Abstract
Background
Microbial culturomics represents an ongoing revolution in the characterization of environmental and human microbiome.
Methods
By using three media containing high salt concentration (100, 150, and 200 g/L), the halophilic microbial culturome of a commercial table salt was determined.
Results
Eighteen species belonging to the Terrabacteria group were isolated including eight moderate halophilic and 10 halotolerant bacteria. Gracilibacillus massiliensis sp. nov., type strain Awa-1T (=CSUR P1441=DSM 29726), is a moderately halophilic gram-positive, non-spore-forming rod, and is motile by using a flagellum. Strain Awa-1T shows catalase activity but no oxidase activity. It is not only an aerobic bacterium but also able to grow in anaerobic and microaerophilic atmospheres. The draft genome of G. massiliensis is 4,207,226 bp long, composed of 13 scaffolds with 36.05% of G+C content. It contains 3,908 genes (3,839 protein-coding and 69 RNA genes). At least 1,983 (52%) orthologous proteins were not shared with the closest phylogenetic species. Hundred twenty-six genes (3.3%) were identified as ORFans.
Conclusions
Microbial culturomics can dramatically improve the characterization of the food and environmental microbiota repertoire, deciphering new bacterial species and new genes. Further studies will clarify the geographic specificity and the putative role of these new microbes and their related functional genetic content in environment, health, and disease.
Keywords: Gracilibacillus massiliensis, taxono-genomics, culturomics, microbial community, salt, halophile
Salt (sodium chloride) is the main mineral constituent of sea water, the oldest and most ubiquitous of food seasonings and an important method of food preservation. Salt was considered hostile to most forms of life; however, it favored the emergence and growth of halophilic bacteria in salty foods (1). Therefore, study on the diversity of hypersaline environmental microorganisms brings important information in the field of environmental microbiology. Recent studies have reported the isolation of new species from salty and/or fermented food (2, 3).
As part of the ongoing microbial culturomics revolution in our laboratory (4), we performed the ‘microbial culturome’ of a table salt isolating a new moderately halophilic bacterial species belonging to the genus Gracilibacillus. First described by Wainø et al. in 1999 (5), the genus Gracilibacillus includes, moderately halophilic or halotolerant, mobile, gram-positive bacteria, most of them forming endospores or filaments containing menaquinone-7 (MK-7) as predominant respiratory quinone (6). This genus includes 12 species (www.bacterio.net) described with valid published names (7). Members of the genus Gracilibacillus are salty environmental bacteria isolated most often from soil (8), food (9), lakes and salty sea water (10, 11).
To extend the halophilic environmental repertoire, we report here the characterization of a new halophilic species using the taxono-genomics strategy. Taxono-genomics integrate proteomic information obtained by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and genomic tests to describe new bacterial species (12, 13). This polyphasic approach overcomes limitations of conventional methods based on genetic, phenotypic, and chemotaxonomic characteristics for new species description (14, 15).
Our new bacterial species Gracilibacillus Awa-1T (=CSUR P1441=DSM 29726, CSUR stands for ‘Collection de Souches de l'Unité des Rickettsies’ and DSM stands for ‘Deutsche Sammlung von Mikroorganismen’), type strain of Gracilibacillus massiliensis sp. nov., was isolated from a sample of commercial table salt, a hand-harvested ‘fleur de sel’, salt from the Camargue natural region. Naturally white, it contains 67% (w/v) NaCl. Fleur de sel is a hand-harvested sea salt collected by workers who scrape only the top layer of salt before it sinks to the bottom of large salt pans. It was harvested in the Saline of Aigues-Mortes in southern France, in a wild, unusual, and unexplored biodiversity habitat. The microbial culturome of this table salt sample and the phenotypic, phylogenetic, and genomic characteristics of the new species isolated in this culturomics approach are reported here.
Materials and methods
Strain isolation
The Camargue sea salt ‘Fleur de Sel de Camargue’ sample was bought in a supermarket. The sample was transported to our laboratory in the same conditions as at the point of sale, at room temperature. The salinity of the sample was measured using a digital refractometer (Fisher Scientific, Illkirch, France) and its pH was measured using a pH-meter (Eutech Instruments, Strasbourg, France). For the cultivation of halophilic microorganisms, we created media containing high salt concentrations (100, 150, and 200 g/L) (16). Gracilibacillus strain Awa-1T was isolated in September 2014 by cultivation under aerobic conditions, on a homemade halophilic culture medium consisting of a Columbia agar (42 g/L) culture medium (Sigma-Aldrich, Saint-Louis, MO, USA) supplemented by the addition of (per liter) MgCl2 6H2O, 10 g; MgSO4 7H2O, 10 g; KCl, 4 g; CaCl2 2H2O, 1 g; NaHCO3, 0.5 g; glucose, 2 g; 100–150 g/L of NaCl and 5 g of yeast extract (Becton Dickinson, Le-Pont-de-Claix, France). The pH was adjusted to 7.5 with 10 M NaOH before autoclaving at 120°C.
Strain identification by MALDI-TOF MS
MALDI-TOF MS protein analysis was performed using a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany), as previously reported (17). Each separate colony selected was deposited in duplicate on a MALDI-TOF target to be analyzed. A matrix solution of 1.5 µL (saturated solution of α-cyano-4-hydroxycinnamic acid diluted in 50% acetonitrile, 2.5% of trifluoroacetic acid, completed with HPLC water) was deposed on each spot. After reading of the plate, the obtained protein spectra were compared with those of the Bruker database (continuously updated with our recent data) in order to obtain a score, which enables, or not, identification of the strain.
Strain identification by 16S rRNA gene sequencing
The colonies unidentified by the MALDI-TOF after three tests were suspended in 200 µL of distilled water for deoxyribonucleic acid (DNA) extraction by EZ1 DNA Tissue Kit (Qiagen, Courtaboeuf, France). The amplification of the 16S rRNA gene was done by standard polymerase chain reaction (PCR), with the use of universal primers pair FD1 and rp2. The amplified DNA was revealed by electrophoresis on 1.5% agarose gel. Once validated, the PCR product was purified and sequenced using the Big Dye Terminator Sequencing Kit and the following internal primers: 536F, 536R, 800F, 800R, 1050F, 1050R, 357F, and 357R, as previously described (4).
Description of a new species by taxono-genomics
Phylogenetic analysis
We performed a phylogenetic analysis based on 16S rRNA of our isolate to identify its phylogenetic affiliations with other isolates of the genus Gracilibacillus. Sequences were aligned using Muscle software (18) and phylogenetic inferences were obtained using the approximately maximum likelihood method within the FastTree software (19). Numbers at the nodes are support local values computed through the Shimodaira–Hasegawa test (20).
Microscopy, sporulation, and motility assays
To observe G. massiliensis strain Awa-1T morphology, transmission electron microscopy was performed after negative staining, using a Tecnai G20 (FEI Company, Limeil-Brevannes, France) at an operating voltage of 60 KV. The gram staining was performed and observed using a photonic microscope Leica DM2500 (Leica Microsystems, Nanterre, France) with a 100X oil-immersion objective. Motility testing was performed by observation of a fresh colony between the blades and slats using DM1000 photonic microscope (Leica Microsystems) at 40x. For the sporulation test, our strain was grown on Chapman agar (Oxoid, Dardilly, France) for 1 week, followed by gram staining and observation for the presence or absence of spores on colonies under the microscope.
Antimicrobial susceptibility and biochemical and atmospheric tests
Sensitivity to antibiotics was determined on a Mueller–Hinton agar in a petri dish (BioMerieux, Marcy-l'Etoile, France). The following antibiotics were tested using Sirscan discs (i2a, Perols, France): doxycycline, rifampicin, vancomycin, amoxicillin, erythromycin, ceftriaxone, ciprofloxacin, gentamicin, penicillin, trimethoprim/sulfamethoxazole, imipenem, and metronidazole. Scan 1200 was used to interpret the results (Interscience, Saint Nom la Bretêche, France).
The commercially available API ZYM, API 50CH, and API 20 NE strips (BioMerieux, Marcy-l'Etoile, France) were used for biochemical tests according to the manufacturer's instructions. The time of incubation was 4 h for API ZYM and 48 h for the others.
Growth of the strain Awa-1T was tested with different growth temperatures (25°C, 30°C, 37°C, 45°C) under aerobic conditions and also in anaerobic and microaerophilic atmospheres, created using AnaeroGenTM (Atmosphere Generation Systems, Dardily, France) and anaerobic jars (Mitsubishi) with GENbag microaer system (BioMerieux), respectively.
Cellular fatty acid analysis
Fatty acid methyl ester (FAME) analysis was performed by Gaz chromatography/mass spectrometry (GC/MS). Two samples were prepared with approximately 40 mg of bacterial biomass, each harvested from several culture plates. FAMEs were prepared as described by Sasser (21). GC/MS analyses were carried out as described before (22). Briefly, FAMEs were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500 – SQ 8 S, Perkin Elmer, Courtaboeuf, France). A spectral database search was performed using MS Search 2.0, operated with the Standard Reference Database 1A (NIST, Gaithersburg, MD, USA) and the FAMEs mass spectral database (Wiley, Chichester, UK).
Genomic DNA preparation
After 48 h of growth of the strain Awa-1T in four petri dishes using our homemade halophilic culture medium, bacteria were resuspended in sterile water and centrifuged at 4°C at 2,000 ×g for 20 min. Cell pellets were resuspended in 1 mL Tris/EDTA/NaCl (10 mM Tris/HCl (pH7.0), 10 mM EDTA (pH8.0), and 300 mM NaCl) and recentrifuged under the same conditions. The pellets were then resuspended in 200 µL Tris-EDTA buffer (TE buffer) and Proteinase K and kept overnight at 37°C for cell lysis. DNA was purified with phenol/chloroform/isoamylalcohol (25:24:1), followed by a precipitation with ethanol at −20°C. The DNA was resuspended in TE buffer and quantified by Qubit fluorometer using the high-sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 112.7 ng/µL.
Genome sequencing and assembly
Genomic DNA (gDNA) of G. massiliensis was sequenced on the MiSeq Technology (Illumina Inc, San Diego, CA, USA) with the mate pair strategy. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina). The mate pair library was prepared with 1.5 µg of gDNA using the Nextera mate pair Illumina guide. The gDNA sample was simultaneously fragmented and tagged with a mate pair junction adapter. The pattern of the fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies Inc, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1.5 up to 11 kb with an optimal size at 6.641 kb. No size selection was performed and 600 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal at 1,309 bp on the Covaris device S2 in T6 tubes (Covaris, Woburn, MA, USA). The library profile was visualized on a high-sensitivity Bioanalyzer LabChip (Agilent Technologies Inc, Santa Clara, CA, USA) and the final concentration library was measured at 47.82 nmol/L. The libraries were normalized at 4 nM and pooled. After a denaturation step and dilution, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. The automated cluster generation and sequencing run were performed in a single 2×251-bp run.
Total information of 7.9 Gb was obtained from an 816 K/mm2 cluster density with cluster passing quality control filters of 91.7% (15,550,000 passing filter paired reads). Within this run, the index representation for G. massiliensis was determined to be 5.41%. The 841,255 paired reads were trimmed then assembled to 13 scaffolds.
Genome annotation and comparison
Prodigal was used for open reading frames (ORFs) prediction (23) with default parameters. Predicted ORFs spanning a sequencing gap region (containing N) were excluded. Bacterial protein sequences were predicted using BLASTP (E-value 1e−03, coverage 0.7 and identity percent 30%) against the clusters of orthologous groups (COG) database. If no hit was found, a search against the non redundant (NR) database (24) was performed using BLASTP with E-value of 1e−03 coverage 0.7 and an identity percent of 30%. If sequence lengths were smaller than 80 amino acids, we used an E-value of 1e−05. PFAM-conserved domains (PFAM-A and PFAM-B domains) were searched on each protein with the hhmscan tools analysis. RNAmmer (25) was used to find ribosomal RNAs genes, whereas tRNA genes were found using the tRNAScanSE tool (26). We predicted the lipoprotein signal peptides and the number of transmembrane helices using Phobius (27). ORFans were identified if all the BLASTP performed had negative results (E-value smaller than 1e−03 for ORFs with sequence size greater than 80 aa or E-value smaller than 1e−05 for ORFs with sequence length smaller than 80 aa). Artemis (28) and DNA Plotter (29) were used for data management and for visualization of genomic features, respectively. We used the MAGI homemade software to estimate the mean level of nucleotide sequence similarity at the genome level. It calculated the average genomic identity of gene sequences (AGIOS) among compared genomes (30). This software combines the Proteinortho software (31) for detecting orthologous proteins in pairwise genomic comparisons, then retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity among orthologous ORFs using the Needleman–Wunsch global alignment algorithm. Genomes from the genus Gracilibacillus and closely related genera were used for the calculation of AGIOS values. The genome of G. massiliensis strain Awa-1T (EMBL-EBI accession number CZRP00000000) was compared with that of Halobacillus halophilus type strain DSM 2266 (HE717023), Amphibacillus jilinensis strain Y1 (AMWI00000000), Halobacillus trueperi strain HT-01 (CCDJ000000000), Gracilibacillus halophilus strain YIM-C55.5 (APML00000000), and Gracilibacillus boraciitolerans strain JCM 21714 (BAVS00000000). Annotation and comparison processes were performed in the Multi-Agent software system DAGOBAH (32), which include Figenix (33) libraries that provide pipeline analysis. We also performed genome-to-genome distance calculator (GGDC) analysis using the GGDC web server as previously reported (34).
Accession numbers
The 16S rRNA and genome sequences are deposited in EMBL-EBI under accession numbers LN626645 and CZRP00000000, respectively.
Results
Description of the table salt microbiota community
The cultivable halophilic and halotolerant bacterial consortia isolated from the fleur de sel of Camargue included 18 bacterial species (Table 1) from 4,303 colonies. MALDI-TOF MS identified 13 species, whereas 16S rRNA gene sequencing identified five other species including a new species (G. massiliensis sp. nov.). Among the four culture conditions used, only three conditions yielded colonies. All colonies were isolated from media with a concentration of 75 g/L (standard Chapman medium), 100 g/L and 150 g/L NaCl (house-made media). Conversely, in the culture medium containing 200 g/L NaCl, no bacterial colonies were isolated. Among the 18 cultured species, 10 were halotolerant and 8 were halophilic species (Table 1).
Table 1.
Description of the table salt microbiota
| Species | Halophile | Salt concentration in the mediuma | |
|---|---|---|---|
| MALDI-TOF identification | |||
| Bacillus firmus | Halotolerant | 75–150 g/L | |
| Bacillus licheniformis | Halotolerant | 75–150 g/L | |
| Gracilibacillus dipsosauri | Moderate halophile | 75–150 g/L | |
| Halobacillus trueperi | Moderate halophile | 75–150 g/L | |
| Micrococcus luteus | Halotolerant | 75–150 g/L | |
| Oceanobacillus picturae | Moderate halophile | 75–150 g/L | |
| Planococcus rifietoensis | Halotolerant | 75–150 g/L | |
| Staphylococcus capitis | Halotolerant | 75–150 g/L | |
| Staphylococcus cohnii | Halotolerant | 75–150 g/L | |
| Staphylococcus haemolyticus | Halotolerant | 75–150 g/L | |
| Staphylococcus hominis | Halotolerant | 75–150 g/L | |
| Staphylococcus epidermis | Halotolerant | 75–150 g/L | |
| Staphylococcus warneri | Halotolerant | 75–150 g/L | |
| 16S identification | |||
| Alkalibacillus halophilus | Moderate halophile | 75–150 g/L | |
| Paraliobacillus quinghaiensis | Moderate halophile | 75–150 g/L | |
| Thalassobacillus devorans | Moderate halophile | 75–150 g/L | |
| Virgibacillus picturae | Moderate halophile | 75–150 g/L | |
| Gracilibacillus massiliensis sp.nov | Moderate halophile | 75–150 g/L |
No colonies grew on the medium with 200 g/L of salt.
Identification and phylogenetic analysis of the new species
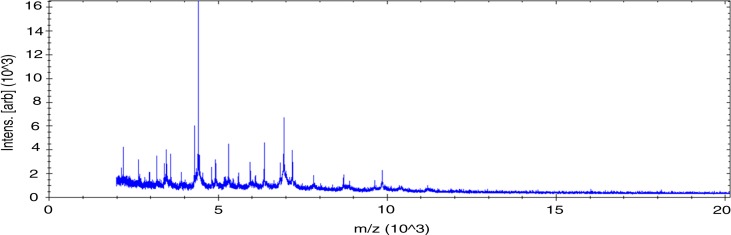
MALDI-TOF score obtained for strain Awa-1T against our database (Bruker database constantly incremented with new data) suggests that our isolate was not a member of a known species. We added the spectrum from strain Awa-1T to our database (Fig. 1).
Fig. 1.
Reference mass spectrum from Gracilibacillus massiliensis strain Awa-1T spectra.
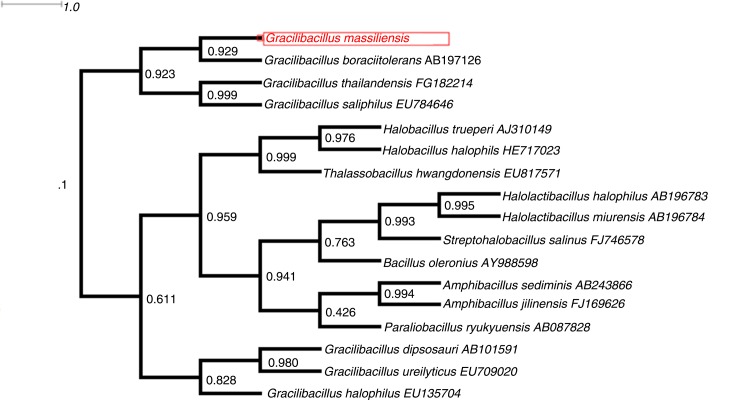
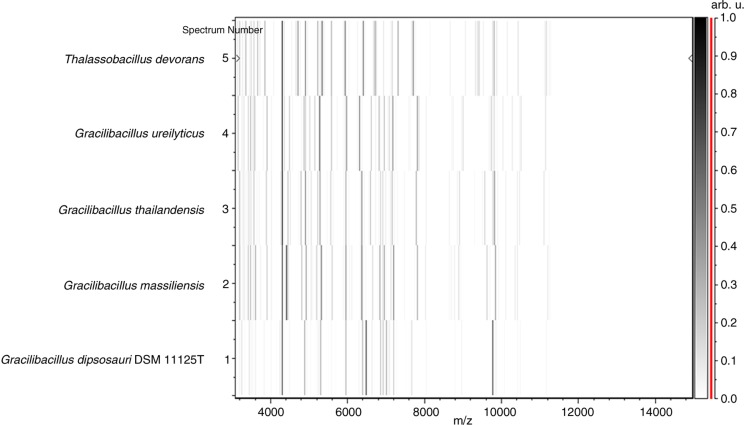
PCR-based identification of the 16S rRNA of G. massiliensis (EMBL-EBI accession number LN626645) yielded 96.9% 16S rRNA gene sequence similarity with the reference Gracilibacillus thailandensis (GenBank accession number NR116568), the phylogenetically closest validated Gracilibacillus species (Fig. 2). This value was lower than the 98.7% 16S rRNA gene sequence threshold advised by Meier-Kolthoff et al. (35) to delineate a new species without carrying out DNA–DNA hybridization. The gel view demonstrated the spectral differences with other members of the genus Gracilibacillus (Fig. 3).
Fig. 2.
Phylogenetic tree highlighting the phylogenetic position of Gracilibacillus massiliensis strain Awa-1T relative to other species. GenBank accession numbers are indicated after the name. Sequences were aligned using Muscle software, and phylogenetic inferences were obtained by using the approximately maximum likelihood method within the FastTree software. Numbers at the nodes are support local values computed through the Shimodaira–Hasegawa test.
Fig. 3.
Gel view comparing Gracilibacillus massiliensis strain Awa-1T to other species within the genera Gracilibacillus and Thalassobacillus.
Physiological and biochemical characteristics
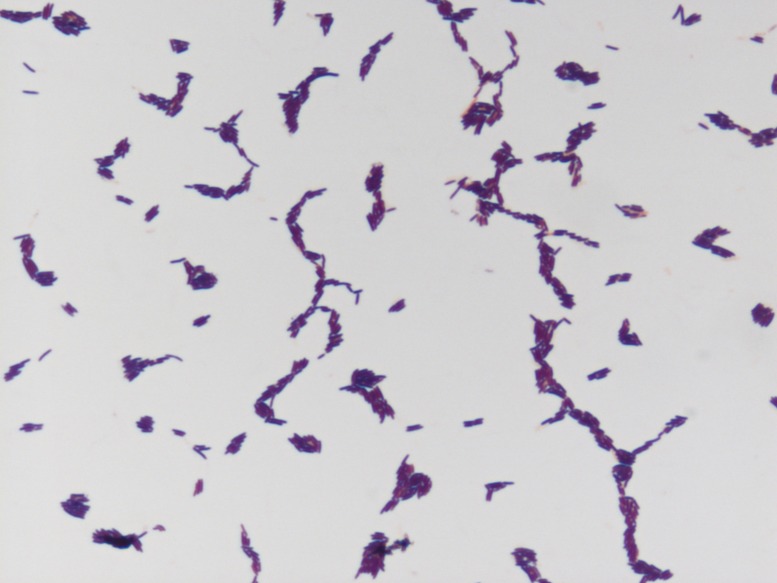
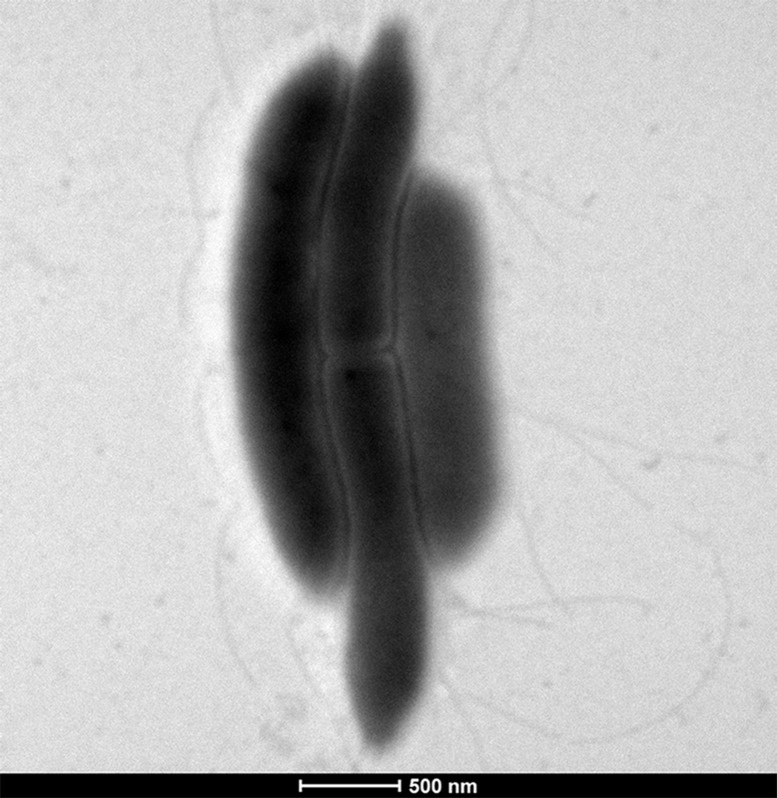
G. massiliensis is a gram-positive (Fig. 4) thin, long rod, with a mean diameter of 0.3 µm and a length of 1.8 µm measured through electron microscopy (Fig. 5). This strain is non-spore-forming, peritrichous, and motile. It grew under aerobic conditions but was also able to grow in anaerobic (at 29°C) and microaerophilic (at 29°C −37°C) atmospheres. The colonies are convex, creamy white, circular, and measured 0.2–0.3 mm in diameter after 2–4 days of growth in our homemade culture medium. Classification and general features are summarized in Table 2.
Fig. 4.
Gram staining of Gracilibacillus massiliensis strain Awa-1T.
Fig. 5.
Transmission electron microscopy of Gracilibacillus massiliensis strain Awa-1T.
Table 2.
Classification and general features of Gracilibacillus massiliensis strain Awa-1T according to the MIGS recommendations (23)
| MIGS ID | Property classification | Term | Evidence codea |
|---|---|---|---|
| Domain: Bacteria | TAS (36) | ||
| Phylum: Firmicutes | TAS (37) | ||
| Class: Bacilli | TAS (36) | ||
| Order: Bacillales | TAS (36) | ||
| Family: Bacillaceae | TAS (36) | ||
| Genus: Gracilibacillus | TAS (5) | ||
| Species: Gracilibacillus massiliensis | IDA | ||
| Type strain: Awa-1T | IDA | ||
| Gram strain | Positive | IDA | |
| Cell shape | Rods | IDA | |
| Motility | Motile | IDA | |
| Sporulation | No sporulating | IDA | |
| Temperature (°C) | Mesophile (25–45) | IDA | |
| Optimum temperature | 37°C | IDA | |
| pH range: optimum | 6.0–9.0: 7.0–8.0 | IDA | |
| Carbon source | Unknown | IDA | |
| MIGS-6 | Habitat | Salt environment | IDA |
| MIGS-6.3 | NaCl range: optimum | 75–150:75 g/L | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Unknown | IDA |
Evidence codes – IDA, inferred from direct assay; TAS, traceable author statement (i.e. a direct report exists in the literature). These evidence codes are from the Gene Ontology project (38).
The strain was catalase test positive and oxidase negative. Using API ZYM, API 20NE, and API 50CH identification strips, positive reactions were observed for esterase, lipase, α-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, urease, and 4-nitrophenyl-βD-galactopyranoside. Acid was not produced from D-glucose, D-mannitol, D-saccharose, D-maltose, D-lactose, L-arabinose, glycerol, D-mannose, D-fructose or D-ribose. Esculin was hydrolyzed, but nitrate was not reduced and indole was negative. Phenotypic characteristics were compared to those of other members of the genus Gracilibacillus (Table 3). Antimicrobial susceptibility tests demonstrated that the isolate was susceptible to doxycycline, rifampicin, vancomycin, erythromycin, ciprofloxacin, gentamicin, trimethoprim/sulfamethoxazole, and imipenem, but resistant to metronidazole, amoxicillin, ceftriaxone, and penicillin G.
Table 3.
Differential characteristics of Gracilibacillus massiliensis compared to other close bacteria of the genus Gracilibacillus
| Properties | G. massiliensis | G. thailandensis | G. saliphilus | G. orientalis | G. ureilyticus | G. halophilus | G. boraciitolerans | G. kekensis | G. halotolerans | G. alcaliphilus |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell diameter (µm) | 0.3–1.8 | 0.3–0.4 | 0.7–0.9 | 0.7–0.9 | 0.7–1 | 0.3–0.5 | 0.5–0.9 | 0.2–1.05 | 0.4–0.6 | 0.5–0.7 |
| Pigmentation | White | White | Creamy white | Creamy | Creamy | White | Dirty white | Creamy white | Creamy white | Creamy white |
| Oxygen requirement | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic |
| Gram stain | + | + | + | + | + | + | + | + | + | + |
| Salt requirement | + | + | + | + | + | + | + | + | + | + |
| Motility | + | + | + | + | + | + | + | + | + | + |
| Sporulation | − | + | + | + | + | + | + | + | + | + |
| Indole | − | − | − | − | − | − | − | − | − | − |
| Production of | ||||||||||
| Alkaline phosphate | − | − | + | NA | + | + | + | NA | + | − |
| Catalase | + | + | + | + | + | + | + | NA | + | + |
| Oxidase | − | + | + | − | + | + | + | − | + | − |
| Nitrate reductase | − | + | + | − | + | + | − | − | + | + |
| Urease | + | − | + | − | + | − | − | − | + | + |
| Arginine dihydrolase | NA | − | − | − | + | − | − | NA | − | − |
| β-galactosidase | − | NA | + | NA | + | + | + | NA | + | NA |
| α-galactosidase | + | NA | − | NA | + | − | + | NA | NA | − |
| N-acetyl-glucosamine | − | NA | + | NA | NA | − | NA | NA | NA | + |
| Acid from | ||||||||||
| L-Arabinose | − | + | + | + | + | − | + | + | + | + |
| Ribose | − | + | + | NA | NA | + | + | + | + | + |
| D-mannose | − | + | + | − | + | − | + | + | − | − |
| D-mannitol | − | + | + | + | + | + | + | + | + | + |
| D-sucrose | NA | + | + | + | + | + | NA | + | − | + |
| D-glucose | − | + | + | + | + | + | + | + | + | + |
| D-fructose | − | + | + | + | NA | + | + | + | + | + |
| D-maltose | − | + | + | + | + | − | + | + | − | + |
| D-lactose | − | − | + | + | + | − | + | + | − | + |
| DNA G+C content (mol%) | 36.05 | 37.6 | 40.1 | 37.1 | 35.3 | 42.3 | 35.8 | 35.8 | 38 | 41.3 |
| Habitat | Cooking salt | Fermented fish | Salt lake | Salt lake | Saline-alkaline soil | Salt soil | Soil | Salt lake | Saline soil | Fermentation liquor for dyeing |
Analysis of the total cellular fatty acid composition of G. massiliensis demonstrated that the fatty acids detected are mainly saturated. The most abundant species (15:0 anteiso, 15:0 iso, and 17:0 anteiso) are branched fatty acids. A few unsaturated fatty acids were detected at low abundances (Table 4).
Table 4.
Total cellular fatty acid composition of Gracilibacillus massiliensis strain Awa-1T
| Fatty acids | IUPAC name | Mean relative (%)a |
|---|---|---|
| 15:0 anteiso | 12-methyl-tetradecanoic acid | 45.6±0.3 |
| 15:0 iso | 13-methyl-tetradecanoic acid | 21.2±0.3 |
| 17:0 anteiso | 14-methyl-hexadecanoic acid | 7.9±0.2 |
| 16:0 | Hexadecanoic acid | 5.7±0.1 |
| 15:0 | Pentadecanoic acid | 5.4±0.1 |
| 16:0 iso | 14-methyl-pentadecanoic acid | 3.4±0.02 |
| 14:0 iso | 12-methyl-tridecanoic acid | 3.0±0.2 |
| 16:1n9 | 7-hexadecenoic acid | 2.5±0.2 |
| 14:0 | Tetradecanoic acid | 1.4±0.1 |
| 16:1n6 iso | 14-methylpentadec-9-enoic acid | 1.2±0.1 |
| 5:0 anteiso | 2-methyl-butanoic acid | TR |
| 16:1n7 | 9-hexadecenoic acid | TR |
| 17:1n7 anteiso | 14-methylhexadec-9-enoic acid | TR |
| 17:0 iso | 15-methyl-hexadecanoic acid | TR |
| 17:0 | Heptadecanoic acid | TR |
| 18:0 | Octadecanoic acid | TR |
Mean peak area percentage calculated from the analysis of FAMEs in two sample preparations±standard deviation (n=3); TR=trace amounts <1%.
Genome properties
The draft genome of G. massiliensis strain Awa-1T is 4,207,226 bp long with 36.05% G+C content (Table 5 and Fig. 6). It is composed of 13 scaffolds with 13 contigs. Of the 3,908 predicted genes, 3,839 were protein-coding genes, and 69 were RNAs (7 genes are 5S rRNA, 1 gene is 16S rRNA, 1 gene is 23S rRNA, and 60 genes are tRNA genes). A total of 2,647 genes (68.95%) were assigned as putative functions (by COGs or by NR blast). A total of 126 genes (3.28%) were identified as ORFans. The remaining genes were annotated as hypothetical proteins (875 genes=22.79%). Genome statistics are summarized in Table 5 and the distribution of the genes into COGs functional categories is presented in Table 6.
Table 5.
Nucleotide content and gene count levels of the genome
| Attribute | Value | % of totala |
|---|---|---|
| Size (bp) | 4,207,226 | 100 |
| G+C content (bp) | 1,516,759 | 36.05 |
| Coding region (bp) | 3,579,496 | 85.07 |
| Total genes | 3,908 | 100 |
| RNA genes | 69 | 1.76 |
| Protein-coding genes | 3,839 | 98.23 |
| Genes with function prediction | 2,647 | 68.95 |
| Genes assigned to COGs | 2,455 | 63.94 |
| Genes with peptide signals | 430 | 11.20 |
| Genes with transmembrane helices | 1,063 | 27.68 |
The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Fig. 6.
Graphical circular map of the chromosome. From outside to the center: Genes on the forward strand colored by clusters of orthologous groups of proteins (COG) categories (only genes assigned to COG), genes on the reverse strand colored by COG categories (only gene assigned to COG), RNA genes (tRNAs green, rRNAs red), GC content, and GC skew.
Table 6.
Number of genes associated with the 25 general COG functional categories
| Code | Value | % value | Description |
|---|---|---|---|
| J | 206 | 5.36 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 205 | 5.33 | Transcription |
| L | 90 | 2.34 | Replication, recombination, and repair |
| B | 1 | 0.026 | Chromatin structure and dynamics |
| D | 51 | 1.32 | Cell cycle control, mitosis, and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 65 | 1.69 | Defense mechanisms |
| T | 140 | 3.64 | Signal transduction mechanisms |
| M | 125 | 3.25 | Cell wall/membrane biogenesis |
| N | 53 | 1.38 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 9 | 0.23 | Extracellular structures |
| U | 32 | 0.83 | Intracellular trafficking and secretion |
| O | 105 | 2.73 | Posttranslational modification, protein turnover, and chaperones |
| X | 46 | 1.19 | Mobilome: prophages and transposons |
| C | 138 | 3.59 | Energy production and conversion |
| G | 328 | 8.54 | Carbohydrate transport and metabolism |
| E | 208 | 5.41 | Amino acid transport and metabolism |
| F | 87 | 2.26 | Nucleotide transport and metabolism |
| H | 148 | 3.85 | Coenzyme transport and metabolism |
| I | 97 | 2.52 | Lipid transport and metabolism |
| P | 144 | 3.75 | Inorganic ion transport and metabolism |
| Q | 70 | 1.82 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 244 | 6.35 | General function prediction only |
| S | 191 | 4.97 | Function unknown |
| – | 1,384 | 36.05 | Not in COGs |
Genome comparison
The G+C content of G. massiliensis strain Awa-1T (36.05%) is smaller than that of H. trueperi, H. halophilus, A. jilinensis, and G. halophilus (41.66, 41.82, 37.27, and 37.92%, respectively) but larger than that of G. boraciitolerans (35.83%). The gene content of G. massiliensis (3,839) is smaller than that of H. trueperi, H. halophilus, and G. boraciitolerans (4,000, 4,135, and 4,450, respectively) but larger than that of A. jilinensis and G. halophilus (3,594 and 2,968, respectively). However, the distribution of genes into COG categories was similar among all compared genomes (Fig. 7). In addition, G. massiliensis shared 1,856 orthologous genes with the most closely related species (G. halophilus): 1,780, 1,614, 1,781, and 1,611 orthologous genes with H. halophilus, A. jilinensis, H. trueperi, and G. boraciitolerans, respectively (Table 7). The average percentage of nucleotide sequence identity ranged from 72.17 to 78.29% at the intraspecies level between G. massiliensis and the two Gracilibacillus species, but it ranged from 52.49 to 68.02% at interspecies level between G. massiliensis and other species. Similar results were obtained for the analysis of the digital DNA–DNA hybridization (dDDH) using GGDC software (Table 8).
Fig. 7.
Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins of Gracilibacillus massiliensis strain Awa-1T among other species.
Table 7.
Numbers of orthologous proteins shared between genomes (upper right) and AGIOS values obtained (lower left)
| GM | HH | AJ | HT | GH | GB | |
|---|---|---|---|---|---|---|
| GM | 3,839 | 1,780 | 1,614 | 1,781 | 1,856 | 1,611 |
| HH | 52.49% | 4,135 | 1,446 | 1,813 | 1,551 | 1,316 |
| AJ | 68.02% | 52.84% | 3,594 | 1,448 | 1,430 | 1,193 |
| HT | 66.14% | 53.12% | 65.43% | 4,000 | 1,560 | 1,316 |
| GH | 72.17% | 52.66% | 67.75% | 65.98% | 2,968 | 1,403 |
| GB | 78.29% | 52.63% | 67.13% | 65.30% | 70.63% | 4,450 |
The numbers of proteins per genome are indicated in bold. GM, Gracilibacillus massiliensis Awa-1T; HH, Halobacillus halophilus DSM 2266; AJ, Amphibacillus jilinensis Y1; HT, Halobacillus trueperi HT-01; GH, Gracilibacillus halophilus YIM-C55.5T; GB, Gracilibacillus boraciitolerans JCM 21714.
Table 8.
dDDH values obtained by comparison of all studied genomes
| HH | AJ | HT | GH | GB | |
|---|---|---|---|---|---|
| GM | 24.4%±0.17 | 20.7%±0.21 | 27.0%±0.16 | 19.0%±0.23 | 22.2%±0.19 |
| HH | 21.9%±0.20 | 21.6%±0.20 | 26.2%±0.16 | 22.7%±0.19 | |
| AJ | 24.2%±0.18 | 18.6%±0.23 | 24.6%±0.17 | ||
| HT | 33.2%±0.12 | 28.7%±0.14 | |||
| GH | 17.4%±0.25 |
dDDH, digital DNA-DNA hybridization. GM, Gracilibacillus massiliensis Awa-1T; HH, Halobacillus halophilus DSM 2266; AJ, Amphibacillus jilinensis Y1; HT, Halobacillus trueperi HT-01; GH, Gracilibacillus halophilus YIM-C55.5T; GB, Gracilibacillus boraciitolerans JCM 21714.
The Awa-1T strain, moderate halophilic bacterium, was isolated from a sample of cooking salt (Sel de Camargue) when studying salt-tolerant bacteria in salty food in the context of the culturomics project. On the basis of the phenotypic characteristics, phylogenetic and genomic analysis, Awa-1T strain is proposed to represent a novel species named G. massiliensis sp. nov.
Description of Gracilibacillus massiliensis sp. nov.
G. massiliensis (mas.si.li.en'sis. L. adj. massiliensis relating to Massilia, the ancient Roman name of Marseille, France, where the type strain was isolated and characterized, like many other species). This bacterium is motile through the use of its peritrichous flagella. It is a moderately halophilic, gram-positive, non-spore-forming rod, with a mean diameter of 0.3 µm and a length of 1.8 µm. The colonies are convex, creamy white, circular and measuring 0.2–0.3 mm in diameter after 2–4 days of growth on our home-made culture medium. Strain Awa-1T is not only aerobic but also able to grow in anaerobic (at 29°C) and microaerophilic (at 29–37°C) atmospheres. Its optimal conditions for growth are 37°C at pH 7.0–8.0 with 75 g/L of NaCl.
Using API identification strips, catalase, urease, esterase, lipase, α-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, and 4-nitrophenyl-βD-galactopyranoside activities are found positive. Oxidase, nitrate reductase, and indole tests are negative. The isolate is susceptible to doxycyclin, rifampicin, vancomycin, erythromycin, ciprofloxacin, gentamicin, trimethoprim/sulfamethoxazole, and imipenem, but resistant to metronidazole, amoxicillin, ceftriaxone, and penicillin G.
The G+C% content of the genome is 36.05%. The 16S rRNA and genome sequences are deposited in EMBL-EBI under accession numbers LN626645 and CZRP00000000, respectively. The type strain of G. massiliensis is strain Awa-1T (=CSUR P1441=DSM 29726) and was isolated from Salt specimen (Salt of Camargue).
Discussion
Because of the concept of ‘microbial culturomics’, which is based on the variation of physicochemical parameters of the culture conditions to explore microbial diversity (4), many new bacterial species have been discovered. As mentioned in our seminal work (4), microbial culturomics provides culture conditions simulating, reproducing, or mimicking the entirety of selective constraints that have shaped natural microbiota for millions of years. Here, the use of hypersaline conditions led to the comprehensive description of the hitherto unknown halophilic repertoire of table salt including a new Gracilibacillus species. All correspond to the Terrabacteria taxonomic group, evidencing the terrestrial adaptation of such microbes with very high resistance to desiccation by salt. The members of Gracilibacillus genus are all gram-positive bacteria, aerobic, motile and peritrichous, moderately halophile, white, and endospore-forming at the terminal position in general. Our strain Awa-1T does not form spores, the first differentiating characteristic compared to other species. It was selected for sequencing based on its phenotypic differences, phylogenetic position, and 16S rRNA sequence similarity with other members of the genus Gracilibacillus. The G+C content of the genomic DNA varies from 35.3 to 42.3 mol% (7). According to the fact that the G+C content deviation within species is at most 1%, these values confirm the classification of strain Awa-1T in a distinct species (42). Furthermore, the values of the AGIOS and dDDH of G. massiliensis compared to all other known species confirm its new species status. Microbial culturomics significantly extend the halophilic repertoire of salty food and/or salt table. This will improve the understanding of the possible involvement of table salt microbiota in human health and disease, with significant contributions to food and environmental microbiology.
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr) for automating the genomic annotation process. They also thank Karolina Griffiths for English reviewing and Claudia Andrieu for administrative assistance.
Authors’ contributions
AD performed the bacterium phenotypic characterization and the genomic analyses and drafted the manuscript. SK participated in its design and helped draft the manuscript. NA performed the cellular fatty acids analysis and helped draft the manuscript. NL performed the genomic sequencing and helped draft the manuscript. PEF and DR conceived the study and helped draft the manuscript. MM conceived the study, participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript.
Conflict of interest and funding
The authors declare that they have no competing interests. This work was supported by the ‘Fondation Méditerranée Infection’.
References
- 1.Cantrell SA, Dianese JC, Fell J, Gunde-Cimerman N, Zalar P. Unusual fungal niches. Mycologia. 2011;103:1161–74. doi: 10.3852/11-108. [DOI] [PubMed] [Google Scholar]
- 2.Hong SW, Kwon SW, Kim SJ, Kim SY, Kim JJ, Lee JS, et al. Bacillus oryzaecorticis sp. nov., a moderately halophilic bacterium isolated from rice husks. Int J Syst Evol Microbiol. 2014;64:2786–91. doi: 10.1099/ijs.0.058768-0. [DOI] [PubMed] [Google Scholar]
- 3.Lo N, Lee SH, Jin HM, Jung JY, Schumann P, Jeon CO. Garicola koreensis gen. nov., sp. nov., isolated from saeu-jeot, traditional Korean fermented shrimp. Int J Syst Evol Microbiol. 2015;65:1015–21. doi: 10.1099/ijs.0.000056. [DOI] [PubMed] [Google Scholar]
- 4.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–93. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 5.Wainø M, Tindall BJ, Schumann P, Ingvorsen K. Gracilibacillus gen. nov., with description of Gracilibacillus halotolerans gen. nov., sp. nov.; transfer of Bacillus dipsosauri to Gracilibacillus dipsosauri comb. nov., and Bacillus salexigens to the genus Salibacillus gen. nov., as Salibacillus salexigens comb. nov. Int J Syst Bacteriol. 1999;49:821–31. doi: 10.1099/00207713-49-2-821. [DOI] [PubMed] [Google Scholar]
- 6.Huo YY, Xu XW, Cui HL, Wu M. Gracilibacillus ureilyticus sp. nov., a halotolerant bacterium from a saline–alkaline soil. Int J Syst Evol Microbiol. 2010;60:1383–6. doi: 10.1099/ijs.0.016808-0. [DOI] [PubMed] [Google Scholar]
- 7.Hirota K, Hanaoka Y, Nodasaka Y, Yumoto I. Gracilibacillus alcaliphilus sp nov, a facultative alkaliphile isolated from indigo fermentation liquor for dyeing. Int J Syst Evol Microbiol. 2014;64:3174–80. doi: 10.1099/ijs.0.060871-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen YG, Cui XL, Zhang YQ, Li WJ, Wang YX, Xu LH, et al. Gracilibacillus halophilus sp. nov., a moderately halophilic bacterium isolated from saline soil. Int J Syst Evol Microbiol. 2008;58:2403–8. doi: 10.1099/ijs.0.65698-0. [DOI] [PubMed] [Google Scholar]
- 9.Chamroensaksri N, Tanasupawat S, Akaracharanya A, Visessanguan W, Kudo T, Itoh T. Gracilibacillus thailandensis sp. nov., from fermented fish (pla-ra) Int J Syst Evol Microbiol. 2010;60:944–8. doi: 10.1099/ijs.0.011981-0. [DOI] [PubMed] [Google Scholar]
- 10.Jeon CO, Lim JM, Jang HH, Park DJ, Xu LH, Jiang CL, et al. Gracilibacillus lacisalsi sp. nov., a halophilic Gram-positive bacterium from a salt lake in China. Int J Syst Evol Microbiol. 2008;58:2282–6. doi: 10.1099/ijs.0.65369-0. [DOI] [PubMed] [Google Scholar]
- 11.Gao M, Liu ZZ, Zhou YG, Liu HC, Ma YC, Wang L, et al. Gracilibacillus kekensis sp. nov., a moderate halophile isolated from Keke Salt Lake. Int J Syst Evol Microbiol. 2012;62:1032–6. doi: 10.1099/ijs.0.030858-0. [DOI] [PubMed] [Google Scholar]
- 12.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, et al. The Genomes OnLine Database (GOLD) v4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–9. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sentausa E, Fournier PE. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19:790–5. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 14.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–38. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–5. [Google Scholar]
- 16.Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–64. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–51. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 18.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–6. [Google Scholar]
- 21.Sasser M. Newark, DE: MIDI lnc; 2006. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME) Technical Note 101. [Google Scholar]
- 22.Dione N, Sankar SA, Lagier JC, Khelaifia S, Michele C, Armstrong N, et al. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43:D30–5. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 29.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–20. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramasamy D, Mishra AK, Lagier JC, Padhmanabhan R, Rossi M, Sentausa E, et al. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–91. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 31.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-) orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouret P, Paganini J, Dainat J, Louati D, Darbo E, Pontarotti P, et al. Berlin: Springer-Verlag; 2011. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH. In: Pontarotti P, ed. Evolutionary biology – concepts, biodiversity, macroevolution and genome evolution; pp. 71–87. [Google Scholar]
- 33.Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, Danchin EG. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–8. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 36.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray RGE. The higher taxa, or, a place for everything? In: Holt JG, editor. Bergey's manual of systematic bacteriology. 1st ed. Vol. 1. Baltimore, MD: The Williams and Wilkins; 1984. pp. 31–4. [Google Scholar]
- 38.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrasco IJ, Márquez MC, Yanfen X, Ma Y, Cowan DA, Jones BE, et al. Gracilibacillus orientalis sp. nov., a novel moderately halophilic bacterium isolated from a salt lake in Inner Mongolia, China. Int J Syst Evol Microbiol. 2006;56:599–604. doi: 10.1099/ijs.0.63971-0. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed I, Yokota A, Fujiwara T. Gracilibacillus boraciitolerans sp. nov., a highly boron-tolerant and moderately halotolerant bacterium isolated from soil. Int J Syst Evol Microbiol. 2007;57:796–802. doi: 10.1099/ijs.0.64284-0. [DOI] [PubMed] [Google Scholar]
- 41.Tang SK, Wang Y, Lou K, Mao PH, Jin X, Jiang CL, et al. Gracilibacillus saliphilus sp. nov., a moderately halophilic bacterium isolated from a salt lake. Int J Syst Evol Microbiol. 2009;59:1620–4. doi: 10.1099/ijs.0.006569-0. [DOI] [PubMed] [Google Scholar]
- 42.Meier-Kolthoff JP, Klenk HP, Göker M. Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–6. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]