvement in transcriptional regulation and splicing in plants is low.10, 38
TSN-mediated regulation of transcription
TSN has been postulated to act as a transcriptional co-activator, interacting with several transcription factors (Figure 2 and Table 1). Thus, TSN functions as a bridge coupling promoter-specific transcription factors and the basal transcription machinery, such as c-Myb and Pim-1 (ref. 39), STAT6 and CREB-binding protein (CBP),40 STAT6 and RNA Helicase A (RHA),41 STAT6 and polycystin-1 (PC1),42 EBNA2 and TFIIE,5 respectively (Figure 2). In addition, TSN interacts with STAT5 (ref. 6), peroxisome proliferator-activated receptor-γ (PPARγ)43 and the nuclear factor kB (NF-kB) (Table 1).44 The interaction with transcription factors is mediated by SN domains, which concurrently bind DNA.6, 7 Accordingly, a set of TSN-bound promoter regions has been recently identified using chromatin immunoprecipitation.26 The interaction of TSN with multiple components of transcription machinery makes it an important factor in the regulation of a plethora of cellular signaling pathways implicated in the pathogenesis of various human diseases.
TSN is critical for assembly of STAT transcriptosome, participating in cell proliferation, cell death, and differentiation.45 Thus, TSN interaction with STAT6 and PC1 activates renal epithelial cell proliferation in autosomal dominant polycystic kidney disease (ADPKD).42 Binding of TSN and STAT6 to CBP stimulates the expression of interleukin-4, an important regulator of immune and anti-inflammatory response. Interaction of TSN with STAT5 facilitates the transcriptional activation of prolactin (PRL) genes.6 STAT proteins undergo phosphorylation and subsequent dimerization upon cytokine stimulation, facilitating their nuclear import and activation of transcription of target genes.46 Given that TSN has been detected in both the cytoplasm and nucleus, the interaction of TSN with STAT proteins might occur already in the cytoplasm, and the pre-formed complex could be translocated to the nucleus. Although the function of TSN as transcriptional co-activator has been only described in human cells, a yeast two-hybrid analysis performed in the malaria parasite P. falciparum suggested, albeit not proved exclusively, a similar role for TSN in protozoa.33
Pim-1 is a serine/threonine protein kinase that cooperates with c-Myc during lymphoid cell transformation in human. Leverson et al.39 revealed that Pim-1 phosphorylates TSN forming a stable complex, which promotes transcriptional activity of c-Myb in a TSN-dependent manner. That was the first evidence of the post-transcriptional regulation of TSN activity by phosphorylation. Importantly, TSN was predicted to be a potential substrate for phosphorylation mediated by Cdk2/4/6 (cyclin-dependent kinase) during the G1/S phase transition.47 This post-transcriptional modification was thought to promote transcriptional activity of E2F-1, which could in turn facilitate activation of transcription of E2F-1-mediated G1/S phase target genes. In plants, a phosphoproteomic analysis revealed phosphorylation of TSN upon stress conditions,48 indicating that this post-transcriptional modification of TSN is conserved between kingdoms. Taken together these findings suggest that phosphorylation of TSN could be an essential step in activation of this protein.
Role of TSN in splicing
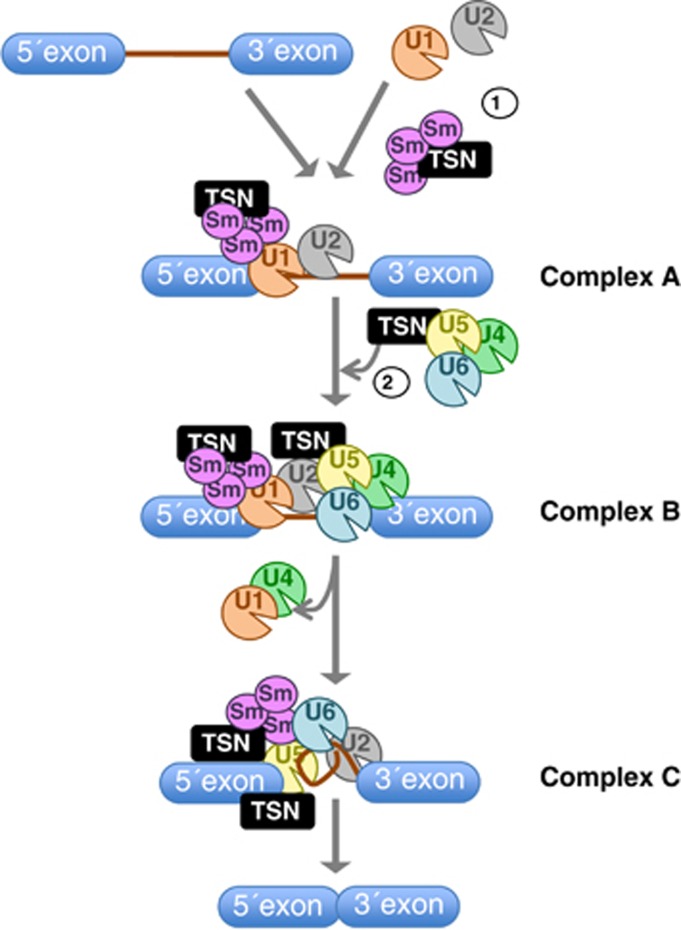
Precursor mRNA (pre-mRNA) splicing is the highly ordered process by which noncoding intronic regions are removed from a pre-mRNA and exons are joined to produce a mature mRNA. This process is carried out by spliceosome composed of five small nuclear ribonucleoprotein (snRNP) particles, each assembled from small nuclear RNAs (snRNAs U1, U2, U4/U6 and U5), seven common Sm proteins and the specific proteins that associate only with a certain snRNP particle.49 Sm proteins function as molecular scaffolds for the stepwise assembly of discrete snRNP particles on the pre-mRNA.50 Briefly, U1 snRNP binds to the 5′-end splice site of the pre-mRNA, and U2 snRNP associates with the branch point sequence of pre-mRNA and other non-snRNP-associated factors to form complex A (Figure 3). The U4, U5 and U6 snRNPs are recruited to the assembling spliceosome to form complex B and, following several rearrangements, complex C (the spliceosome) is activated for catalysis.51
Figure 3.
A model for a dual role of TSN in splicing. The spliceosome assembly starts with the binding of U1 snRNP to the 5′ splice site followed by the recruitment of U2 snRNP together with small Sm proteins to give rise to the complex A. Subsequent recruitment of the U4/U6-U5 tri-snRNP leads to the formation of the complex B, which contains all spliceosomal subunits that carry out pre-mRNA splicing. This is followed by conformational changes and remodeling, including the loss of U1 and U4 snRNPs, resulting in the formation of complex C, which is the catalytically active spliceosome. The interaction of TSN with several Sm proteins leads to the recruitment of Sm proteins to the U1 snRNP particles and the subsequent formation of the complex A (1). In addition, the interaction of TSN with U5 snRNP facilitates the formation of the complex B (2)
TSN has been shown to interact with U1, U2, U4, U5 and U6 snRNPs and Sm proteins SmB and SmD1/D3, thereby facilitating spliceosome complex formation and enhancing splicing rate in vitro.30, 32 Considering that these interactions are mediated by the Tudor domain and that recombinant SN domains abrogate the in vitro splicing reaction and the spliceosome complex formation, both domains might have a coordinate role in the regulation of pre-mRNA splicing. These findings led to postulate a model where TSN may have two successive roles in splicing (Figure 3). First, TSN may facilitate the recruitment of Sm proteins to the U1 snRNPs, thereby promoting the formation of spliceosomal complex A (Figure 3; role 1). Thereafter, TSN may facilitate the transition from complex A to complex B via association with U5 snRNP (Figure 3; role 2).30 A recent finding that P. falciparum TSN interacts with SmD1 protein suggests that the role of TSN in pre-mRNA splicing may also be conserved between protozoa and animals.33
Misregulation of splicing contributes to an increasing number of human pathologies, including genetic and neurodegenerative diseases, as well as cancer.52 These disorders can be caused by mutations that disrupt splicing of specific genes or by a general loss of a spliceosomal function affecting entire networks of downstream genes. Recent work has connected the function of TSN in splicing regulation to oncogenesis by demonstrating interaction of TSN with a splicing factor SAM68 in prostate cancer cells.31 The authors have revealed that TSN can promote SAM68-dependent splicing of specific exons in DC44 mRNA, an event associated with tumor progression and metastasis. However, whether this phenomenon is specific for prostate cancer or is a more general event requires additional investigations.
Role of TSN in post-transcriptional regulation of gene expression
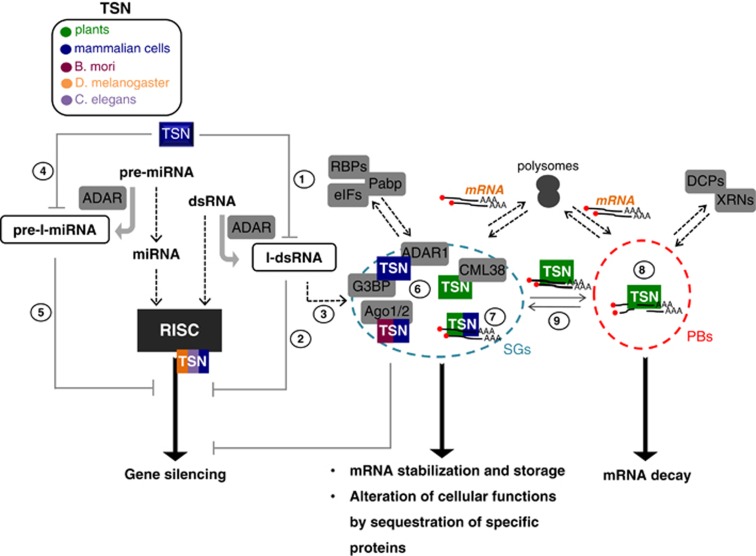
Post-transcriptional processes, such as RNAi, stabilization, decay and editing allow cells and the whole organism to timely and specifically alter the proteome in order to embark on a developmental transition or respond to changing environmental conditions. RNAi silencing pathway is mediated by the ribonucleoprotein complex known as RISC, which possesses the endonuclease activity resulting in target mRNA degradation.53 The basic RNAi machinery contains Dicer RNase III endoribonuclease and Ago proteins that process long dsRNA into small interfering RNAs (siRNAs), which are then used as templates for recognizing complementary mRNA. TSN was initially found to be associated with several components of RNAi machinery, such as the fragile X mental retardation protein (FMRP), Vasa Intronic Gene product (VIG), Fragile X-Related protein (FXR), AEG-1, Ago1 and Ago2 in such evolutionary distant organisms as Drosophila melanogaster, Caenorhabditis elegans and Bombyx mori, as well as in mammalian cells (Table 1 and Figure 4).3, 9, 12, 54 Given that RISC activity was inhibited by 3′,5′-Deoxythymidine bisphosphate (pdTp), an inhibitor of staphylococcal nuclease, there is a high probability that TSN might contribute to the nucleolytic activity of RISC.9
Figure 4.
Role of TSN in the cross-talk between different pathways post-transcriptionally regulating gene expression. Degradation of both hyper-edited double-stranded RNAs (I-dsRNA, 1) and miRNA precursors (pre-I-miRNA, 4) by mammalian TSN can lead to the inhibition of the RNAi silencing pathway (2 and 5), and in this way will antagonize the nucleolytic function of TSN in RISC complex. Concurrently, the enhanced accumulation of I-dsRNA molecules in the cell promotes inhibition of translation and reduction of mRNA levels via formation of SGs under stress conditions (3). In the same, stress scenario, TSN might act in promoting sequestration of specific proteins to SGs (6) thus altering their cellular function. In addition, the localization of TSN in both SGs and PBs is required for mRNA stabilization and degradation under stress conditions (7–9)
dsRNA can further undergo extensive covalent editing by ADAR enzymes, resulting in the conversion of adenosine residues to inosine (I) (I-dsRNA).55 In mammals, TSN has been implicated in the degradation of I-dsRNA molecules (Figure 4; pathway 1).21, 56 Although various RNAs can be extensively edited, the cellular function of hyper-edited dsRNA remains elusive. Several lines of evidence suggest that editing of dsRNA may antagonize RNAi pathway by inhibiting the production of siRNAs, the key intermediates in the pathway (Figure 4; pathway 2). Accumulation of I-dsRNA in the cell leads to both inhibition of translation and reduction of mRNA levels via formation of SGs (Figure 4; pathway 3).57 At the same time, I-dsRNAs were found to specifically bind SG components, including TSN. Apart from the cleavage of I-dsRNA, TSN is involved in the selective degradation of hyper-edited miRNA precursors (pre-I-miRNAs) (Figure 4; pathway 4).56 After maturation of miRNA precursors, miRNAs are integrated into RISC complex in order to block translation of specific mRNAs containing partially complementary targets or to guide degradation of target mRNAs (similarly to siRNAs).58, 59 It has recently been reported that pre-I-miRNAs might affect miRNA-processing and target mRNA binding (Figure 4; pathway 5).60 Therefore, the available data suggest that editing of dsRNA or miRNA molecules by ADARs with subsequent TSN-dependent degradation of I-dsRNAs and pre-I-miRNAs could provide an efficient means of switching off the dsRNA/miRNA-induced silencing pathway.
SGs typically contain poly(A)+ RNA, translation initiation factors (eIFs), poly(A)-binding protein (Pabp), RNA-binding proteins (RBPs) and ribosomal proteins (Figure 4), and have a major role in translational repression by sequestering, stabilizing and storing mRNAs and translation factors.61 The interaction of TSN with proteins forming the core of SGs, such as Pabp1, eIF4E, eIF5A, TIAR (TIA1-related protein) and TIA1 (T-cell-restricted intracellular antigen) in different organisms indicates that TSN is a key component of these cytoplasmic foci (Table 1). In addition to proteins forming their core, SGs recruit additional proteins involved in numerous cellular pathways. Thus, Ago2 and protein kinase interferon-inducible double-stranded RNA-dependent activator (PACT), a Dicer cofactor, are recruited to SGs.62, 63 It has been suggested that the translocation of these proteins to SGs may reflect a dynamic change in their activity.63, 64 Notably, TSN has been found to interact with Ago1 and Ago2 in B. mori and mammals, with ADAR1 and G3BP in mammalian cells and with the calmodulin-like protein 38 (CML38) in Arabidopsis thaliana (Figure 4; pathway 6).11, 12, 22, 35, 37 Experimental evidence indicates that TSN may act as a scaffolding factor recruiting other proteins to the cytoplasmic foci.10 In this context, one can also speculate that TSN might sequester different proteins into SGs thus repressing their physiological function.
During stress, cytoplasmic mRNAs can be stored in SGs or subjected to degradation in the processing bodies (PBs),61 which contain proteins involved in mRNA decay, including subunits of decapping (DCPs), exosome complexes (XNRs) and deadenylases (Figure 4).61 Stabilization of specific mRNAs by RBPs in SGs was shown to protect them from degradation in PBs during stress.65, 66 Accordingly, TSN as a SG-associated protein appears to have an essential role in binding and stabilization of specific mRNAs during stress in mammals and plants (Figure 4; pathway 7).36, 37, 38 TSN-mediated mRNA stabilization was shown to be accompanied by enhanced translational efficiency of AGTR1 (angiotensin II receptor, type 1 mRNA), resulting in an elevated protein levels.3 It has also been revealed that TSN is required for the proper aggregation and stabilization of poly(A)+ mRNAs, such as those encoding AGTR1 and IGFBP2 (insulin-like growth factor-binding protein 2), in SGs during stress in mammals.37 In plants, TSN was consistently shown to be required for stabilization of stress-responsive mRNAs under various experimental settings.36, 38, 67 These findings demonstrate that TSN-dependent stabilization of specific mRNA molecules is crucial for stress adaptation.
In addition to SGs, Arabidopsis TSN protein is also localized to PBs, suggesting its involvement in the mRNA decay process. Indeed, stress-induced accumulation of uncapped mRNAs (intermediates of the mRNA decay pathway) was abrogated in TSN knockout plants (Figure 4; pathway 8).10 Although mRNA decay typically occurs in PBs, a number of studies in mammalian systems demonstrated that mRNA decay enzymes, such as XRN1, can be also found in SGs, pointing to the possibility that TSN could potentially act in mRNA degradation in animal SGs in a manner similar to plant PBs.68 Furthermore, shuttling of cytoplasmic mRNAs between PBs and SGs has been well documented in stressed mammalian cells.69 There is a long-standing notion that mRNAs stalled at a stage of translation initiation are either stored in SGs or re-directed to PBs for degradation. Accordingly, TSN might function as a key regulator of cytoplasmic mRNA fate during stress conditions, having two seemingly antagonistic roles: facilitating global mRNA decapping and thus degradation and at the same time stabilizing a subset of mRNAs that are required for survival (Figure 4; pathway 9). However, underlying molecular mechanisms as well as how these two roles of TSN are co-regulated remain unknown.
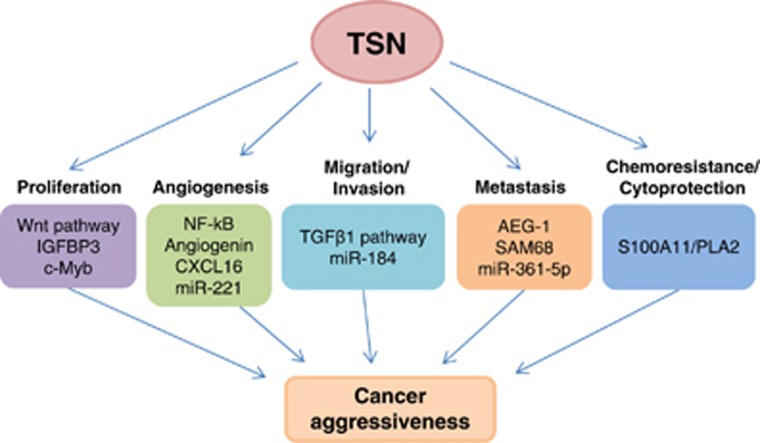
Role of TSN in carcinogenesis and cell death
Recent findings show that TSN is overexpressed in various types of cancers, and its elevated levels are associated with poor prognosis for cancer patients.14, 70, 71 Indeed, TSN has been linked to various oncogenic pathways through interaction with a broad spectrum of signaling proteins (Figure 5).44, 70, 71 Thus, TSN was shown to promote tumor cell proliferation through activation of the Wnt signaling pathway.72 Overexpression of TSN suppressed the levels of adenomatous polyposis coli protein, which is a ‘gatekeeper-type' tumor suppressor gene in colon cancer development.71 Similarly, stable overexpression of recombinant TSN in rat intestinal epithelial cells led to the enhancement of cell proliferation and anchorage-independent growth activity in a soft agar colony formation assay, accompanying the subcellular translocation of E-cadherin to the cytoplasm that influenced the loss of contact inhibition.72 Furthermore, TSN could affect hepatocellular carcinoma (HCC) progression by two distinct pathways.70, 73 First, as a member of RISC complex, TSN interacts with AEG-1 leading to the increased degradation of tumor suppressor mRNAs.70 On the other hand, a negative regulator of insulin-like growth factor (IGF) pathway, an insulin-like growth factor-binding protein 3 (IGFBP3), was reported to be significantly overexpressed in TSN knockdown HCC cells, suggesting the contribution of TSN to cell proliferation, colony and tumor formation.73
Figure 5.
Role of TSN in carcinogenesis. TSN is implicated in a subset of cancer-related processes. In particular, TSN can promote tumor cell proliferation through activation of the Wnt and IGF signaling pathways. Furthermore, TSN interacts with c-Myb, sustaining breast cancer proliferation. TSN supports angiogenesis by activating NF-kB, resulting in the induction of the oncomiR-221, angiogenin and CXCL16. TSN is involved in the regulation of migration and invasion. Interaction of TSN with AEG-1, SAM68 and miR-361-5p promotes tumor metastasis. TSN is a direct target of miR-184 in the regulation of malignant glioma invasion. TSN was also reported to support epithelial–mesenchymal transition (EMT) via association with the TGFβ1 signaling pathway in breast cancer. In addition, TSN is involved in the regulation of chemoresistance in lung cancer acting through TSN-S100A11-PLA2 axis
Another important process regulated by TSN is angiogenesis (Figure 5). TSN promotes angiogenesis in HCC by activating the NF-kB, resulting in the induction of the oncomiR-221, angiogenin and CXCL16.44 Consequently, the interplay between the TSN and the NF-kB activation status might provide an important link in coordinating the liver response to tumorigenic and angiogenic stimuli.
More recently, TSN via association with the TGFβ1 signaling pathway was reported to promote epithelial–mesenchymal transition (EMT) in breast cancer. The TSN promoter region contains several Smad-specific recognition domains (RD motifs), which were recognized and bound by the Smad complex that enhanced the transcriptional activation of TSN.74 Since TSN is involved in the regulation of migration and invasion in different cancers its expression level could be used as a potent autonomous poor-prognosis marker.
TSN is considered as a good marker for breast cancer metastasis.14 It was demonstrated that the expression level of TSN was higher in the invasive/metastatic breast cancer cell line (MDAMB-231) compared to poorly invasive/non-metastatic lines (MCF-7 and T47D). Likewise, TSN has a regulatory role in malignant glioma invasion, which is a major determinant of its aggressiveness.75 In this type of cancer, TSN is a downstream target of miR-184, and either knockdown of TSN or overexpression of miR-184 significantly decreased cell viability and glioma invasion (Figure 5).75 In addition to miR-184, TSN is also a target of miR-361-5p, whose expression level is negatively correlated with lung metastasis and prognosis in clinical colorectal cancer patients.76 Overexpression of miR-361-5p markedly suppressed proliferation, migration and invasion of cancer cells and could be partially rescued by the ectopic expression of TSN. Importantly, TSN was able to bind pre-miR-361-5p thereby suppressing the expression of miR-361-5p through feedback regulation. Interestingly, in vivo studies showed that restoration of miR-361-5p significantly inhibited tumor growth and especially the lung metastasis in nude mice. Thus, miR-184 and miR-361-5p function as tumor-suppressive miRNAs through direct inhibition of TSN, highlighting TSN's potential as a novel target for the treatment of patients with glioma, lung or colorectal cancers.
TSN was shown to have a critical role in mammary tumorigenesis by regulating oncogene-induced expansion and activities of tumor-initiating cells (TICs). Mechanistically, through its interaction with AEG-1, TSN can support the survival of mammary epithelial cells under oncogenic/stress conditions. Silencing TSN or AEG-1 individually or disrupting their interaction compromises tumorigenenic potential of TICs in vivo.77 Interestingly, AEG-1 knockout cells are characterized by low level of tumor development. This effect is completely rescued by ectopic expression of AEG-1 in breast cancer cells. Nevertheless, downregulation of TSN in these cells completely eliminated the rescued effect of AEG-1 expression. Moreover, downregulation of TSN in AEG-1+/+ cells is resulted in reduced ability of tumor formation in vivo, resembling the phenotype of AEG-1−/− cells and signifying the role of TSN for oncogenic properties of AEG-1 in breast cancer. Analysis of samples obtained from breast cancer patients revealed simultaneous overexpression of c-Myb and TSN.78 The authors estimated that the TSN promoter is one of the target sites of c-Myb, proposing the presence of a positive regulatory mechanism to maintain c-Myb and TSN at sufficiently high levels to sustain breast cancer progression.
Accumulated evidence suggests that TSN can be used as an efficient diagnostic and/or prognostic marker of various tumor types. Thus, detailed analysis revealed that the level of expression of TSN correlates with the histological grade of the prostate tumor.79 Moreover, the level of TSN expression was compatible with the level of another protein marker of prostate cancer, α-methylacyl-CoA racemase (AMACR). Mass spectrometry analysis of proteins interacting with splicing factor SAM68 in prostate cancer cells showed significant accumulation of both SAM68 and TSN.31 Upregulation of these proteins exerted interdependent synergic effect on exon v5 inclusion in the CD44 mRNA, providing further evidence for TSN as a novel regulator of alternative splicing, promoting prostate cancer cell growth and survival.
One of the hallmarks of human cancers is the ability to evade cell death.80 TSN was demonstrated to contribute to the progression of apoptosis as a primary substrate for caspase-3 (ref. 20). Intriguingly, cleavage of TSN appears to represent evolutionary conserved cell death mechanism, since ancestors of animal caspases, metacaspases,20, 81, 82 have also been shown to cleave TSN in plant cells during developmentally regulated and stress-induced cell death.20 Furthermore, high expression of TSN in cancer cells might contribute to malfunction of apoptotic machinery and chemotherapeutic response. Indeed, recently we demonstrated that TSN overexpression suppresses activity of pro-apoptotic genes that eventually compromises tumor ability to undergo apoptosis promoting chemoresistance of non-small cell lung adenocarcinoma cells (NSCLC).83 Silencing of TSN was accompanied by a significant decrease in S-100 calcium-binding protein A11 (S100A11) expression and consequently, downregulation of S100A11 resulted in enhanced sensitivity of NSCLC cells to cisplatin, oxaliplatin and 5-fluouracil. A phospholipase A2 (PLA2) inhibitor as well as its silencing strongly abrogated chemosensitization upon downregulation of S100A11 suggesting that PLA2 inhibition by S100A11 governs the chemoresistance of NSCLC. These findings highlight the importance of TSN as an essential mediator of chemoresistance and the novel TSN-S100A11-PLA2 axis in the regulation of apoptosis triggered by chemotherapeutic agents in NSCLC cells (Figure 5). Targeting TSN or S100A11 may be particularly beneficial for anti-cancer treatment and may have important therapeutic implications.
Genome-scale analysis of DNA methylation in colorectal cancer identified TSN among 14 highly robust candidates showing methylation marks.84 Importantly, a CpG site located in TSN gene was identified with highest discriminative accuracy, emphasizing role of this gene/protein in development of prostate cancer and the possibility to be a potential diagnostic candidate gene. Examination of 72 SNPs in 21 miRNA-processing genes in a total of 99 osteosarcoma patients and 387 controls resulted in interesting finding of association of three SNPs in TSN within RISC complex with the osteosarcoma risk.85
Immunohistochemical staining of samples from human colon cancer and their corresponding non-cancerous colon tissues revealed simultaneous increase of expression of AEG-1 and TSN in tumor samples compared with paired non-cancerous colon tissues.77 Importantly, their positive expression was both significantly associated with nodal status, pathological stage and differentiation. Moreover, the overall survival rate in colon cancer patients with positive expression of both AEG-1 and TSN was significantly reduced proposing the co-expression of AEG-1 and TSN as a potent marker for prediction of the prognosis in colon cancer.
Conclusions
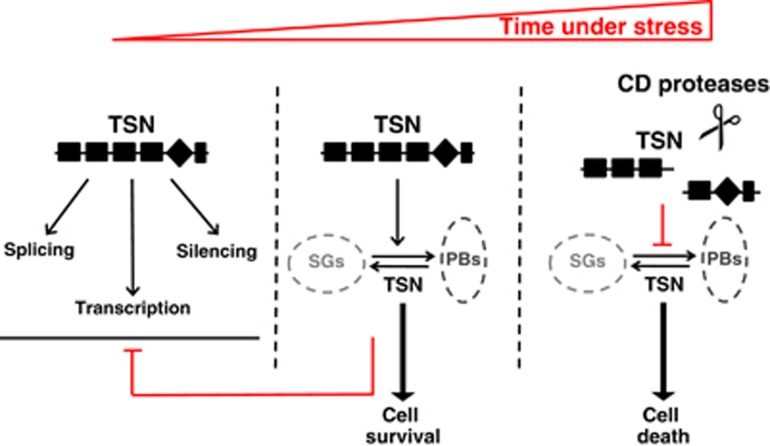
Considering growing evidence for the tight involvement of TSN in the control of cell viability under stress and oncogenic transformation, we propose a hypothetical model for TSN-mediated gene regulation under increasing level of stress (Figure 6). Under normal conditions, TSN has housekeeping role by stimulating transcription, mRNA splicing and/or silencing, depending on the phylogenetic position of the organism. After stress perception, TSN is re-localized to cytoplasmic foci contributing to (i) global translational suppression and (ii) repression of specific pathways due to re-distribution of TSN away from its partner proteins. Given that SGs and PBs confer stress resistance,86 re-localization of TSN to cytoplasmic foci may help to protect cells from death (Figure 6). This is consistent with our recent observation of increased cell death in the roots of Arabidopsis TSN knockout mutants under prolonged stress.10 Considering that TSN is a common target of caspases and metacaspases in animals and plants, respectively20 we propose that protease-mediated cleavage of TSN abrogates its pro-survival role and triggers cell death, presumably through disrupting cytoplasmic foci (Figure 6). This possibility is supported by recent works demonstrating disruption of SGs under prolonged exposure to stress.10, 87 More research in different organisms is required to validate and complement our hypothetical model.
Figure 6.
Hypothetical model for TSN-dependent regulation of cell viability under normal and stress conditions. Under normal condition, TSN has its housekeeping role by stimulating transcription, mRNA splicing or silencing, depending on phylogenetic position of the organism. Following stress perception, TSN is re-localized to cytoplasmic foci, SGs and PBs, contributing to cell survival. Finally, the pro-survival role of TSN is abrogated by caspase- or metacaspase-mediated cleavage, triggering cell death. CD proteases, CD clan of cysteine proteases
Future prospects in TSN research are also associated with its role in tumorigenesis. Although expression of TSN is linked to various steps of tumor progression, it is still unclear whether overexpression of TSN alone is sufficient to transform cells. It is important to understand whether tissue-specific expression of TSN is associated with its oncogenic properties. Moreover, we still do not know whether involvement of TSN in carcinogenesis is associated with its molecular functions in the regulation of gene expression described above.
Surprisingly, in addition to cancer TSN was recently linked to the susceptibility to autism and autism spectrum disorders.88 Association analysis showed strong coupling of MKL2 (MKL/myocardin-like 2) and TSN with autism in at least two European populations, Finnish and Northern Dutch ones. This raises the question: does TSN have a direct role in the development of this or any other diseases and if so, what are potential mechanisms of its function? Full and/or conditional knockout of TSN in mammalian models should help to address these and other questions about this multifunctional protein.
Acknowledgments
This work was supported by grants from Knut and Alice Wallenberg Foundation (to PVB) and the Russian Science Foundation (14-25-00056; to BZ). The work in our laboratories is also supported by grants from the Olle Engkvist Foundation (to PVB), Pehrssons Fund (to PVB), the Russian Foundation for Basic Research (to TVD) and the Russian President Fund (to BZ), as well as the Stockholm and Swedish Cancer Societies (to BZ), the Swedish Childhood Cancer Foundation (to BZ), the Swedish Foundation for Strategic Research (to PVB) and the Swedish Research Council (to PVB and BZ). We apologize to those authors whose primary works could not be cited owing to space limitations.
Glossary
- dsRNA
double-stranded RNA
- I-dsRNA
inosine containing double-stranded RNA
- PB
processing body
- pre-mRNA
precursor mRNA
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- SG
stress granule
- siRNA
small interfering RNA
- snRNA
small nuclear RNA
- snRNP
small ribonucleoprotein
- STAT
signal transducer and activator of transcription
- TSN
Tudor staphylococcal nuclease
The authors declare no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Callebaut I, Mornon JP. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. Biochem J 1997; 321: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. P100, a transcriptional coactivator, is a human homologue of staphylococcal nuclease. Protein Sci 1997; 6: 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Shi X, Fu X, Ge L, Zhang Y, Su C et al. Human Tudor staphylococcal nuclease (Tudor-SN) protein modulates the kinetics of AGTR1-3'UTR granule formation. FEBS Lett 2014; 588: 2154–2161. [DOI] [PubMed] [Google Scholar]
- Arretxe E, Armengol S, Mula S, Chico Y, Ochoa B, Martinez MJ. Profiling of promoter occupancy by the SND1 transcriptional coactivator identifies downstream glycerolipid metabolic genes involved in TNFalpha response in human hepatoma cells. Nucleic Acids Res 2015; 43: 10673–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol 1995; 15: 4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol Endocrinol 2003; 17: 1805–1814. [DOI] [PubMed] [Google Scholar]
- Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N et al. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J 2002; 21: 4950–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst MK, Wheeler TT. The p100 coactivator is present in the nuclei of mammary epithelial cells and its abundance is increased in response to prolactin in culture and in mammary tissue during lactation. J Endocrinol 2001; 171: 329–337. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature 2003; 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015; 27: 926–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach R, Scadden AD. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA 2012; 18: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Tatsuke T, Mon H, Li Z, Xu J, Lee JM et al. Characterization of Tudor-sn-containing granules in the silkworm, Bombyx mori. Insect Biochem Mol Biol 2013; 43: 664–674. [DOI] [PubMed] [Google Scholar]
- Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK et al. Novel breast cancer metastasis-associated proteins. J Proteome Res 2009; 8: 583–594. [DOI] [PubMed] [Google Scholar]
- Blanco MA, Aleckovic M, Hua Y, Li T, Wei Y, Xu Z et al. Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J Biol Chem 2011; 286: 19982–19992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest 2009; 119: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw N, Zhao M, Cheng C, Xu H, Saarikettu J, Li Y et al. The multifunctional human p100 protein 'hooks' methylated ligands. Nat Struct Mol Biol 2007; 14: 779–784. [DOI] [PubMed] [Google Scholar]
- Li CL, Yang WZ, Chen YP, Yuan HS. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res 2008; 36: 3579–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 2003; 32: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MJ, Korde R, Singh S, Mohmmed A, Dasaradhi PV, Chauhan VS et al. Tudor domain proteins in protozoan parasites and characterization of Plasmodium falciparum tudor staphylococcal nuclease. Int J Parasitol 2008; 38: 513–526. [DOI] [PubMed] [Google Scholar]
- Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E et al. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 2009; 11: 1347–1354. [DOI] [PubMed] [Google Scholar]
- Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 2005; 12: 489–496. [DOI] [PubMed] [Google Scholar]
- Gao X, Ge L, Shao J, Su C, Zhao H, Saarikettu J et al. Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions. FEBS Lett 2010; 584: 3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol 2003; 327: 507–520. [DOI] [PubMed] [Google Scholar]
- Friberg A, Corsini L, Mourao A, Sattler M. Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J Mol Biol 2009; 387: 921–934. [DOI] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci USA 2010; 107: 18398–18403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dong L, Zhang X, Wang B, Wang X, Li H et al. Identification of p100 target promoters by chromatin immunoprecipitation-guided ligation and selection (ChIP-GLAS). Cell Mol Immunol 2011; 8: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Jia J, Gao Y, Zhang B, Han Y. The AtTudor2, a protein with SN-Tudor domains, is involved in control of seed germination in Arabidopsis. Planta 2010; 232: 197–207. [DOI] [PubMed] [Google Scholar]
- Pek JW, Anand A, Kai T. Tudor domain proteins in development. Development 2012; 139: 2255–2266. [DOI] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 2011; 18: 1414–1420. [DOI] [PubMed] [Google Scholar]
- Gao X, Zhao X, Zhu Y, He J, Shao J, Su C et al. Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins. J Biol Chem 2012; 287: 18130–18141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Saarikettu J et al. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene 2014; 33: 3794–3802. [DOI] [PubMed] [Google Scholar]
- Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON et al. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res 2007; 35: 4485–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MJ, Korde R, Singh PK, Kanodia S, Ranjan R, Ram G et al. Plasmodium falciparum Tudor staphylococcal nuclease interacting proteins suggest its role in nuclear as well as splicing processes. Gene 2010; 468: 48–57. [DOI] [PubMed] [Google Scholar]
- Ku HY, Gangaraju VK, Qi H, Liu N, Lin H. Tudor-SN Interacts with Piwi Antagonistically in regulating spermatogenesis but synergistically in silencing transposons in Drosophila. PLoS Genet 2016; 12: e1005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokdarshi A, Conner WC, McClintock C, Li T, Roberts D. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 2016; 170: 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Yan Z, Wang Y, Yan X, Han Y. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Bot 2014; 65: 5933–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Fu X, Song J, Zhang Y, Cui X, Su C et al. Poly(A)(+) mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics. FEBS J 2015; 282: 874–890. [DOI] [PubMed] [Google Scholar]
- dit Frey NF, Muller P, Jammes F, Kizis D, Leung J, Perrot-Rechenmann C et al. The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant cell 2010; 22: 1575–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson JD, Koskinen PJ, Orrico FC, Rainio EM, Jalkanen KJ, Dash AB et al. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell 1998; 2: 417–425. [DOI] [PubMed] [Google Scholar]
- Valineva T, Yang J, Palovuori R, Silvennoinen O. The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J Biol Chem 2005; 280: 14989–14996. [DOI] [PubMed] [Google Scholar]
- Valineva T, Yang J, Silvennoinen O. Characterization of RNA helicase A as component of STAT6-dependent enhanceosome. Nucleic Acids Res 2006; 34: 3938–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT et al. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev Cell 2006; 10: 57–69. [DOI] [PubMed] [Google Scholar]
- Duan Z, Zhao X, Fu X, Su C, Xin L, Saarikettu J et al. Tudor-SN, a novel coactivator of peroxisome proliferator-activated receptor gamma protein, is essential for adipogenesis. J Biol Chem 2014; 289: 8364–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhekadur PK, Das SK, Gredler R, Chen D, Srivastava J, Robertson C et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J Biol Chem 2012; 287: 13952–13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci 2004; 117: 1281–1283. [DOI] [PubMed] [Google Scholar]
- Bromberg J. Stat proteins and oncogenesis. J Clin Invest 2002; 109: 1139–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Zhang C, Tecle A, Fu X, He J, Song J et al. Tudor staphylococcal nuclease (Tudor-SN), a novel regulator facilitating G1/S phase transition, acting as a co-activator of E2F-1 in cell cycle regulation. J Biol Chem 2015; 290: 7208–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenman M, Sorensson C, Andreasson E. Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated Arabidopsis. Mol Plant Microbe Interact 2008; 21: 1275–1284. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 2001; 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Mura C, Randolph PS, Patterson J, Cozen AE. Archaeal and eukaryotic homologs of Hfq: a structural and evolutionary perspective on Sm function. RNA Biol 2013; 10: 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Koester T, Staiger D. Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins implicated in the splicing process. Biomolecules 2015; 5: 1717–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 2010; 24: 2343–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol 2005; 6: 127–138. [DOI] [PubMed] [Google Scholar]
- Milochau A, Lagree V, Benassy MN, Chaignepain S, Papin J, Garcia-Arcos I et al. Synaptotagmin 11 interacts with components of the RNA-induced silencing complex RISC in clonal pancreatic beta-cells. FEBS Lett 2014; 588: 2217–2222. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 2002; 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 2006; 13: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol Cell 2007; 28: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Cui Y, Huang T, Zhang X. RNA editing of microRNA prevents RNA-induced silencing complex recognition of target mRNA. Open Biol 2015; 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell 2009; 20: 3273–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA 2006; 103: 18125–18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 2013; 38: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH et al. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 2006; 175: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N, Kohn M, Lederer M, Glass M, Reinke C, Singer RH et al. IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes Dev 2012; 26: 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Bozhkov PV, Moschou PN. Tudor staphylococcal nuclease plays two antagonistic roles in RNA metabolism under stress. Plant Signal Behav 2015; 10: e1071005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005; 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harbor Perspect Biol 2012; 4: a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Santhekadur PK, Gredler R, Chen D, Emdad L, Bhutia S et al. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 2011; 53: 1538–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya N, Ochiai M, Nakashima K, Ubagai T, Sugimura T, Nakagama H. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res 2007; 67: 9568–9576. [DOI] [PubMed] [Google Scholar]
- Tsuchiya N, Nakagama H. MicroRNA, SND1, and alterations in translational regulation in colon carcinogenesis. Mutat Res 2010; 693: 94–100. [DOI] [PubMed] [Google Scholar]
- Yin J, Ding J, Huang L, Tian X, Shi X, Zhi L et al. SND1 affects proliferation of hepatocellular carcinoma cell line SMMC-7721 by regulating IGFBP3 expression. Anat Rec (Hoboken) 2013; 296: 1568–1575. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu X, Cui K, Di Y, Xin L, Sun X et al. SND1 acts downstream of TGFbeta1 and upstream of Smurf1 to promote breast cancer metastasis. Cancer Res 2015; 75: 1275–1286. [DOI] [PubMed] [Google Scholar]
- Emdad L, Das SK, Dasgupta S, Hu B, Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC: signaling pathways, downstream genes, interacting proteins, and regulation of tumor angiogenesis. Adv Cancer Res 2013; 120: 75–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Song H, Guo B, Zhang Y, Zheng Y, Lin C et al. MiR-361-5p inhibits colorectal and gastric cancer growth and metastasis by targeting staphylococcal nuclease domain containing-1. Oncotarget 2015; 6: 17404–17416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Du X, Zang L, Song N, Yang T, Dong R et al. Prognostic impact of Metadherin-SND1 interaction in colon cancer. Mol Biol Rep 2012; 39: 10497–10504. [DOI] [PubMed] [Google Scholar]
- Quintana AM, Liu F, O'Rourke JP, Ness SA. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancer 2011; 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruma H, Kamata Y, Takahashi H, Igarashi K, Kimura T, Miki K et al. Staphylococcal nuclease domain-containing protein 1 as a potential tissue marker for prostate cancer. Am J Pathol 2009; 174: 2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Smertenko AP, Zhivotovsky B. Aspasing out metacaspases and caspases: proteases of many trades. Sci Signal 2010; 3: pe48. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV. Metacaspases. Cell Death Differ 2011; 18: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagryazhskaya A, Surova O, Akbar NS, Allavena G, Gyuraszova K, Zborovskaya IB et al. Tudor staphylococcal nuclease drives chemoresistance of non-small cell lung carcinoma cells by regulating S100A11. Oncotarget 2015; 6: 12156–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, Chernyshov SV et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics 2013; 8: 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbao-Aldaiturriaga N, Gutierrez-Camino A, Martin-Guerrero I, Pombar-Gomez M, Zalacain-Diez M, Patino-Garcia A et al. Polymorphisms in miRNA processing genes and their role in osteosarcoma risk. Pediatr Blood Cancer 2015; 62: 766–769. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta 2015; 1849: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet 2011; 20: 1400–1410. [DOI] [PubMed] [Google Scholar]
- Holt R, Barnby G, Maestrini E, Bacchelli E, Brocklebank D, Sousa I et al. Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Hum Genet 2010; 18: 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]