Abstract
Eukaryotic elongation factor 2 kinase (eEF2K) inhibitors may aid in the development of new therapeutic agents to combat cancer. Purified human eEF2K was obtained from an Escherichia coli expression system and a luminescence-based high-throughput screening (HTS) assay was developed using MH-1 peptide as the substrate. The luminescent readouts correlated with the amount of adenosine triphosphate remaining in the kinase reaction. This method was applied to a large-scale screening campaign against a diverse compound library and subsequent confirmation studies. Nine initial hits showing inhibitory activities on eEF2K were identified from 56,000 synthetic compounds during the HTS campaign, of which, five were chosen to test their effects in cancer cell lines.
Key words: eEF2K, Inhibitors, High-throughput screening, Luminescence, MH-1 peptide
Graphical abstract
In this paper, we describe a luminescence-based high-throughput screening (HTS) assay for eukaryotic elongation factor 2 kinase (eEF2K) which represents a potential target for anti-cancer agents. Our HTS campaign against 56,000 small molecule compounds identified 9 initial hits which were further studied in cancer cell lines.
1. Introduction
Protein is the most important structural and functional macromolecule in the cell, therefore, the process of translation (also called protein synthesis), which translates a messenger RNA (mRNA) into a protein, is essential in this regard. Protein synthesis is usually divided into three main stages: initiation, elongation and termination1. The initial stage involves a variety of translation initiation factors that work coordinately to provide stringent regulatory mechanisms of translation. The elongation stage consumes a large amount of energy and amino acids within cells to synthesize new polypeptide chain, which will undergo further post-translational modification and fold into correct quaternary structure to function in the cell. Indeed, more than 99% of the energy and nutrients used in protein synthesis are consumed during elongation. Studies on translation termination mainly focus on abnormal signals and their effects on mRNA degradation while the understanding of other aspects of this step is rather limited2.
Human eukaryotic elongation factor 2 (eEF2) is a member of the GTP-binding translation factor family that promotes the movement of ribosome along mRNA from one codon to the next3, 4. During the elongation stage, eEF2 facilitates the translocation of peptidyl-tRNA from ribosome A site to P site. eEF2 can be phosphorylated and thus inactivated by human eEF2K5. The phosphorylation of eEF2 plays an important role in the regulation of protein synthesis. It was widely recognized that under stress conditions such as nutrient starvation, hypoxia or acidosis, the activity of eEF2K in tumor cells was upregulated which allows tumor cells to adapt and survive in such an adverse microenvironment. Thus, eEF2K has been proposed as a potential target for cancer therapy.
Among the human kinome, the majority (90%) of protein serine/threonine and tyrosine kinases share a similar architecture within their catalytic domain, so they are classified as conventional protein kinases (CPKs). The remainder (about 10%), which displays little sequence homology with CPKs, are named as atypical protein kinases (APKs). APKs and CPKs lack sequence identity, but share a related catalytic core6 as well as a remarkably similar N-terminal lobe that predominantly folds into a curved β-sheet and contains the phosphate binding loop (P-loop). The inter-lobe cleft serves as ATP binding pocket and possesses conserved key catalytic residues7. Despite having similar ATP binding pockets, the spatial location of their conserved region GXGXXG is different. eEF2K is such an atypical Ser/Thr-protein kinase that it was also known as ‘Ca2+/CaM-kinase III’. Human eEF2K is composed of 725 amino acids.
It is well established that phosphorylation at Thr56 inactivates eEF2 thereby regulating protein synthesis at the elongation step of translation8, 9. This mechanism appears crucial for cancer cell survival as it reduces energy and amino acid consumption to help them live through adverse conditions10. This point of view is supported by studies on cellular microenvironment11 and metabolic stress10 with breast cancer cells12, 13. Since eEF2K is neither a conventional protein kinase nor has other known substrates, blockage of its action does not significantly affect normal biological processes. eEF2K inhibitors can become new drug candidates for cancer therapy.
It was thought that an eEF2 peptide (amino acid sequence: SARAGETRFTDTRKDE) containing the phosphorylation site was the substrate of eEF2K. When tested in vitro, however, it did not work efficiently. Scientists subsequently found two protein kinases MHCK (myosin heavy chain kinase from Dictyostelium discoideum) A and B, whose structures had considerable similarity to eEF2K, could phosphorylate a 16 amino acid peptide MH-1 (amino acid sequence: RKKFGESEKTKTKEFL) in vitro, an action that can be replicated by eEF2K14. Thus, MH-1 peptide has been used as a surrogate substrate for eEF2K.
In this study, we developed and validated a luminescence-based high-throughput screening (HTS) assay for the identification of eEF2K inhibitors by use of MH-1 peptide. Traditional method to detect the catalytic activities of protein kinase is to quantify the incorporation of radioactive γ-phosphate form [γ-32P] adenosine triphosphate (ATP) into a peptide or protein substrate by using a scintillation counter. This is impracticable for HTS application. Immunoblotting assay can also be employed in screening, but the sample preparation is complex and costs time and money: it is therefore not suitable for HTS and could be readily replaced by fluorescence and luminescence approaches. MH-1 peptide substrates have been commercialized. For instance, ADP-GloTM luminescent kinase assay kit can eliminate redundant ATP in the assay system and transform ADP into ATP effectively. This became the key technology used in this study.
2. Materials and methods
2.1. Reagents
A484954, eEF2K enzyme produced and purified from Escherichia coli according to the method previously described15 and calmodulin were provided by Professor C. Proud at South Australia Health and Medical Research Institute. MH-1 peptide was purchased from China Peptides Co., Ltd. (Shanghai, China). EDTA, EGTA, CaCl2 and MgCl2 were bought from Shanghai Chemical Reagents Co., Ltd. (Shanghai, China). 3-(N-morpholino) propanesulfonic acid (MOPS), DMSO and β-mercaptoethanol (β-ME) were obtained from Sigma (St Louis, MO, USA). The assay plates were the products of PerkinElmer (Boston, MA, USA). ADP-GloTM luminescent kinase assay kit was procured from Promega (Wisconsin, MA, USA). The anti-eEF2 and anti-phospho-eEF2 antibodies were purchased from Cell Signaling Technologies (Boston, MA, USA). L15 medium, fetal bovine serum, penicillin, streptomycin, sodium pyruvate, geneticin and 0.5% trypsin–EDTA were bought from Life Technologies (Carlsbad, CA, USA). Cancer cell lines used in this study were obtained from ATCC (Manassas, VA, USA).
2.2. Cell culture
MDA-MB-453 cells were maintained in L15 medium containing 10% fetal bovine serum (FBS), 100 units/mL penicillin/streptomycin, 1 mmol/L sodium pyruvate and 0.5 mg/mL geneticin without CO2. H1299 cells and HCT116 cells were maintained in RPMI-1640 medium and McCoy׳s 5a medium containing 10% FBS, respectively, 100 U/mL penicillin/streptomycin, 1 mmol/L sodium pyruvate and 0.5 mg/mL geneticin in a 5% CO2 incubator.
2.3. Compound library
The compound library used for the screening of eEF2K inhibitors is consisted of 56,000 pure synthetic compounds. The stock was presolubilized in 100% DMSO prior to application in the HTS campaign with an average concentration of 1 mg/mL for each compound. The purity (95% minimum) of all compounds was documented and randomly verified by the Chinese National Compound Library.
2.4. Luminescence-based HTS campaign
Before the primary screening, assays were performed manually in solid white 384 square well plates (BD Bioscience, NJ, USA). Serial dilutions of each reagent (enzyme, ATP and unlabeled peptide MH-1) were made to determine the optimal experimental conditions, which led to the selection of 6 ng/µL and 100 µmol/L for eEF2K and MH-1, respectively. In detail, we selected a 10 µL reaction volume. 4 μL of assay mixture 2 (MH-1 and ATP diluted in H2O) was first dispensed by Bravo liquid handler (Agilent, CA, USA) to an assay plate. 2 µL of compounds (1 mg/mL) were diluted with 18 µL 1× kinase buffer in 96 square well plates, then transferred (2 µL) to the 384 square well plates as described above. 4 μL of assay mixture 1 (eEF2K, β-ME and calmodulin diluted in 1× kinase buffer) was then dispensed to each well of the 384 square well plates. Enzyme was first mixed with compounds to initiate the reaction prior to incubation at room temperature (RT) for 2 h. The reaction was stopped by 10 µL of ADP-GloTM reagent from the ADP-GloTM luminescent kinase assay kit followed by incubation at RT for at least 40 min. At last, 10 µL of kinase detection reagent was introduced and incubated at RT for 30 min. Luminescence was measured with an EnVision plate reader and the emission filter used was luminescence 700 (PerkinElmer).
Hits identified from the primary screening were hand-picked (5 mg/mL in DMSO) and serially diluted (1:3) seven times to give a total of eight different concentrations. Each compound was tested in duplicate and the dose-response characteristics were analyzed using GraphPad Prism software (GraphPad, San Diego, CA, USA) for nonlinear regression analysis.
2.5. Western blot
Cells were seeded in 24-well plates at 2×105 cells/well. Twenty-four hours later, they were treated with A484954 and hit compounds for 6 h, rinsed once with Dulbecco׳s phosphate-buffered saline (D-PBS) and harvested in 2× SDS-PAGE loading buffer (TakaRa, Dalian, China). Cell lysates were incubated at 95 °C for 10 min and then resolved in Criterion XT 10% gels (Bio-Rad, Hercules, CA, USA) for at least 60 min. Proteins were transferred to an Immobilon FL PVDF membrane (Millipore, Merck KGaA, Darmstadt, Germany) in 1× transfer buffer for 40 min. The membranes were rocked gently in blocking buffer (5% skimmed milk) and probed with primary antibodies to phospho-eEF2 at Thr56 and eEF2 at 4 °C overnight. The antibody against β-actin was from Life Technologies. Detection of primary antibodies was performed using Alexa Fluor 680 against rabbit secondary antibodies from Invitrogen. Image acquisition and analysis were performed using the ChemiDoc™ MP System (Bio-Rad).
2.6. Statistical analysis
All results were analyzed by using Prism 5 (GraphPad). Z′ factor is a simple statistical parameter for HTS assay, whereas signal-to-background ratio (S/B) is to assess assay quality. Data variation is analyzed by standard deviation (SD) or coefficient of variation (CV) 16. The statistical characteristics and percentage inhibition in this study were calculated as below.
| (1) |
| (2) |
| (3) |
| (4) |
where PC represents positive control wells; NC represents negative control wells; SD represents standard deviation, M represents mean.
3. Results
3.1. Assay optimization
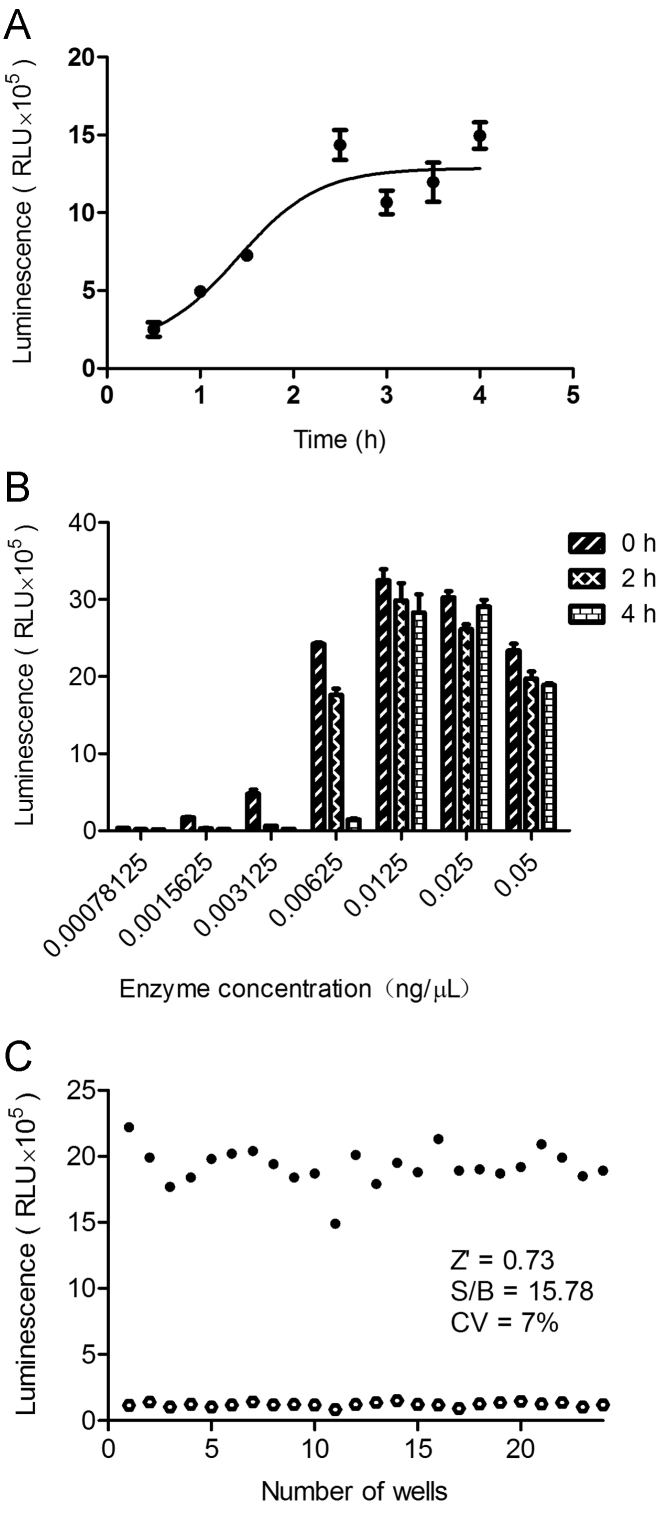
A number of assay parameters that may affect signals evoked by kinase reaction and background noises were studied. In order to ascertain the optimal incubation time, we detected the final luminescence signal at different times after initiation of the assay with appropriate enzyme and superfluous substrate. Since a reaction time of 2 h gave a good signal window it was set as an optimized incubation period (Fig. 1A). Considering that there is a time postponement between first and last batch of samples during the HTS process and kinase at RT may be unstable, we diluted the enzyme (initial concentration was 0.05 mg/mL) by 1:2–1:7 different concentration gradients and placed them at RT for 0, 2 and 4 h, respectively, before adding to the assay plate to activate the reaction. In this condition, according to the luminescence signal shown in Fig. 1B, 0.00625 mg/mL (or 6 ng/µL) enzyme displayed acceptable response which was thus set as an optimal concentrations. Other reagents such as β-ME, ATP and calmodulin were introduced according to the amounts described above. Finally, DMSO (blank) and compound A484954 (positive control) were employed to calculate Z′ factor (0.73), S/B ratio (15.78) and CV (7%) values (Fig. 1C), which suggest that our assay system is of high quality and suitable for HTS.
Figure 1.
Optimization of a luminescence-based HTS assay against eEF2K. (A) Reaction time. (B) Enzyme concentration and room temperature storage time. (C) Z′ factor of the assay was determined at the optimized assay conditions. The background (closed circles) represents luminescence in 2% DMSO (negative control) and the signal (open circles) indicates that in 25 µmol/L of A484954 (positive control). Values given in A and B are means±SEM. RLU, relative luminescence unit.
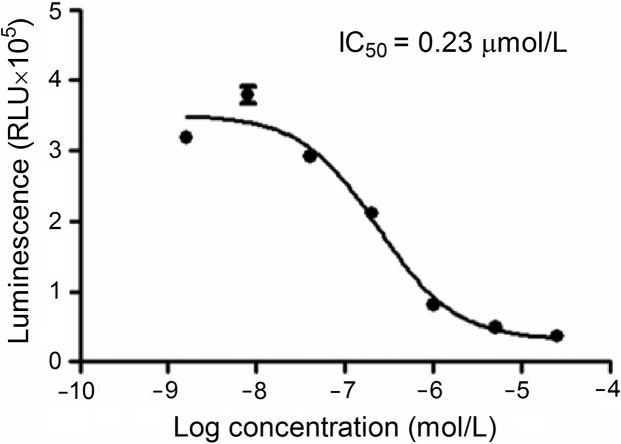
After optimizing the assay conditions, we studied concentration-dependent response features of A484954. An IC50 value of 0.23 µmol/L, consistent with that reported in the literature17 (0.28 µmol/L), was obtained (Fig. 2).
Figure 2.
The experiment was performed under the optimized assay conditions and the IC50 calculated from the concentration–response curve of A484954 is 0.23 µmol/L. RLU, relative luminescence unit.
3.2. HTS campaign
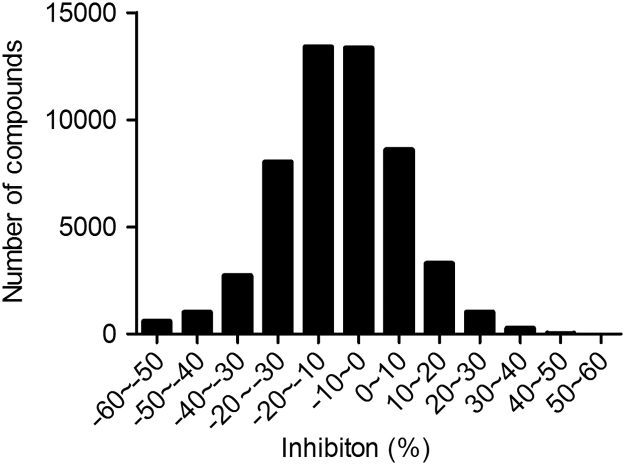
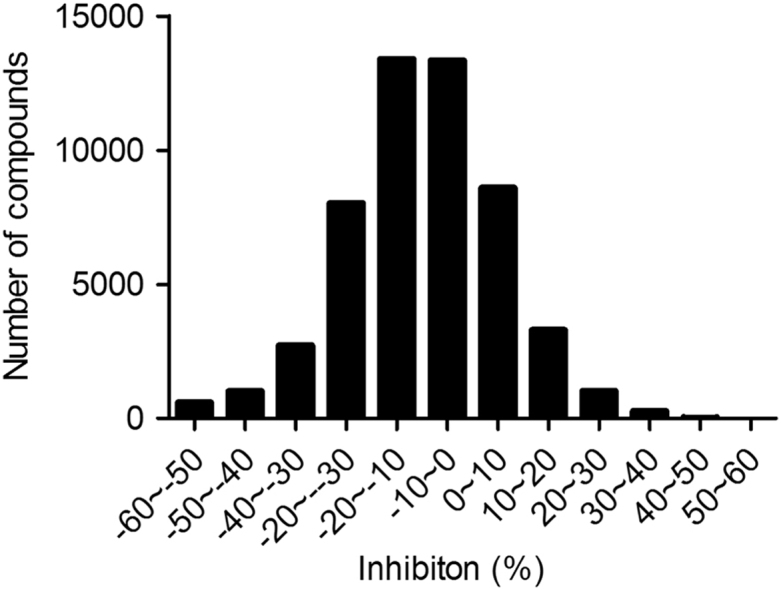
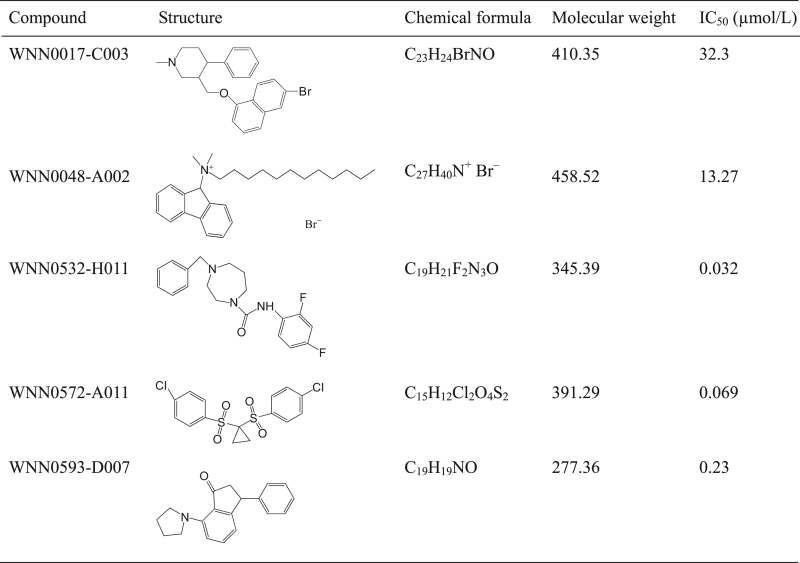
We applied 56,000 compounds from the Chinese National Compound Library to a large-scale random screening against eEF2K. Distribution of inhibition is illustrated in Fig. 3. Only nine initial hits showing inhibition over 25% were identified. They were hand-picked and rescreened using the same assay in duplicates. Five compounds (WNN0017-C003, WNN0048-A002, WNN0532-H011, WNN0572-A011 and WNN0593-D007) displaying consistent inhibition on eEF2K (>30%) were confirmed and further studied for their concentration–response characteristics (Table 1) and effects on cancer cell lines.
Figure 3.
Results of high-throughput screening campaign against eEF2K. Distribution of potential hits identified from 56,000 small molecule compounds is expressed as percentage inhibition of each sample compared with the positive control (A484954).
Table 1.
Confirmed hits showing consistent inhibition of eEF2K activity in vitro.
 |
3.3. Effects on cells
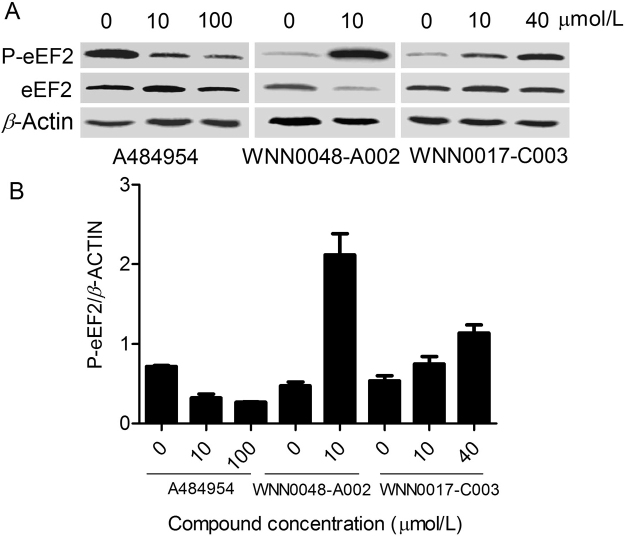
A484954 is a highly selective eEF2K inhibitor with an IC50 value of 0.28 µmol/L in an enzymatic assay while having little activity against a wide panel of other serine/threonine and tyrosine kinases17. Our Western blot data (Fig. 4) demonstrate that A484954 was capable of inhibiting eEF2 phosphorylation in MDA-MB-453, H1299 and HCT116 cells. Due to limited compound availability, we selected WNN0048-A002 (purity>95%) as a representative to investigate its activity in MDA-MB-453 cells. WNN0017-C003 was used as a negative control. Surprisingly, both compounds increased the phosphorylation of eEF2 (p-eEF2) in the cell and the total protein level of eEF2 was not affected by either A484954 or hit compound treatment.
Figure 4.
Western blot analysis of hit compounds in cancer cells. (A) Effect of WNN0048-A002 on eEF2K in MDA-MB-453 cells. The breast cancer cell line was treated with different concentrations of the compound for 6 h. Cell lysates were analyzed by western blotting using antibodies specific for phospho-eEF2 (Th-56). β-Actin was used as a loading control. P-eEF2, eEF2 phosphorylation. (B) Histogram of the western blotting analysis data shown as mean ±SEM from three independent experiments in duplicate.
4. Discussion
In this paper, we presented a robust a luminescence-based enzymatic assay applicable to an HTS setting (Fig. 1). When used to screen a large collection of 56,000 synthetic compounds, it only identified 9 initial hits and 5 of them were subsequently confirmed (Fig. 3 and Table 1). It appears that the hit rate against eEF2K in our screening system is rather low although the assay parameters investigated such as Z′ factor, S/B ratio and CV values are all optimal. The underlying reasons responsible for this phenomenon may be explained by data from additional HTS campaigns with more diverse compound libraries.
Another interesting observation relates to inhibition of eEF2 phosphorylation by the newly discovered eEF2K inhibitors. Unlike the positive control (A484954), both WNN0048-A002 and WNN0017-C003 were unable to show such an effect, but instead, they promoted eEF2 phosphorylation in MDA-MB-453 breast cancer cells (Fig. 4). At this time we do not understand why this happened and whether it was due to a non-specific effect of these compounds or they hit other targets in the signaling pathways upstream of eEF2K. Extending the investigation to other confirmed hits may provide some clues. It should be noted that eEF2K is activated under a wide range of stress conditions.
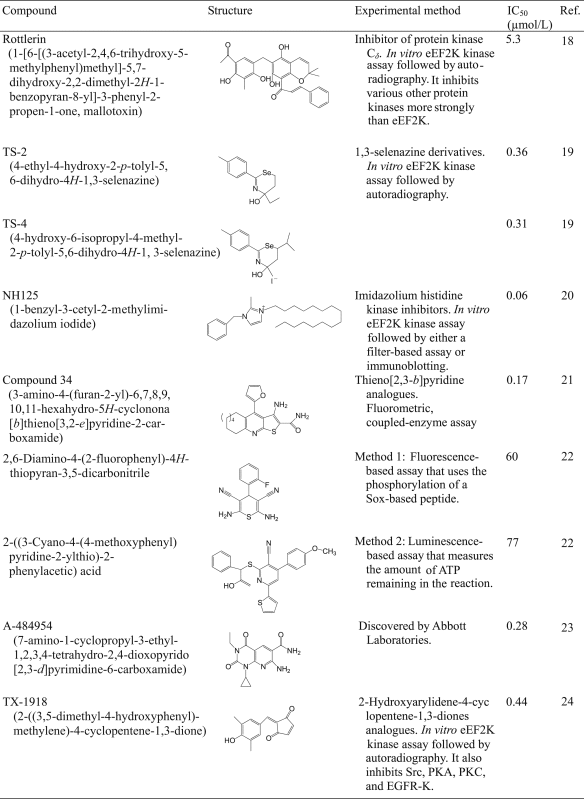
To date, some small molecule eEF2K inhibitors have been reported. They are summarized and compared in terms of the experimental methods led to their discovery in Table 218, 19, 20, 21, 22, 23, 24. Rottlerin is mainly used as a selective inhibitor of protein kinase Cδ. It was found to activate calcium and potassium ion channels and to influence the function of mitochondria, thus can also be used as an eEF2K inhibitor18, 25. 1,3-selenazine analogs react with cysteine residues in cells thereby preventing eEF2 phosphorylation19. NH125 is a widely accepted eEF2K inhibitor: it inhibits eEF2K activity in vitro but promotes eEF2 phosphorylation in vivo11, 20. Thieno [2,3-b] pyridine is an ATP-competitive inhibitor with weak inhibitory effects on eEF2K in cells22. Thiopyran dicarbonitrile analogs are a class of eEF2K inhibitors discovered by HTS23. Finally, A484954 employed in this study is also an ATP-competitive inhibitor23 possessing a good inhibitory effect on eEF2K in vitro but rather low potency in cells. It is also cytotoxic at high concentrations26.
Table 2.
Summary of identified eEF2K inhibitors.
 |
Clearly, there exists an unmet demand for novel eEF2K inhibitors which can inhibit eEF2K effectively but have low or none cell cytotoxicity. In this regard, our work with eEF2K is just a beginning on the road to validate if it is an ideal target for cancer therapy.
Acknowledgments
We are indebted to Claire Moore, Liang Qiu and Caihong Zhou for technical assistance. This work was partially supported by the National Health and Family Planning Commission of China (2012ZX09304-011, 2013ZX09401003-005, 2013ZX09507001 and 2013ZX09507-002), Shanghai Science and Technology Fund (15DZ2291600) and the Thousand Talents Program in China.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Merrick W.C. Eukaryotic protein synthesis: still a mystery. J Biol Chem. 2010;285:21197–21201. doi: 10.1074/jbc.R110.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czaplinski K., Ruiz-Echevarria M.J., González C.I., Peltz S.W. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays. 1999;21:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Rapp G., Klaudiny J., Hagendorff G., Luck M.R., Scheit K.H. Complete sequence of the coding region of human elongation factor 2 (EF-2) by enzymatic amplification of cDNA from human ovarian granulosa cells. Biol Chem Hoppe Seyler. 1989;370:1071–1075. doi: 10.1515/bchm3.1989.370.2.1071. [DOI] [PubMed] [Google Scholar]
- 4.Kaneda Y., Yoshida M.C., Kohno K., Uchida T., Okada Y. Chromosomal assignment of the gene for human elongation factor 2. Proc Natl Acad Sci U S A. 1984;81:3158–3162. doi: 10.1073/pnas.81.10.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenney J.W., Moore C.E., Wang X., Proud C.G. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul. 2014;55:15–27. doi: 10.1016/j.jbior.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi H., Matsushita M., Nairn A.C., Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- 7.Middelbeek J., Clark K., Venselaar H., Huynen M.A., van Leeuwen F.N. The α-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci. 2010;67:875–890. doi: 10.1007/s00018-009-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryazanov A.G., Shestakova E.A., Natapov P.G. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 9.Redpath N.T., Price N.T., Severinov K.V., Proud C.G. Regulation of elongation factor-2 by multisite phosphorylation. Eur J Biochem. 1993;213:689–699. doi: 10.1111/j.1432-1033.1993.tb17809.x. [DOI] [PubMed] [Google Scholar]
- 10.Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A.R., Kool M. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorovkov M.V., Pavur K.S., Petrov A.N., Ryazanov A.G. Regulation of elongation factor-2 kinase by pH. Biochemistry. 2002;41:13444–13450. doi: 10.1021/bi026494p. [DOI] [PubMed] [Google Scholar]
- 12.Parmer T.G., Ward M.D., Yurkow E.J., Vyas V.H., Kearney T.J., Hait W.N. Activity and regulation by growth factors of calmodulin-dependent protein kinase III (elongation factor 2–kinase) in human breast cancer. Br J Cancer. 1999;79:59–64. doi: 10.1038/sj.bjc.6690012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russnes H.G., Caldas C. eEF2K—a new target in breast cancers with combined inactivation of p53 and PTEN. EMBO Mol Med. 2014;6:1512–1514. doi: 10.15252/emmm.201404683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavur K.S., Petrov A.N., Ryazanov A.G. Mapping the functional domains of elongation factor-2 kinase. Biochemistry. 2000;39:12216–12224. doi: 10.1021/bi0007270. [DOI] [PubMed] [Google Scholar]
- 15.Abramczyk O., Tavares C.D., Devkota A.K., Ryazanov A.G., Turk B.E., Riggs A.F. Purification and characterization of tagless recombinant human elongation factor 2 kinase (eEF-2K) expressed in Escherichia coli. Protein Expr Purif. 2011;79:237–244. doi: 10.1016/j.pep.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J.H., Chung T.D., Oldenburg K.R. A Simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 17.Edupuganti R., Wang Q., Tavares C.D., Chitjian C.A., Bachman J.L., Ren P. Synthesis and biological evaluation of pyrido [2,3-d] pyrimidine-2,4-dione derivatives as eEF-2K inhibitors. Bioorgan Med Chem. 2014;22:4910–4916. doi: 10.1016/j.bmc.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gschwendt M., Kittstein W., Marks F. Elongation factor-2 kinase: effective inhibition by the novel protein kinase inhibitor rottlerin and relative insensitivity towards staurosporine. FEBS Lett. 1994;338:85–88. doi: 10.1016/0014-5793(94)80121-5. [DOI] [PubMed] [Google Scholar]
- 19.Cho S.I., Koketsu M., Ishihara H., Matsushita M., Nairn A.C., Fukazawa H. Novel compounds, ‘1,3-selenazine derivatives’ as specific inhibitors of eukaryotic elongation factor-2 kinase. Biochim Biophys Acta. 2000;1475:207–215. doi: 10.1016/s0304-4165(00)00061-1. [DOI] [PubMed] [Google Scholar]
- 20.Arora S., Yang J.M., Kinzy T.G., Utsumi R., Okamoto T. Identification and characterization of an inhibitor of eukaryotic elongation factor 2 kinase against human cancer cell lines. Cancer Res. 2003;63:6894–6899. [PubMed] [Google Scholar]
- 21.Lockman J.W., Reeder M.D., Suzuki K., Ostanin K., Hoff R., Bhoite L. Inhibition of eEF2-K by thieno [2,3-b] pyridine analogues. Bioorg Med Chem Lett. 2010;20:2283–2286. doi: 10.1016/j.bmcl.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Devkota A.K., Warthaka M., Edupuganti R., Tavares C.D., Johnson W.H., Ozpolat B. High-throughput screens for eEF-2 kinase. J Biomol Screen. 2014;19:445–452. doi: 10.1177/1087057113505204. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z., Gopalakrishnan S.M., Bui M.H., Soni N.B., Warrior U., Johnson E.F. 1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) induces phosphorylation of eukaryotic elongation factor-2 (eEF2): a cautionary note on the anticancer mechanism of an eEF2 kinase inhibitor. J Biol Chem. 2011;286:43951–43958. doi: 10.1074/jbc.M111.301291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hori H., Nagasawa H., Ishibashi M., Uto Y., Hirata A., Saijo K. TX-1123: an antitumor 2-hydroxyarylidene-4-cyclopentene-1,3-dione as a protein tyrosine kinase inhibitor having low mitochondrial toxicity. Bioorg Med Chem. 2002;10:3257–3265. doi: 10.1016/s0968-0896(02)00160-8. [DOI] [PubMed] [Google Scholar]
- 25.Stephen S.P. Rottlerin: an inappropriate and ineffective inhibitor of PKCδ. Trends Pharmacol Sci. 2007;28:453–458. doi: 10.1016/j.tips.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Devkota A.K., Tavares C.D., Warthaka M., Abramczyk O., Marshall K.D., Kaoud T.S. Investigating the kinetic mechanism of inhibition of elongation factor 2 kinase by NH125: evidence of a common in vitro artifact. Biochemistry. 2012;51:2100–2112. doi: 10.1021/bi201787p. [DOI] [PMC free article] [PubMed] [Google Scholar]