Abstract
We determined prognostic impact of KRAS, BRAF, PIK3CA and TP53 mutation status and mutation heterogeneity among 164 colorectal cancer (CRC) patients undergoing liver resections for metastatic disease. Mutation status was determined by Sanger sequencing of a total of 422 metastatic deposits. In univariate analysis, KRAS (33.5%), BRAF (6.1%) and PIK3CA (13.4%) mutations each predicted reduced median time to relapse (TTR) (7 vs. 22, 3 vs. 16 and 4 vs. 17 months; p < 0.001, 0.002 and 0.023, respectively). KRAS and BRAF mutations also predicted a reduced median disease‐specific survival (DSS) (29 vs. 51 and 16 vs. 49 months; p <0.001 and 0.008, respectively). No effect of TP53 (60.4%) mutation status was observed. Postoperative, but not preoperative chemotherapy improved both TTR and DSS (p < 0.001 for both) with no interaction with gene mutation status. Among 94 patients harboring two or more metastatic deposits, 13 revealed mutation heterogeneity across metastatic deposits for at least one gene. Mutation heterogeneity predicted reduced median DSS compared to homogeneous mutations (18 vs. 37 months; p = 0.011 for all genes; 16 vs. 26 months; p < 0.001 analyzing BRAF or KRAS mutations separately). In multivariate analyses, KRAS or BRAF mutations consistently predicted poor TRR and DSS. Mutation heterogeneity robustly predicted DSS but not TTR, while postoperative chemotherapy improved both TTR and DSS. Our findings indicate that BRAF and KRAS mutations as well as mutation heterogeneity predict poor outcome in CRC patients subsequent to liver resections and might help guide treatment decisions.
Keywords: heterogeneity, mutations, colorectal cancer, liver metastases, chemotherapy
Short abstract
What's new?
Preliminary evidence suggests that poor outcome after liver resection in metastatic colorectal cancer (CRC) is predicted by mutations in KRAS and BRAF and by intra‐individual heterogeneity involving copy number alterations that vary from one metastatic lesion to the next. Little is known, however, about the clinical implications of intra‐individual mutation heterogeneity in CRC. Here, in a comparison of KRAS and BRAF wild‐type status, mutational homogeneity, and mutational heterogeneity, mutation heterogeneity was found to be the strongest predict or of reduced disease‐specific survival following liver resection in metastatic CRC. Knowledge of intra‐individual mutation heterogeneity in KRAS and BRAF in CRC could facilitate therapeutic decisions.
Abbreviations
- 5FU
fluorouracil
- ASCAT
allele‐specific copy number analysis of tumors;
- CEA
carcinoembryonic antigen
- CI
confidence interval
- CR
complete response
- CRC
colorectal cancer
- CT
computer tomography
- DSS
disease‐specific survival
- HR
hazard ratio
- PR
partial response
- RECIST 1.1
Response Evaluation Criteria in Solid Tumors
- SD
stable disease
- TTR
time to relapse
Approximately 40% of all patients diagnosed with distant metastases from colorectal cancer (CRC) have metastatic deposits confined to the liver.1 Partial liver resection has been shown to improve long‐term survival for selected CRC patients,2, 3, 4 with a 10‐year disease‐free survival approaching 20% reported even for patients suffering from multiple liver deposits.3
Selection of patients eligible for surgery has traditionally been based on clinicopathological prognostic factors.5, 6, 7 Recent work, however, has focused on the response to chemotherapy and various molecular alterations in the tumor as more appropriate prognostic factors after liver resections. Response to neoadjuvant chemotherapy has been related to improved survival,8 whereas KRAS and BRAF mutations have been associated with more aggressive disease and a poor prognosis.9, 10, 11, 12 While liver resection for metastatic CRC (mCRC) has become routine practice,13 the fact that the majority of patients undergoing surgery subsequently relapse14 underlines the need for better predictive markers guiding treatment decisions.
In addition to individual molecular markers, tumor heterogeneity has become a major issue with respect to tumor evolution and, potentially, therapy outcome. Preliminary evidence indicates that clonal evolution may cause acquired resistance to EGFR blockade.15 Moreover, in a parallel study in collaboration with others, we found intraindividual heterogeneity with respect to copy number alterations to predict outcome after liver resections (Sveen et al., manuscript submitted). While several studies have compared genetic alterations between primary tumors and their corresponding metastatic deposits,16, 17, 18 the biological and clinical implications of intraindividual metastatic heterogeneity remain poorly understood.
Sampling of multiple synchronous metastatic deposits represents a challenge in most solid tumors. However, the fact that liver resections for CRC metastases in general contain multiple lesions makes this a suitable model for studying intermetastatic mutation heterogeneity.
The aim of this study was twofold. First, we aimed to assess the influence of mutations in four major driver genes (KRAS, BRAF, PIK3CA and TP53) considered as potential prognostic factors in CRC in patients treated with liver resections. Second, as genetic heterogeneity may characterize a distinct tumor phenotype,19 we analyzed gene mutation status across multiple metastatic deposits from each patient in order to determine intraindividual metastatic heterogeneity and the potential prognostic impact of such mutation heterogeneity.
Material and Methods
Patients
Metastatic liver deposits were collected from 164 mCRC patients undergoing partial liver resection between August 2006 and March 2013. The total number of deposits sampled and analyzed was 428, including six primary samples and 36 samples collected at re‐resections from 23 of these patients.
Seventy‐six patients had synchronous while 88 patients had metachronous liver metastases. Metastases were considered metachronous if diagnosed 3 months or more after diagnosis of the primary tumor.
All patients had a computer tomography (CT) scan performed before surgery. In addition, patients who received preoperative chemotherapy (n = 72) had CT scans performed within 4 weeks before start of chemotherapy and after three and six treatment cycles to evaluate response to therapy.
The treatment regimen administered most often included oxaliplatin and fluorouracil, administered as FOLFOX or Nordic FLOX.2, 20 Fourteen patients were treated with irinotecan before liver resection, and 13 patients were also exposed to EGFR (seven patients) or VEGF inhibitors (six patients), 12 of these in combination with irinotecan. Fifty‐three patients received chemotherapy after liver resection (postoperative chemotherapy).
Response to preoperative chemotherapy was determined according to the Response Evaluation Criteria in Solid Tumors (RECIST 1.1).21 Clinical data were retrieved from electronic patient's charts. Baseline patient characteristics and treatment are summarized in Table 1 with further details in Supporting Information Table S1. The study was approved by the Regional Ethical Committee, and all patients provided written informed consent.
Table 1.
Patient characteristics and treatment for 164 metastatic colorectal cancer patients treated with partial liver resection between August 2006 and March 2013 at Haukeland University Hospital, Bergen, Norway
| Age (years) at diagnosis, mean (range) | 63 (22–83) |
| Sex (male, female), n | 91, 73 |
| Months of follow‐up, median (range) | 57 (20–98) |
| Primary tumor site (colon, rectum, multiple), n | 110, 48, 6 |
| Nodal status of primary cancer (pN0, pN+), n | 56, 108 |
| Disease‐free interval (months) between primary and metastases, mean (range) | 3 (0–54) |
| Synchronous or metachronous liver metastasesa, n | 76, 88 |
| CEA before liver surgery (<200, >200, missing), n | 108, 15, 41 |
| WHO performance status at start of treatment (0, I, II, missing), n | 102, 58, 3, 1 |
| Number of sampled and analyzed liver metastases (first liver resection): | |
| ‐Mean (range) | 2 (1–8) |
| ‐Patients harboring multiple metastases, n | 102 |
| ‐Patients harboring single metastasis, n | 62 |
| Chemotherapy exposure: | |
| ‐Adjuvant chemotherapy (primary setting), n | 26 |
| ‐Chemotherapy before liver resectionb, n | 72 |
| ‐Chemotherapy after liver resection (postoperative chemotherapy) | 53 |
| Best response on chemotherapy (CR, PR, SD) c , n | 1, 41, 28 |
Metastases were considered metachronous if diagnosed ≥ 3 months after diagnosis of the primary tumor.
≥Three courses of treatment (except three patients who had to stop treatment after one or two cycles, due to side effects).
Response Evaluation Criteria in Solid Tumors (RECIST 1.1). Not calculated for the patients receiving only one treatment course.
Abbreviations: CEA: carcinoembryonic antigen; CR: complete response; PR: partial response; SD: stable disease.
Sample collection and evaluation
Following removal, the resected liver specimens were subjected to immediate dissection in the operating theater, and tissue samples were snap‐frozen in liquid nitrogen. In addition, a small part of each sample was formalin fixed and paraffin embedded to allow for histopathological confirmation of tumor cell content. The amount of tumor cells in the whole section slides was on average 76% (range 10–100%). A total of 14 samples from ten patients were excluded due to poor genomic DNA (gDNA) quality and/or a high degree of normal tissue contamination, leaving samples from 408 metastases and six primaries available for mutation analysis. For 45 of the patients SNP6 data including ASCAT calculations22 of the aberrant cell fraction for each sample were also available (Sveen et al., manuscript submitted).
DNA isolation
Genomic DNA from all patient samples was extracted from fresh frozen tissue using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RNA isolation
RNA was extracted from fresh frozen tissue using the RNeasy Mini Kit (Qiagen), and complimentary DNA (cDNA) was synthesized using qScript cDNA SuperMix (Quanta, BioSciences, Gaithersburg, MD), with 320 ng total RNA as input. Both procedures were performed according to the manufacturer's instructions.
DNA preamplification
Genomic DNA from all samples was globally amplified with a phi29 polymerase using the REPLI‐g Midi Kit (Qiagen) according to the manufacturer's instructions. In brief, gDNA was incubated in denaturation buffer for 3 min before addition of neutralization buffer and a master mix containing phi29 DNA polymerase with exonuclease proofreading activity. The isothermal amplification reaction was carried out for 16 hr at 30°C.
Mutation screening
BRAF V600 status was analyzed for as previously described.23 KRAS exon 2 (harboring codon 12 and 13) and PIK3CA exon 9 and 20 were amplified by PCR (detailed in Supporting Information Methods) and Sanger sequenced. Capillary gel electrophoresis, data collection and sequence analyses were performed on an automated ABI 3700 DNA sequencer (Applied Biosystems, Carlsbad, CA).
Mutation screening was performed on preamplified gDNA in order to save patient material. All alterations detected were verified in an independent amplification using original gDNA as template. For cases with intraindividual heterogeneity samples with wild‐type results were also reanalyzed using original gDNA.
TP53 mutation analysis was performed for the whole coding region of the gene on cDNA from all liver metastatic deposits as previously described24 (for further details, see Supporting Information Methods). In short, TP53 was amplified in a nested PCR and the PCR products were purified and sequenced on an automated ABI 3700 DNA sequencer (Applied Biosystems).
All mutations identified were verified by exon‐wise sequencing of gDNA. For cases with intraindividual heterogeneity, wild‐type results based on cDNA were also reanalyzed using gDNA.
Statistics
For descriptive statistics we used the mean, median, counts and proportions (percents). The time to event or censoring was calculated from the start of treatment for liver metastases (liver resection or start of preoperative chemotherapy). Patients dying of causes other than CRC with no evidence of relapse were treated as censored observations. As for patients dying subsequent to having a relapse from their CRC, these patients were considered as dying from CRC unless dying for reasons obviously unrelated to their CRC.
Unadjusted survival curves for time to relapse (TTR) and disease‐specific survival (DSS) were estimated using the Kaplan–Meier plot25 and compared between subgroups with the log‐rank test.26
To adjust for possible confounders the Cox proportional hazards model was applied.27 Due to collinearity multivariate analyses were performed adjusting for different variables in the Cox models. In the Supporting Information Tables results are presented by hazard ratios (HRs) and their 95% confidence intervals (CIs).
All statistical analyses were performed using the SPSS v.22 software. All p‐values are given as two‐sided.
Results
Mutation frequencies and intraindividual heterogeneity
Overall, the proportions of patients revealing a mutation in KRAS, BRAF, PIK3CA or TP53 were 33.5, 6.1, 13.4 and 60.4%, respectively. Detailed information on individual mutations is listed in Supporting Information Table S2. As expected, most of the KRAS mutations detected were point mutations localized to codons 12 and 13 (104 and 25 samples, respectively). In addition, we found one insertion between these two codons in two samples and a point mutation localized at codon 19 in one sample. All but two of the BRAF‐mutated samples harbored the V600E mutation. All PIK3CA mutations were point mutations, the most frequent ones being E542K and E545K in exon 9. As for TP53, 226 and 27 samples harbored point mutations and indels, respectively. Notably, in five of the patients more than one TP53 mutation was detected in each mutated sample, and for one patient the second TP53 mutation was found in one out of three metastases only.
Among 94 patients from whom two or more metastatic deposits collected at the same resection were examined, 13 patients revealed mutation heterogeneity for at least one of the four genes examined across their metastatic deposits (Fig. 1). For each individual gene, the number of patients revealing heterogeneous mutation results for KRAS, BRAF, PIK3CA and TP53 was 10, 1, 5 and 9, respectively. Notably, in five patients, KRAS or TP53 mutations seemed to evolve over time either between the primary and the metastases or between the first and second liver resection (see details in Supporting Information Table S2).
Figure 1.
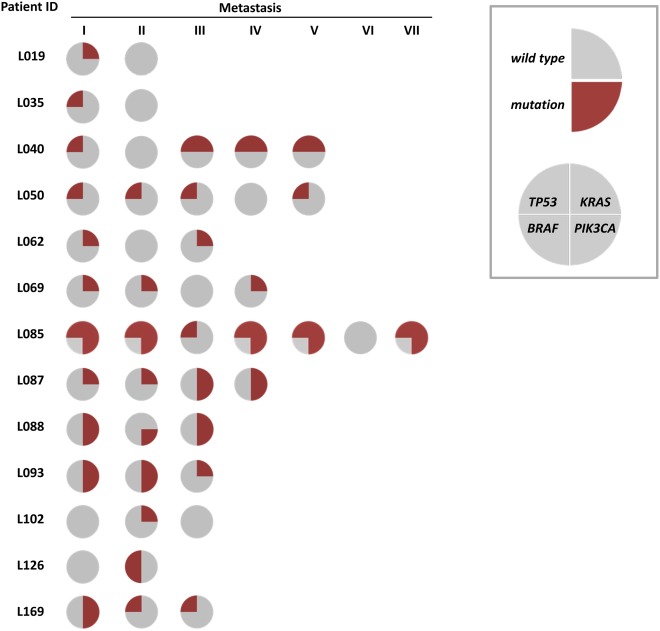
Intraindividual heterogeneity between liver metastases as determined by different mutation status for one or more of the genes TP53, KRAS, BRAF and PIK3CA. Each row represents one patient. Each circle represents a metastatic sample harvested at the first liver surgery for each patient. Sectors indicate mutational status as mutated (red) or wild‐type (gray) for each of the four genes.
Mutation frequencies in subgroups of patients with different chemotherapy exposure are listed in Supporting Information Table S3.
Mutation status and prognosis after liver resection
To evaluate the prognostic impact of the mutations described above in patients treated with liver resections, we excluded patients who had undergone a previous liver resection (n = 13) before inclusion in the present study, leaving a total of 151 patients.
In univariate analyses (Fig. 2), we found KRAS and BRAF mutations both to be associated with reduced median TTR (7 vs. 22 and 3 vs. 16 months; p < 0.001 and p = 0.002, respectively) and reduced median DSS (29 vs. 51 and 16 vs. 49 months; p = 0.008 and p < 0.001, respectively). Because KRAS and BRAF mutations are known to be mutually exclusive,28, 29 we performed a combined analysis in addition (Fig. 2). Patients harboring either a KRAS or a BRAF mutation revealed an inferior median TTR (7 vs. 24 months) as well as DSS (26 vs. 56 months, p < 0.001 for both comparisons) compared to patients with deposits wild‐type for both genes. Mutations in PIK3CA were associated with a reduced TTR (4 vs. 17 months; p = 0.023), but not with reduced DSS, while TP53 mutation status revealed no significant relation to either TTR or DSS. Considering the number of metastatic deposits, patients with multiple liver deposits had a somewhat shorter TTR compared to patients with one deposit only (p = 0.008) while no significant effect on DSS was recorded (Supporting Information Table S4).
Figure 2.
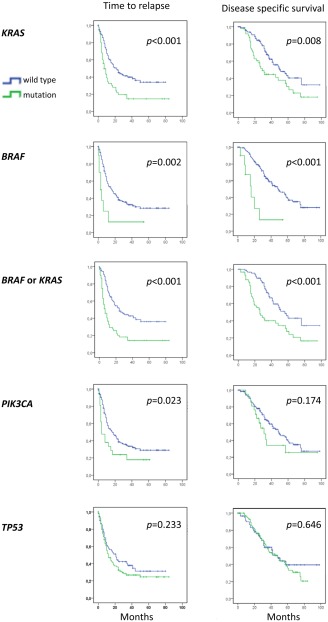
Kaplan–Meier survival curves illustrating time to relapse (left panels) and disease‐specific survival (right panels) after liver surgery for metastatic colorectal cancer with respect to mutation status for KRAS, BRAF, KRAS and BRAF combined, PIK3CA and TP53 (n = 151 in each panel). A patient was classified as harboring a gene mutation as long as it was present in at least one lesion. p‐values are from log‐rank tests.
Interestingly, KRAS mutations seemed to be associated with a shorter median TTR in patients harboring synchronous (p = 0.002) as well as metachronous (p = 0.019) disease, but with DSS only in synchronous disease (p = 0.001; Supporting Information Fig. S1a). Similar findings were observed for KRAS and BRAF mutations combined (Supporting Information Fig. S1b).
Influence of chemotherapy exposure
Among the 151 patients, 33 patients received preoperative chemotherapy only, 17 patients had chemotherapy after surgery only, 36 patients received both presurgical and postsurgical chemotherapy, while 65 patients received no chemotherapy neither before nor after their liver surgery. Comparing all the four treatment groups together, a significant effect on TRR (p < 0.001) as well as DSS (p = 0.001) was recorded, with a clear preference for postoperative chemotherapy (Fig. 3). Analyzing separately for preoperative or postoperative therapy as variables in univariate analysis, interestingly postoperative therapy influenced TTR as well as DSS for the total group of patients (p < 0.001 for both) (Fig. 3), while presurgical chemotherapy improved TTR among patients harboring a KRAS mutation only (Supporting Information Figs. S2c and S2d; p = 0.018). However, except for a weak interaction between KRAS and BRAF wild‐type status and benefit from preoperative chemotherapy (p = 0.02), no statistically significant interaction between mutation status for any of the genes analyzed and benefit from chemotherapy administered in the preoperative or postoperative setting was recorded (Supporting Information Figs. S2 and S3). Notably chemotherapy response (partial response vs. stable disease) per se did not influence survival after liver resection in our dataset (Supporting Information Table S4); neither did we observe any difference in TTR or DSS between patients commencing postsurgical chemotherapy within a time frame of 4 weeks vs. patients commencing on chemotherapy later than 4 weeks after surgery (Supporting Information Fig. S4).
Figure 3.
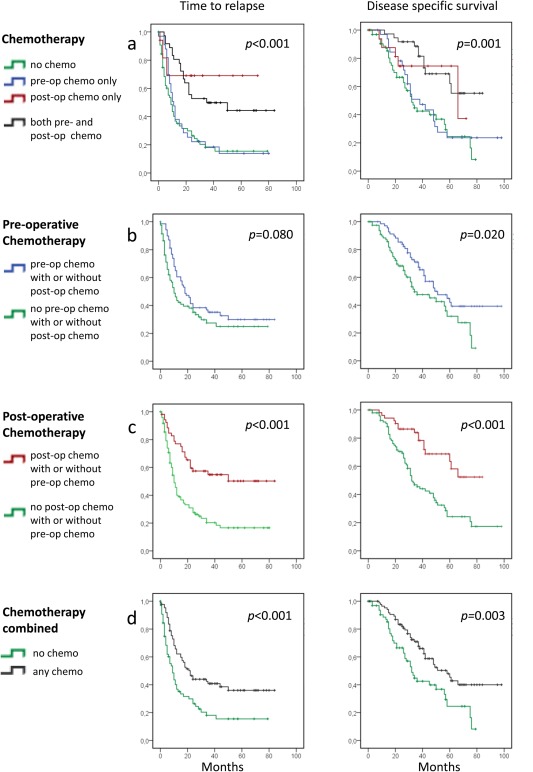
Kaplan–Meier survival curves illustrating differences in time to relapse (left panels) and disease‐specific survival (right panels) after liver surgery for metastatic colorectal cancer, based on chemotherapy regimens administered at different times in relation to liver surgery. p‐values are from log‐rank tests. (a) All four treatment groups presented. (b) All patients receiving preoperative chemotherapy (alone or combined with postoperative chemotherapy) vs. patients receiving no preoperative chemotherapy (with or without postoperative chemotherapy). (c) Patients receiving postoperative chemotherapy (alone or in concert with preoperative therapy) vs. patients receiving no postoperative chemotherapy (with or without preoperative chemotherapy). (d) All patients receiving chemotherapy for liver metastases at any time vs. patients who did not receive any chemotherapy. n = 151 in each panel.
Mutation heterogeneity and prognosis after liver resection
The potential impact of mutation heterogeneity (defined as different mutation status between metastases harvested from the same patient at the same surgical procedure) on outcome was evaluated in the subgroup of patient harboring two or more liver deposits (n = 94).
First, we confirmed the prognostic impact of KRAS, BRAF and PI3K mutation status revealed in the total patient cohort in the subgroup of patients harboring multiple deposits (Supporting Information Table S5). Next, we compared outcome between (i) patients with mutation heterogeneity affecting either KRAS, BRAF, PI3K or TP53 across metastatic deposits (n = 13), (ii) patients harboring a homogeneous mutation in at least one gene across all metastatic deposits and with no heterogeneous mutation and (iii) patients with no mutations across either gene. Comparing all three groups, univariate analysis revealed a significant different TTR (median of 4 vs. 5 vs. 15 months, p < 0.001) as well as DSS (median of 18 vs. 37 vs. 58 months, p < 0.001) between the groups (Fig. 4a). Notably, the difference in DSS between patients in Group 1 (harboring heterogeneous mutation) vs. patients in Group 2 (harboring homogeneous mutations only), 18 vs. 37 months, was of statistical significance (p = 0.011). Similar analysis restricted to patients being (i) heterogeneous vs. (ii) homogeneous for BRAF or KRAS mutations or (iii) wild type for both genes revealed similar results (median DSS of 16, vs. 26 vs. 49 months, p < 0.001 comparing all three groups; p < 0.035 comparing patients heterogeneous vs. homogenous for BRAF and KRAS mutations; Fig. 4b).
Figure 4.
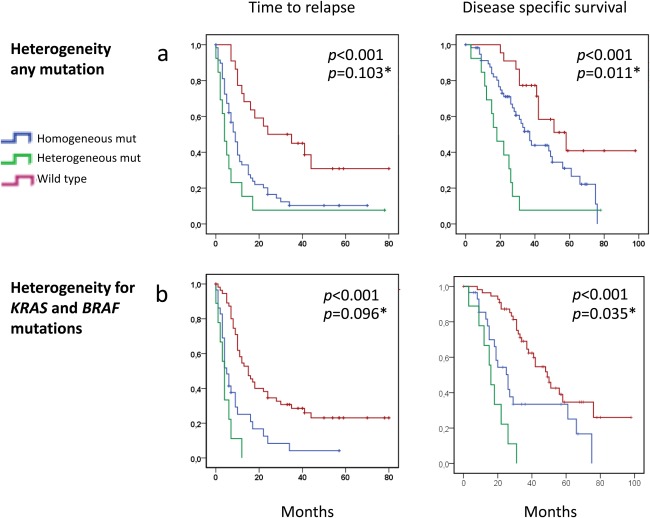
Kaplan–Meier survival curves illustrating differences in time to relapse (left panels) and disease‐specific survival (right panels) after liver surgery for metastatic colorectal cancer comparing patients harboring no mutations to patients harboring intraindividual mutation heterogeneity across either KRAS, BRAF, TP53 or PI3K and patients revealing at least one homogenous but no heterogeneous mutation in either gene (a). Similar analyses comparing patients harboring no mutations, heterogeneous or homogeneous mutations in either KRAS or BRAF without attention to TP53 or PI3K status (b). p‐values are from log‐rank tests. p‐values relate to comparison between all three groups. p* values relate to the difference between patients harboring heterogeneous vs. homogenous mutations.
Multivariate analyses
As KRAS and BRAF mutations were mutually exclusive, these parameters (BRAF or KRAS mutation vs. wild‐type status for both) were combined into one parameter for the multivariate analyses. In addition, as the use of different treatment regimens depended on stage of disease (meta‐ vs. synchronous metastases, nodal status of the primary) as well as previous therapy, repeated multivariate analyses were performed entering different variables into the models.
The detailed results of these analyses are summarized in Table 2 and Supporting Information Table S6. For the total cohort (n = 151) as well as for the subcohort harboring multiple metastatic deposits (n = 94), mutations affecting KRAS or BRAF in a homogeneous or heterogeneous manner consistently predicted a poor TRR as well as a poor DSS across all models. In addition, postoperative chemotherapy (with or without prior preoperative therapy) improved TTR as well as DSS. Notably, in the subgroup of patients with multiple deposits (n = 94), mutation heterogeneity independently predicted DSS but not TTR across models (Supporting Information Table S6).
Table 2.
Results from Cox regression for all patients (n = 151)
| Time to relapse (TTR) | Disease‐specific survival (DSS) | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p values | HR | 95% CI | p values | |
| Age (< or > 65 years) | 1.08 | (0.71, 1.65) | 0.720 | 0.72 | (0.45, 1.17) | 0.183 |
| Sex (male vs. female) | 0.93 | (0.62, 1.40) | 0.726 | 0.98 | (0.62, 1.57) | 0.942 |
| Nodal status of primary (N+ vs. N0) | 1.55 | (0.97, 2.48) | 0.064 | 1.99 | (1.13, 3.53) | 0.018 |
| Synchronous vs. metachronous mets. | 2.00 | (1.21, 3.29) | 0.007 | 1.58 | (0.90, 2.77) | 0.110 |
| Multiple vs. single mets. | 1.73 | (1.11, 2.68) | 0.015 | 1.55 | (0.93, 2.58) | 0.095 |
| TP53 mutations | 0.92 | (0.61,1.42) | 0.720 | 0.78 | (0.47,1.28) | 0.318 |
| PIK3CA mutations | 1.12 | (0.63, 1.97) | 0.700 | 0.78 | (0.40, 1.51) | 0.451 |
| KRAS or BRAF mutations (mut vs. double wt) | 2.34 | (1.50, 3.66) | <0.001 | 2.47 | (1.49, 4.10) | <0.001 |
| Chemotherapy a | 0.39 | (0.23, 0.64) | <0.001 | 0.46 | (0.26, 0.80) | 0.005 |
p‐values are from the first step of the likelihood ratio (LR) test. Bold indicates significant p‐values.
Administered before surgery, after surgery or both.
Abbreviations: HR: hazard ratio; CI: confidence interval; wt: wild type; mets: metastases; muts: mutations.
Discussion
Liver resections improve outcome in CRC patients harboring liver metastases, and some patients become long‐term survivors.3, 4 It represents a major surgical intervention however, and the fact that many patients only gain limited benefit14 calls for identification of improved prognostic and predictive factors.5
The aim of this study was twofold; first, we assessed the influence of mutations in four key driver genes (KRAS, BRAF, PIK3CA and TP53) determined on metastatic deposits on outcome after liver resections for CRC. Second, we analyzed gene mutation status across multiple deposits from each patient in order to determine intraindividual mutation heterogeneity and its potential impact on prognosis.
The frequencies of KRAS, PIK3CA and TP53 mutations resemble what has been detected by others in metastatic liver deposits, but for BRAF the incidence was slightly higher than in most other series analyzing liver metastases.16, 30, 31, 32 While previous studies have revealed BRAF mutations to be associated with a poor outcome following liver resection for CRC,11, 12 the results for KRAS mutation have been at variance.9, 32, 33
Here, BRAF as well as KRAS mutation status (each one individually as well as combined) was associated with a poor prognosis in univariate analysis in the total patient cohort as well as in the subgroup of patients harboring multiple liver deposits. Moreover, both mutations consistently predicted a poor outcome across different multivariate models. Considering PI3KCA and TP53 mutation status, conflicting results have linked mutations in these genes to outcome in CRC patients.34, 35, 36, 37, 38 Here, we found PI3KCA mutations to be associated only with a reduced TTR but not DSS in univariate analysis; in contrast, no association between TP53 mutation status and outcome (either TTR or DSS) was recorded.
Chemotherapy administered in relation to liver resections has been reported to improve TTR but not DSS.2 Patients included in this study were selected for chemotherapy based on demographic and medical conditions including age, general health condition and previously chemotherapy exposure.39 The fact that chemotherapy was not administered in a randomized setting means that the results need to be interpreted carefully. Our finding that postoperative but not preoperative chemotherapy seems to be of significant benefit however is of interest. If confirmed in independent studies, this will have clinical implications. This observation contrasts results obtained with use of primary medical therapy (neoadjuvant chemotherapy) in other cancer forms like breast cancer.40 However, liver resection normally is followed by a rapid liver tissue regeneration, to be accompanied by secretion of multiple growth factors.41 This setting may have detrimental effects on subclinical metastatic deposits, offering a potential explanation for a particular benefit of chemotherapy treatment during this time period. The finding of no interaction (except for a weak association between KRAS and BRAF wild‐type status and benefit from preoperative therapy) between gene mutation status and mutation heterogeneity on the one hand and benefit from chemotherapy on the other hand further supports a prognostic role of these mutations, suggesting mechanisms related to tumor cell growth, invasion, metastatic propensity and others and not resistance to chemotherapy to explain our observations.
Heterogeneity within primary tumors is well recognized as an important challenge to personalized medicine19, 42, 43, 44, 45 Recent studies have revealed clonal heterogeneity for KRAS mutations within primary CRCs,46, 47 and several studies have examined concordance in mutation status between primary tumors and metastatic deposits.16, 17, 18 Mutation heterogeneity across synchronous deposits, however, is not well described.
We observed heterogeneous mutation results in respect to the four driver genes analyzed in 13 of 94 patients (12.2%) harboring multiple liver deposits. Comparing DSS between tumors revealing a heterogeneous mutation affecting any of the four genes analyzed and tumors harboring a homozygous mutation for one (or more) of these genes revealed an inferior DSS for tumors harboring heterogeneous mutations. Notably, mutation heterogeneity was found to be an independent prognostic factor in multivariate analysis.
Ten patients harbored a heterogeneous KRAS mutation, and one revealed a heterogeneous BRAF mutation. Similar to what was recorded in respect to mutation heterogeneity across all four genes analyzed, mutation heterogeneity for BRAF or KRAS revealed an inferior prognosis as compared to homozygous mutations across the two genes. While we may not exclude the possibility that heterogeneity for BRAF and KRAS mutations could explain the findings obtained in respect to heterogeneity across all four genes, our findings in metastatic CRC are consistent with observations by others revealing tumor heterogeneity to predict a poor prognosis across different forms of primary solid tumors.48, 49, 50
In conclusion, we confirmed KRAS and BRAF mutations to be a robust prognostic parameter in patients treated with partial liver resections for metastatic CRC. These findings indicate that KRAS/BRAF mutation status should be taken into account together with other prognostic parameters when selecting patients for surgery. Second, our results point to an important role of postsurgical but probably not presurgical chemotherapy in relation to liver resections; confirmation in independent studies is warranted. Finally, our data support the hypothesis that mutation heterogeneity could be a marker associated with an aggressive clinical behavior. If confirmed by others, these findings may be of importance to patient treatment and also improve our understanding of the clinical and biological importance of mutation heterogeneity in metastatic disease.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgments
This work was performed in the Mohn Cancer Research Laboratory, Haukeland University Hospital/University of Bergen. The authors appreciate the skilled technical assistance from Dagfinn Ekse, Nhat Kim Duong, Christine Eriksen and Silje Bjørneklett. They thank Geir Egil Eide at Centre for Clinical Research, Western Norway Regional Health Authority and the Department of Global Public Health and Primary Care, University of Bergen, Norway for excellent advice and statistical review.
References
- 1. Grossmann I, Doornbos PM, Klaase JM, et al. Changing patterns of recurrent disease in colorectal cancer. Eur J Surg Oncol 2014;40:234–9. [DOI] [PubMed] [Google Scholar]
- 2. Nordlinger B, Sorbye H, Glimelius B, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long‐term results of a randomised, controlled, phase 3 trial. Lancet Oncol 2013;14:1208–15. [DOI] [PubMed] [Google Scholar]
- 3. Tomlinson JS, Jarnagin WR, DeMatteo RP, et al. Actual 10‐year survival after resection of colorectal liver metastases defines cure. J Clin Oncol 2007;25:4575–80. [DOI] [PubMed] [Google Scholar]
- 4. Rees M, Tekkis PP, Welsh FK, et al. Evaluation of long‐term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg 2008;247:125–35. [DOI] [PubMed] [Google Scholar]
- 5. Kanas GP, Taylor A, Primrose JN, et al. Survival after liver resection in metastatic colorectal cancer: review and meta‐analysis of prognostic factors. Clin Epidemiol 2012;4:283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fong Y, Fortner J, Sun RL, et al. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis. Of 1001 consecutive cases. Ann Surg 1999;230:309–18; discussion 18‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nordlinger B, Guiguet M, Vaillant JC, et al. Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Association Francaise de Chirurgie. Cancer 1996;77:1254–62. [PubMed] [Google Scholar]
- 8. Gruenberger B, Scheithauer W, Punzengruber R, et al. Importance of response to neoadjuvant chemotherapy in potentially curable colorectal cancer liver metastases. BMC Cancer 2008;8:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brudvik KW, Kopetz SE, Li L, et al. Meta‐analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg 2015;102:1175–83. [DOI] [PubMed] [Google Scholar]
- 10. Nash GM, Gimbel M, Shia J, et al. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol 2010;17:572–8. [DOI] [PubMed] [Google Scholar]
- 11. Teng HW, Huang YC, Lin JK, et al. BRAF mutation is a prognostic biomarker for colorectal liver metastasectomy. J Surg Oncol 2012;106:123–9. [DOI] [PubMed] [Google Scholar]
- 12. Yaeger R, Cercek A, Chou JF, et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer 2014;120:2316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abdalla EK, Adam R, Bilchik AJ, et al. Improving resectability of hepatic colorectal metastases: expert consensus statement. Ann Surg Oncol 2006;13:1271–80. [DOI] [PubMed] [Google Scholar]
- 14. Kopetz S, Chang GJ, Overman MJ, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol 2009;27:3677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diaz LA, Jr. , Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brannon AR, Vakiani E, Sylvester BE, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 2014;15:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mao C, Wu XY, Yang ZY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 2015;5:8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan IB, Malik S, Ramnarayanan K, et al. High‐depth sequencing of over 750 genes supports linear progression of primary tumors and metastases in most patients with liver‐limited metastatic colorectal cancer. Genome Biol 2015;16:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15–26. [DOI] [PubMed] [Google Scholar]
- 20. Sorbye H, Glimelius B, Berglund A, et al. Multicenter phase II study of Nordic fluorouracil and folinic acid bolus schedule combined with oxaliplatin as first‐line treatment of metastatic colorectal cancer. J Clin Oncol 2004;22:31–8. [DOI] [PubMed] [Google Scholar]
- 21. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 22. Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele‐specific copy number analysis of tumors. Proc Natl Acad Sci USA 2010;107:16910–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loes IM, Immervoll H, Angelsen JH, et al. Performance comparison of three BRAF V600E detection methods in malignant melanoma and colorectal cancer specimens. Tumour Biol 2014;36:1003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chrisanthar R, Knappskog S, Lokkevik E, et al. CHEK2 mutations affecting kinase activity together with mutations in TP53 indicate a functional pathway associated with resistance to epirubicin in primary breast cancer. PLoS One 2008;3:e3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaplan EL, Meier P. Nonparametric‐estimation from incomplete observations. J Am Stat Assoc 1958;53:457–81. [Google Scholar]
- 26. Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep 1966;50:163–70. [PubMed] [Google Scholar]
- 27. Cox DR. Regression models and life‐tables. J R Stat Soc B 1972;34:187. [Google Scholar]
- 28. Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 29. Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch‐repair status. Nature 2002;418:934. [DOI] [PubMed] [Google Scholar]
- 30. Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012;30:2956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vermaat JS, Nijman IJ, Koudijs MJ, et al. Primary colorectal cancers and their subsequent hepatic metastases are genetically different: implications for selection of patients for targeted treatment. Clin Cancer Res 2012;18:688–99. [DOI] [PubMed] [Google Scholar]
- 32. Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013;119:4137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Samowitz WS, Curtin K, Schaffer D, et al. Relationship of Ki‐ras mutations in colon cancers to tumor location, stage, and survival: a population‐based study. Cancer Epidemiol Biomarkers Prev 2000;9:1193–7. [PubMed] [Google Scholar]
- 34. Iida S, Kato S, Ishiguro M, et al. PIK3CA mutation and methylation influences the outcome of colorectal cancer. Oncol Lett 2012;3:565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res 2012;18:2257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pietrantonio F, Biondani P, Perrone F, et al. TP53 mutations in advanced colorectal cancer: the dark side of the moon. Oncology 2014;86:289–94. [DOI] [PubMed] [Google Scholar]
- 37. Russo A, Bazan V, Iacopetta B, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol 2005;23:7518–28. [DOI] [PubMed] [Google Scholar]
- 38. Geisler S, Lonning PE, Aas T, et al. Influence of TP53 gene alterations and c‐erbB‐2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res 2001;61:2505–12. [PubMed] [Google Scholar]
- 39. Sorbye H, Mauer M, Gruenberger T, et al. Predictive factors for the benefit of perioperative FOLFOX for resectable liver metastasis in colorectal cancer patients (EORTC Intergroup Trial 40983). Ann Surg 2012;255:534–9. [DOI] [PubMed] [Google Scholar]
- 40. Hanrahan EO, Hennessy BT, Valero V. Neoadjuvant systemic therapy for breast cancer: an overview and review of recent clinical trials. Expert Opin Pharmacol 2005;6:1477–91. [DOI] [PubMed] [Google Scholar]
- 41. Shi JH, Line PD. Effect of liver regeneration on malignant hepatic tumors. World J Gastroenterol 2014;20:16167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338–45. [DOI] [PubMed] [Google Scholar]
- 43. Bedard PL, Hansen AR, Ratain MJ, et al. Tumour heterogeneity in the clinic. Nature 2013;501:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hiley C, de Bruin EC, McGranahan N, et al. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol 2014;15:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015;21:751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Normanno N, Rachiglio AM, Lambiase M, et al.; on the behalf of the C‐Gi. Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann Oncol 2015;26:1710–4. [DOI] [PubMed] [Google Scholar]
- 47. Ciardiello F, Normanno N, Maiello E, et al. Clinical activity of FOLFIRI plus cetuximab according to extended gene mutation status by next‐generation sequencing: findings from the CAPRI‐GOIM trial. Ann Oncol 2014;25:1756–61. [DOI] [PubMed] [Google Scholar]
- 48. Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 2006;38:468–73. [DOI] [PubMed] [Google Scholar]
- 49. Almendro V, Kim HJ, Cheng YK, et al. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res 2014;74:1338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mroz EA, Tward AD, Pickering CR, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013;119:3034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


