Main Text
By the early 1990s, most biophysicists and cell biologists agreed that polymerization of actin filaments at the leading edge of a motile cell pushes the plasma membrane forward, creating a protrusion that extends the border of the cell (Fig. 1). For cells on a flat substrate, like a microscope slide, this leading edge is a relatively flat lamella less than 1 μm thick. To balance protrusion at the front end, contractility of the cytoplasm was believed to pull the rear of the cell forward toward attachments that form continuously behind the leading edge.
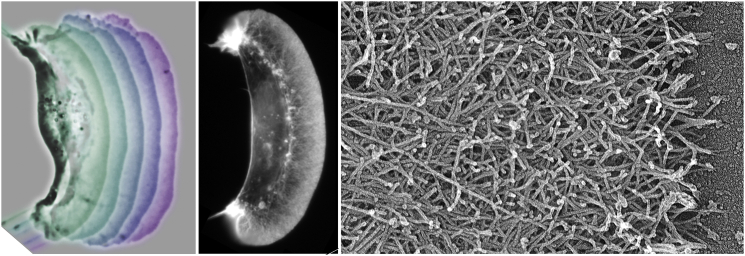
Figure 1.
Actin filaments in motile keratocytes. (left) Superimposed series of phase contrast micrographs at 15 s intervals of fish keratocytes moving on a glass slide. (middle) Fluorescence micrograph of a cell stained with rhodamine-phalloidin to label actin filaments. (right) Transmission electron micrograph of the leading edge of a cell fixed while moving to the right and prepared by extraction, critical point drying and shadowing with platinum. (Courtesy of Gary Borisy and Tanya Svitkina). To see this figure in color, go online.
Multiple lines of evidence supported the idea that actin polymerization drives cellular movements. Early electron micrographs of thin sections showed microfilaments (subsequently confirmed to be actin) in the cytoplasm next to the leading edge (1). Even better, electron micrographs of negatively stained specimens showed dense arrays of filaments at angles to the plasma membrane (2). Drugs that interfere with actin polymerization stop the protrusion of the leading edge (3), and two elegant fluorescence microscopy experiments established that actin polymerizes near the leading edge. First, Yu-Li Wang injected live cells with fluorescent actin, which incorporated into cellular actin structures (4). When he bleached a spot in the fluorescent leading lamella of the stationary cell, the spot moved away from the leading edge as new fluorescent actin was incorporated next to the plasma membrane. Over time, the fluorescence recovered, showing that the filaments turned over. Julie Theriot and Tim Mitchison used photoactivation to learn more (5). They injected motile cells with actin coupled to a “caged” fluorescent dye. After the tagged actin molecules incorporated into cellular actin structures, they uncaged the fluorescent dye with a light pulse near the leading edge. As the cell moved forward, the spot of fluorescent actin was stationary relative to the substrate and turned over on a time scale of tens of seconds. Electron microscopy (2) showed that these leading edge actin filaments are mostly oriented with their faster growing “barbed ends” (6) toward the leading edge of motile cells.
Many of these ideas were reinforced by parallel experiments on the movements of certain bacteria through the cytoplasm of host animal cells (7). Actin polymerization next to the bacterium assembles a comet tail of actin filaments that propels the bacterium through the cytoplasm (8, 9). This work culminated in reconstitution of bacterial motility from purified actin and a few other proteins (10).
Knowing the equilibrium constant for subunit addition to actin filaments, Hill and Kirschner made solid thermodynamic arguments for how polymerization might produce the force to push the membrane forward (11). However, Peskin, Odell, and Oster (12) pointed out “such arguments provide no mechanistic explanation of how the free energy of polymerization is actually transduced into directed mechanical force.”
A series of three classic papers in Biophysical Journal by George Oster with Charles Peskin, Gary Odell, and Alex Mogilner provided the theoretical basis for this energy transduction mechanism. The first paper (12) established the feasibility of a Brownian ratchet to convert polymerization into force that displaces a barrier, such as the plasma membrane or a bacterium. The second paper (13) explained how thermal fluctuations of the tips of growing filaments contribute to the production of force. In such an elastic Brownian ratchet, rapid bending of stiff filaments is rectified rather than Brownian movements of the load (plasma membrane or bacterium). This paper also worked out the optimal geometrical conditions for such a Brownian ratchet to displace the barrier. The third paper (14) showed how the elastic Brownian ratchet works even if some of the actin filaments are transiently linked to the load.
These papers were successful for two reasons. First, the authors were on firm ground in their elegant mathematics and creative intuition about the physics. Equally important, they were also masters of the experimental literature, having absorbed and appreciated previous work on the kinetics and thermodynamics of actin assembly as well as the key experiments on motile cells. This combination of assets allowed the authors to formulate and test plausible physical mechanisms for actin-driven protrusion. Remarkably, their theory not only accounted for prior observations, but also predicted several features of the process that were only later confirmed by experiment. This is an early example of theory leading experiment in biology.
Peskin, Odell, and Oster, 1993: Brownian ratchet mechanism
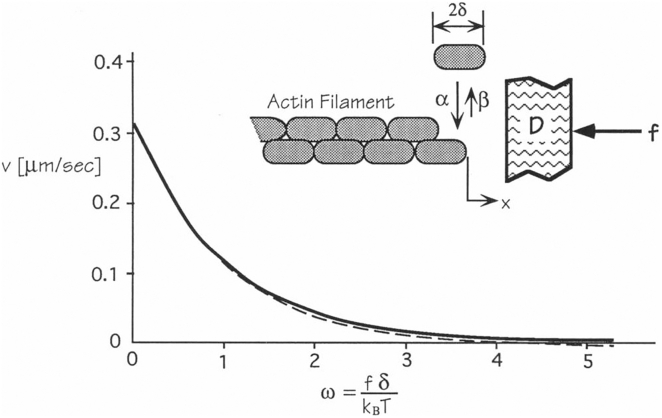
Peskin, Odell, and Oster (12) proposed a “model for how chemical reactions generate protrusive forces by rectifying Brownian motion” of the object that is being moved (Fig. 2). They imagined that growing actin filaments were embedded in a rigid actin filament network with their barbed ends against a barrier, such as a membrane or bacterium. If the barrier underwent thermal fluctuations, space would open up between the end of the filament and the barrier for some fraction of time. If these fluctuations produced a gap as large as an actin subunit, one could bind to the end of the polymer. Each new actin subunit prevented the barrier from diffusing backward, but not from continuing to diffuse forward. Polymerization did not “actually push the bacterium: propulsion is simply Brownian diffusion rendered unidirectional by the polymerization of the actin tail.” In this “imperfect Brownian ratchet” “the concentration of monomers and the binding energy of polymerization supply the free energy to implement the ratchet.” Calculation of force-velocity curves using reasonable parameters showed that the mechanism was feasible for moving bacteria and the plasma membrane at the leading edge of a cell.
Figure 2.
Brownian ratchet model from Peskin et al. (12). Diffusive fluctuations in the position of the barrier (D) open up space for insertion of an actin subunit at the end of the filament. The graph shows the dependence of the velocity on load on the barrier at 10 μM actin monomers. The continuous and dashed lines are the results using two different sets of assumptions (12).
Mogilner and Oster, 1996: Elastic Brownian ratchet mechanism
The simple Brownian ratchet model predicted that the velocity would depend on the size of the particle, but bacteria of different sizes moved at similar rates (15), so random movements of something other than the bacterium must drive the Brownian ratchet. In fact, mathematics graduate student Alex Mogilner realized that thermal fluctuations of something as large as Listeria in cytoplasm are too small to make the simple mechanism work. (See his personal recollections in this issue of Biophysical Journal.) Fortunately, he was able to join Oster’s lab for a one year postdoc before starting a faculty position. During this year, they worked out the elastic Brownian ratchet mechanism.
Mogilner and Oster (13) explored a range of assumptions, including the length, stiffness, and orientations of the filaments and formulated a statistical mechanical model for propulsion by polymerizing actin filaments. They applied their model to the motion of Listeria through the cytoplasm and protrusion of the leading edge.
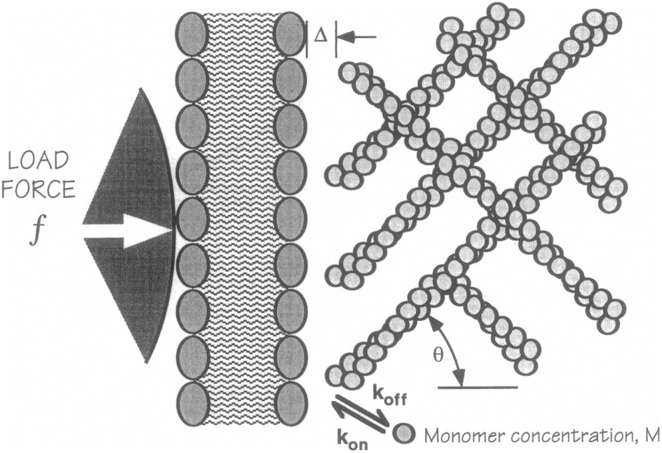
They assumed that the growing filaments were anchored in a stiff network of preexisting filaments and that their tips underwent rapid thermal bending motions, as expected from their known stiffness (Fig. 3). These fluctuations of the filaments produced gaps between the tip and the load that allowed new subunits to elongate the filament. Their key insight was that Brownian ratchet rectifies the motions of the filaments rather than the motions of the bacterium. They calculated how a rapidly fluctuating filament acted as a spring to apply force to the load as it grew in length. They explored how the angle of incidence with the load and the length of the filament influenced the force and velocity produced by polymerization. For the leading lamella, the optimum angle was ∼50° for transducing polymerization into displacement of the membrane.
Figure 3.
Elastic Brownian ratchet model from Mogilner and Oster (13) showing an orthogonal network of flexible actin filaments fluctuating as they grow near the inside of the plasma membrane. The optimal angle (θ) for transducing polymerization into displacement of the membrane is ∼50°.
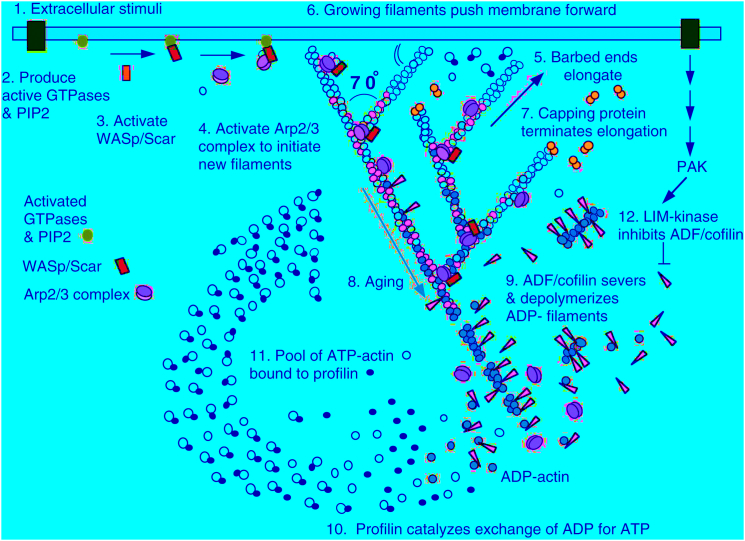
Remarkably, subsequent work (16, 17, 18) showed that Arp2/3 complex produces filaments with the properties of this model (Fig. 4). Arp2/3 complex nucleates branches on the sides of existing filaments (providing the rigid framework assumed in the model) and the branches grow at their free barbed ends at a 70° angle to the mother filament. This sets up an orthogonal network of filaments oriented ±35° relative to the membrane. This was ∼55° in the coordinates of Mogilner and Oster, close to the optimal angle predicted by their model. Selective pressures during evolution of the system ∼1 billion years ago arrived at the physically optimal angle of the branches. Reconstitution experiments (10) showed that movement of bacteria by actin comets depends on capping the filaments so their lengths are limited. Mogilner and Oster showed that short stiff filaments are favorable for the elastic Brownian ratchet. Loisel et al. (10) also showed that cofilin is required for robust comet tails. Cofilin promotes the turnover of the filaments so comet tails maintain a constant length as the bacterium moves forward. Disassembly of the comet tail cycles the proteins to continue the processes. The actin filament polymerase, VASP, and a crosslinking protein, such as alpha-actinin, both enhance movement of bacteria.
Figure 4.
Dendritic nucleation model from Pollard and Borisy (21). Note the 70° branches formed by Arp2/3 complex, fluctuating filaments pushing at an angle against the inside of the plasma membrane as they elongate, and capping to keep the lengths of the branches short enough to push effectively. (Reproduced with permission from Cell). To see this figure in color, go online.
Mogilner and Oster, 2003
Subsequent experimental work showed that components of the actin polymerization machinery (N-WASP and VASP) link growing filaments to the bacterium or the plasma membrane at least transiently (19, 20). These observations raised concerns that such tethers would prevent actin polymerization from moving the barriers, so Mogilner and Oster expanded the elastic Brownian ratchet model to take these tethers into account (14). In their revised model, Arp2/3 complex and newly formed filaments are linked briefly to the load, but they detach and generate force by the elastic Brownian ratchet mechanism until they are capped. Then other growing actin filaments push the barrier away from the capped filaments. Using measured parameter values and reasonable estimates of the missing parameter values, simulations of their model produced movements of bacteria similar to those observed in experiments.
Long-term impact
This theoretical work by Oster and colleagues had a major impact on thinking in biophysics and cell biology despite initial skepticism on the part of some reviewers (see Mogilner’s personal account in this issue of Biophysical Journal). The dendritic nucleation/array treadmilling model (Fig. 4) for force production by actin filament networks (21) stands on three foundations. 1) Quantitative biochemical and biophysical analysis of the proteins that assemble and recycle the actin filament network has provided the mechanistic details. 2) The elastic Brownian ratchet mechanism established the theoretical basis for the fundamental physical principles of the process. 3) Observations of live cells inspired the biophysical and theoretical work and have confirmed the general features of this model for protrusion of the leading edge.
Twenty years after the pivotal 1996 paper, investigators are still busy discovering elegant new features of the system. For example, recent work revealed that force on a branching network of actin filaments increases the density of the network and its force and power (22). Such “force-feedback” presumably enhances the capacity of the self-assembling proteins to adapt to physical conditions during cellular motility.
Editor: Brian Salzberg.
References
- 1.Abercrombie M., Heaysman J.E., Pegrum S.M. The locomotion of fibroblasts in culture. IV. Electron microscopy of the leading lamella. Exp. Cell Res. 1971;67:359–367. doi: 10.1016/0014-4827(71)90420-4. [DOI] [PubMed] [Google Scholar]
- 2.Small J.V., Isenberg G., Celis J.E. Polarity of actin at the leading edge of cultured cells. Nature. 1978;272:638–639. doi: 10.1038/272638a0. [DOI] [PubMed] [Google Scholar]
- 3.Forscher P., Smith S.J. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y.L. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theriot J.A., Mitchison T.J. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- 6.Woodrum D.T., Rich S.A., Pollard T.D. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J. Cell Biol. 1975;67:231–237. doi: 10.1083/jcb.67.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilney L.G., Portnoy D.A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilney L.G., DeRosier D.J., Tilney M.S. How Listeria exploits host cell actin to form its own cytoskeleton. I. Formation of a tail and how that tail might be involved in movement. J. Cell Biol. 1992;118:71–81. doi: 10.1083/jcb.118.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theriot J.A., Mitchison T.J., Portnoy D.A. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 10.Loisel T.P., Boujemaa R., Carlier M.F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 11.Hill T.L., Kirschner M.W. Subunit treadmilling of microtubules or actin in the presence of cellular barriers: possible conversion of chemical free energy into mechanical work. Proc. Natl. Acad. Sci. USA. 1982;79:490–494. doi: 10.1073/pnas.79.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peskin C.S., Odell G.M., Oster G.F. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mogilner A., Oster G. Cell motility driven by actin polymerization. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogilner A., Oster G. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys. J. 2003;84:1591–1605. doi: 10.1016/S0006-3495(03)74969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg M.B., Theriot J.A. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc. Natl. Acad. Sci. USA. 1995;92:6572–6576. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svitkina T.M., Verkhovsky A.B., Borisy G.G. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullins R.D., Heuser J.A., Pollard T.D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch M.D., Rosenblatt J., Mitchison T.J. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 19.Kuo S.C., McGrath J.L. Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature. 2000;407:1026–1029. doi: 10.1038/35039544. [DOI] [PubMed] [Google Scholar]
- 20.Co C., Wong D.T., Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 22.Bieling P., Li T.D., Mullins R.D. Force Feedback Controls Motor Activity and Mechanical Properties of Self-Assembling Branched Actin Networks. Cell. 2016;164:115–127. doi: 10.1016/j.cell.2015.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]