Abstract
Traditional microbiological methods tend to be labor-intensive and time-consuming. Rapid and novel methods in microbiological tests provide more sensitive, precise and reproducible results compared with conventional methods. In microbiology, the most rapid testing methods belong to the field of biotechnology such as PCR, ELISA, ATP bioluminescence and etc. Nevertheless impedance microbiology, biosensors and analytical procedures to determine microbial constituents are of significance. The present review article was conducted using internet databases and related scientific literatures and articles that provide information on developments in the rapid methods in microbiology. The main focus is on the application of rapid methods in microbial quality control of pharmaceutical products. Reviewed literature showed that rapid methods and automation in microbiology is an advanced area for studying and applying of improved methods in the early detection, and characterization of microorganisms and their products in food, pharmaceutical and cosmetic industrials as well as environmental monitoring and clinical applications. It can be concluded that rapid methods and automation in microbiology should continue as potent and efficient technologies to develop the novel tests to be performed in the future because of the ever-increasing concerns about the safety of food and pharmaceutical products. However the main issues to be considered are the scale up of developed methods and the regulatory requirements.
Keywords: Microbial quality control, Sampling, Detection, Identification, Rapid method
Introduction
Microbial infection is acquired as a result of the existence and proliferation of microorganisms.1 The ability of microorganisms to grow in food, pharmaceutical and cosmetic products has been identified for many years and it has been the subject of debates for years. From the infectious point of view, the existence of pathogenic microbes in pharmaceutical and food products makes them hazardous and would objectionable.2 The microbial infections can also alter the physicochemical and biological properties of active ingredients or even change them into toxic materials.3 Some diseases such as diarrhea, acute gastroenteritis and abdominal pain are consequence of microbial toxin. Nevertheless depending on the individual sensitivity to toxin, symptoms are different from mild distress to stomach death.4
Many technologies have been developed in food and pharmaceutical microbiology laboratories in the recent century to sensitive, precise, and quick microbial detection. Generally, rapid methods include some forms of automation to obtain data of quantity and quality of microbes present in the sample.5-8 Although, it is important to know that sometimes the word “rapid” is used to define the range of techniques employed. Indeed, some of the new methods do not provide a more rapid result compared with traditional methods. They present a more accurate, precise, or detailed result and therefore the term “alternative” is better to use for them.5
Applying rapid methods would help companies for saving time and cost. To better and faster control of raw materials and final products, rapid microbiological methods are essential. These rapid methodologies can also offer a better reactivity throughout the manufacturing procedure. Due to the inexpensivity and simplicity of the traditional techniques, they are still widely used in different microbiological experiments. However, these methods involve incubation time between 2 to 7 days for product (in liquid or solid culture media) before the final result of microbial contamination. The long incubation time that is necessary here is mainly attributable to the fact that harassed microorganisms found in complex environment of pharmaceutical and food products need several days to grow to visible colonies to be detected. Additionally, in specific situations like sterility testing, this incubation period can be increased up to 14 days for the release of pharmaceutical formulations. Therefore, the major problem in traditional process is the length of time taken to get microbiological results.
The pharmaceutical and medical industry has also benefited from rapid techniques. Introduction of rapid microbiological methods in medical areas occurred from mid-1960s and then accelerated in the 1970s and has continued to grow up to now.7,9 Choosing the best and optimal method is difficult and sometimes expensive because of several different technologies available on the market. This review provides information on developments in the rapid methods in microbiology. The main focus is on the pharmaceutical products. However, general information of article is common between pharmaceutical, food and cosmetic industries.
Sampling processes
Routine microbiological test is a vital part of analysis of any product in which microorganisms can survive and grow. Indeed, microbiological tests are significant process to determine the safety and quality of these products. One of the most important steps for this purpose is sampling process. Sample collection is the original stage of a process whereby data on the characteristics of a batch are collected for evaluation. Practically, only a fraction of the batch is sampled for testing; and therefore, that fraction must be representative of the batch under consideration. Since the fate of the batch depends upon the results generated from that first sample, sample selection must be regarded as a critical process.10 Appropriate sampling process is necessary to confirm that manufacturing control mechanisms are effective. The main purpose of the sample preparation procedures is to diminish growth or death of the microorganisms exists in the product.5,7,9,11 However, in microbiological sampling, microorganisms may not necessarily be randomly distributed throughout the whole batch or product and random sampling scheme should not therefore work there. Sampling schemes employed in microbial quality control are based on an analysis of susceptible points in a system, known as hazard analysis and control of critical points (HACCP). The seven principles of HACCP are listed in Figure 1.10,12
Figure 1.
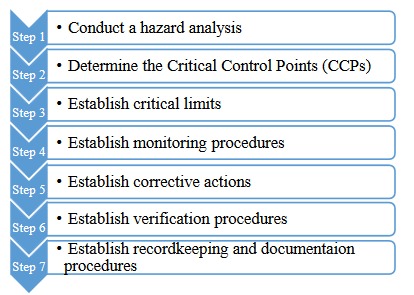
The seven principles of HACCP
Microbiological samples can be categorized in four groups including solid samples, liquid samples, surface samples, and air samples.
Liquid samples
Liquid samples of foods (like water, fruit juices, milk, coffee, teas), liquid pharmaceuticals (like syrups, aqueous preparations, suspensions and emulsions), liquid cosmetic products (like shampoos and liquid soaps) can be categorized in this group. As we know, preparation of liquid samples is easier than solid samples. After collection of sample and its transportation to laboratory site, appropriate mixing to obtain a homogenous mix is required. This can be done by vigorous hand shaking or by instrument. Then, aseptically dilution process by a sterile diluent (a known volume of liquid sample and a desired volume of sterile diluent) should be performed.7,13 The addition of a surfactant such as lecithin or polysorbate 80 in the diluent may also aid to reach a homogenous sample.10
Solid samples
Solid foods (like fruits, vegetables, bread, and meat), solid pharmaceutical dosage forms (like tablets, capsules) and solid cosmetic products (like solid bathroom products) belong to this category. Solid pharmaceutical dosage forms are disposed to microbial spoilage or degradation. There are some common processes for preparing of solid samples including aseptic techniques to collect sample, rapid transportation of samples (less than 24 h) to laboratory site and aseptically removal of the subsamples. Dilution in a proper sterile diluent is the next stage that can be operated in 1:10, 1:25, 1:50, and so on. Further dilutions can be done as necessary. After dilution, the sample should be homogenized by a diversity of methods such as homogenizers or blenders. The addition of surfactant and use of heat can be also used in aseptic condition. After homogenization stage, the sample is ready to analyze.7,9
Environmental samples
Surface sample
The detection microbial flora on the surfaces of foods, pharmaceuticals, cosmetic products or their environments are performed by taking surface samples.14 The main conventional method for surface sampling is swap method. A sterile moisten swab (often cotton) is needed to mild collection of microbes from the surface.15,16 Then, the swab is placed into a diluent (with known volume), shaken and then plated on an agar plate. Using a sterile knife, especially for food products or soft tissues, is another method for surface sampling. It is assume that all the organisms are on the surface and the product itself is sterile.7,9 Using of adhesive tapes, sterile gauges and sterile sponge is also possible for surface sampling. A main adhesive tape method named “Hands-free, Pop-up" developed by Fung and coworkers.17 In this method, a tape unit is placed on the wrist of an analyst, while both hands are free to operate other experimental procedures.7,9,17
For surface sampling, the analyst should decide on the appropriate unit to report the results of sampling. These results can be reported based on number of bacteria per inch square, per cm square, or other units. The shape of the sample unit is also important for surface sampling. In order to easier sampling, a sterile template can be useful. Rarely, some analysts use unfamiliar shapes for surface sampling which calculation and quantification of these unfamiliar shape areas will be complex.9
Air samples
Since air can play a main role as a reservoir for microorganisms, then monitoring of manufacturing environments of products is necessary. Because of recent concerns about environmental air pollution and public health there is also a rapid need to check and control of microbes and their toxins in the air.18
There are two main monitoring methods in air sampling: passive and active.19 The most common passive method for air sampling is using of air plates (without lid) which are standard “Petri dishes” containing culture media. The plates are exposed to the air for a determined time in order to obtain biological particles. It can be taken for 10 minutes, 30 minutes, or four hours. Then, the plate is covered and incubated for counting of colonies. The air quality is considered unacceptable if the colony numbers exceed a certain value. The information of passive method is not too quantitative. Results of passive monitoring are reported in Colony Forming Units (CFU)/plate/time or in CFU/m2/hour.7,9,18,19
Active monitoring method can be performed using an air suction sampler for drawing of air through or by a particle collection device. The device can be a liquid or a solid culture media or a nitrocellulose membrane. The result of quantity is presented in CFU/m3 of air. However, active monitoring methods are applicable in low concentration of microorganisms for example in clean rooms and controlled-environment hospitals.7,9,18,19
Enumeration methods
Counting the number of living organisms exist in the pharmaceutical and food products is very important as an indicator of product microbial quality. Colony Forming Unit (CFU) stands for the number of microbial colonies. Numerous methods have been developed and applied for total viable cell count so far.
"Standard Plate Count" has been used as a main and the most conventional method in applied microbiology for many years. In this method, after the preparation and dilution stages, the sample is mixed with a general agar media, incubated (at 35 °C or 25 °C) and then the colonies is counted after 48 h.7,9,20 There are some shortcomings for this methodology despite of its simple and easy operation. This process is time-consuming and also employs a massive amount of culture media and large number of sterile test materials (such as tubes, pipettes, sterile plates) as well as large incubation spaces. Cleaning of the reusable glass wares and re-sterilizing them is also needed.19-21 Some efforts have been done for semi automation and easiness of the standard method such as mechanical spreading of the sample on the surface of a preformed agar plate,21 application of membranes to trap microorganisms and then standard plate culture22 and the methods based on nutrient stored in films or strips.7,9 These methods have been developed as alternatives for performing viable cell counting in pharmaceutical industry.
Most Probable Number (MPN) system (often the three or five tube) has been applied as one of the most common procedures in this context for more than one century23 especially in food industries. As standard plate method, MPN system also has been endured to automation. A mechanized and automated system was presented in 2007 for operation of viable cell count process. It was a hands-off 16 tube system for operation of MPN procedure and then was applied in pharmaceutical and food laboratories around the world.7,9,24
Some methods based on real-time viable cell counting have been tried during the recent years. These real time tests are based on using “vital” stains to stain “live” cells to count fluorescing viable cells under microscope which are really rapid techniques. A viable cell count can be done by the eye or by an automated system in less than an hour. In Direct Epifluorescent Filter Techniques (DFET) the viable microbial cells (contain predominantly RNA) are stained red with acridine orange while non-viable cells (contain predominantly DNA) stain green. The main disadvantage of this method is that RNA may be stable in preservative or heat-damaged cells and then stain red even in non-viable cells.6 A modified DEFT method named the micro-colony technique (DEFT-MEM) method reported by Newby in 1991. In the modified process, at first cells filtered via a membrane, then incubated to produce micro-colonies and finally stained and counted.25
Nakajima et al tested fluorescent staining method for assessment of microbial quality of herbal medicines using 6-carboxyfluorescein diacetate (6CFDA) and 4,6-diamidino-2-phenylindole (DAPI). The results were compared with conventional colony count method. Their results showed that with the conventional colony count method most of physiologically active bacteria could not detect in the medical samples. They concluded that 6CFDA-DAPI double staining method can be used for daily bacteriological quality control because of its simplicity and short time (<1 h) for staining and enumeration.26
Other technique in this area is ATP detection of live cells that trapped in special membranes. Presence of somatic ATP in food products, produce some difficulties in microbial detection of food samples. However, this method is valuable in pharmaceutical and cosmetics. ATP detection methods can provide viable cell counts in about one to four hours.7 Generally, the level of ATP is detected using the amount of light emitted during the enzymatic reaction (bioluminescence process) by ATP detection devices.27 In the field of rapid microorganism’s detection, ATP bioluminescence based on luciferine/luciferase reaction has shown great interest. Indeed, adenosine triphosphate (ATP) is found in all living organisms and is an excellent marker for viability and cellular contamination (Figure 2). Detection of ATP through ATP-luminescence technology is therefore a method of choice to replace traditional method and significantly shorten time to detection without losing reliability.28-31
Figure 2.
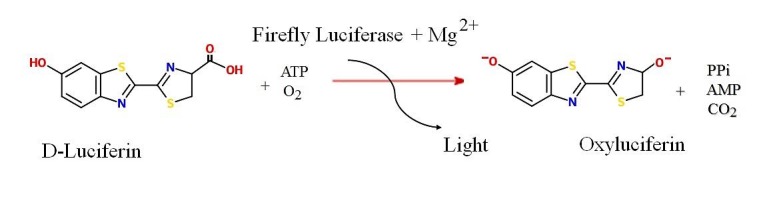
ATP bioluminescence based on luciferine/luciferase reaction
Impedance microbiology is a main branch of rapid methodologies that is useable for qualitative and quantitative tracing of microorganisms by measuring the change in the electrical conductivity. There are two type of impedance technology: direct and indirect. The alteration in the conductivity of a liquid culture medium act as a measuring parameter in a direct impedance technology, while in an indirect type, the alteration in the electrical conductivity of a reaction solution is measured.8,11
Connolly et al studied three rapid microbiological methods including impedance, DEFT and ATP bioluminescence for their application in pharmaceuticals and cosmetic products (like dextrose, potassium dihydrogen phosphate, magnesium phosphate heptohydrate, ammonium sulphate) against Staphylococcus aureus, Pseudomonas Aeruginosa, Aspergillus niger and Candida albicans. They reported a good correlation between these methods and total colony counts for untreated suspensions of Staphylococcus aureus, Pseudomonas aeruginosa and Candida albicans. For Aspergillus niger correlation was good only with impedance technique. Their results showed that neither ATP bioluminescence nor DEFT methods gave a satisfactory dose/response curve while impedance method presented a satisfactory dose/response results.6
Diagnostic methods
Different diagnostic systems in pharmaceutical microbiology have been used to obtain reliable results in as short as time. Systems such as miniaturized diagnostic kits, ELISA tests, immuno-magnetic separation (IMS), and Polymerase Chain Reaction (PCR), microarray and biochip technologies have been widely used during the recent years as diagnostic systems with ability to handle several isolates at the same time.7,9,14,32,33
Miniaturized systems are mainly developed to reduce the volume of reagents and media for microbiological tests. Basically, a miniaturized system is composed of sterilized Microtiter plates, a multiple inoculation device and containers to house solid media and liquid media. Using diagnostic kits leads to rapid and accurate identification due to saving many lives pathogens. In a diagnostic kit, the color reaction of each well is read and then via a manual identification code the organism is identified (key-out). In the recent years, automatic readers are also developed to rapid and accurate detection of the unknown cultures.34 Cox et al and also Fung et al evaluated comparative analysis of diagnostic kits and selection criteria for miniaturized systems. According to their research, miniaturized systems are cheap, accurate and efficient systems. These methods are also labor-saving and space-saving methods compare to conventional methods. There are some marketed miniaturized systems including enterics (Salmonella, Shigella, Proteus, Enterobacter) and non-fermentors, anaerobes, gram positives and even yeasts and molds.7,9,34
Immunological tests based on antigen and antibody reaction has been used for years to detect microorganisms in medical and pharmaceutical microbiology. ELISA test is the most common immunological method. For ELISA tests the overnight incubation is necessary to obtain a detectable level of the target organism. This method is the main procedure for detecting bacteria such as Salmonella, Escherichia coli and Listeria monocytogenes. In recent years, some entirely automated ELISA systems have been developed that can perform a test from 45 minutes to 2 hours (after overnight incubation time).7,9,34-36
Polymerase Chain Reaction (PCR) has been used to detect microorganisms (for example Salmonella) by amplification of the target DNA and detecting the target PCR products. PCR products can be detected by gel electrophoresis, fluorescent probes, special dyes or molecular beacon. In some PCR based systems screening of 4 different targets in the same sample can be performed which is known as multiplexing process.9,37,38 Real-time quantitative PCR is a rapid methodology for sensitive and reliable quantifying of DNA, and then it can be used for accurate detection of microorganisms. This procedure provides accurate detection of nucleic acids in a mixture even for nucleic acids with a low concentration of starting amount. Our group used the RT-PCR method for microbial determination of the amikacin with the test organism of S. aureus. The results showed that the utilized RT-PCR method was accurate and precise in whole concentration range while the hand turbidimetric method showed precision at all of the used concentration, but accuracy only in middle and low concentrations. RT-PCR method can be used as a precise and accurate method for detection and quantification of amikacin. However, we found some problems for RT-PCR method comparing with turbidimetric method. It is an indirect method for detection of amikacin. It is also a complex method that involves expensive equipments, as well as very labor-intensive procedures.39
In another research Multiplex PCR method was applied by our team as an rapid method to control the microbial quality of pharmaceutical products.40 The authors reported to develop a Multiplex PCR assay for simultaneous detection and identification of four indicator pathogenic bacteria of Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa and Salmonella, in a single reaction. Specific primers for the indicator bacteria were reported to apply, and the sensitivity and specificity of each primer pairs were determined. In the mPCR with mixed DNA samples, specific bands for corresponding bacteria were simultaneously detected. Low levels of microbial contamination less than 10 cfu per milliliter or gram of product were detected using mPCR assay. The detection of all four indicator pathogenic bacteria were completed in less than 8 h with this novel mPCR method, whereas the conventional United States Pharmacopeia methods for completion. Using mPCR assay, the microbial quality control of nonsterile pharmaceutical products can be performed in a cost-effective and timely manner in pharmaceutical industry.
Furthermore multiplex PCR method was applied by our group in several studies to rapidly identify the entrococci in clinical, dairy and poultry samples.41-44
In IMS systems, paramagnetic beads are coated with a variety of molecules (like antibodies, antigens and DNA) to capture target cells (like E. coli O157, Listeria). Then, the beads are plated on agar. This method can also combine with ELISA test or PCR procedures. Such combinations can decrease incubation time and also increase sensitivity of the total process.9,45,46
A biosensor is an analytical device, used for the detection of an analyte that combines a biological component with a physicochemical detector.47,48 It can be a molecule or a group of molecules of biological origin attached to a signal recognition material. Any contact between analyte and the biosensor initiate a recognition signal that can be detected via an instrument. Analytes can be including toxins, pathogens, carbohydrates, insecticides and herbicides and ATP. Electrochemical, optical and miscellaneous transducers can use as the recognition signals. The technology of biochip and microchip has also used to detect a variety of molecules including pathogens such as Salmonella, Listeria, Escherichia coli, Staphylococcus aureus, etc.9,11,47,48
Another rapid method to detect bacteria, based on analysis of their fatty acids membrane, has been applied. Phospholipid fatty acid analysis (PLFA) has been used to characterize and monitor broad changes in the "living" soil microbiota for more than 30 years. The Sherlock MIS is a commercially-available system that can be used for both microbial ID and PLFA. This MIS software includes many tools including Peak Categorization, Library Generation (LGS), Principal Component Analysis (PCA), and Electronic Records & Signatures (ERS).49,50 Advantages and some shortcomings of the discussed methods are gathered in Table 1.
Table 1. Advantages and shortcomings of the rapid methods discussed in the paper.
| Method | Sample | Advantages and/or disadvantages that was reported | Reference |
| mechanized and automated MPN method | Pharmaceutical and food samples | Precise and automation in test, lowering human interventions and contaminations | 7,9 |
| Fuorescent staining (used as enumeration method) |
Herbal medicines | Advantages: Simplicity and short time for staining and enumeration. | 26 |
| DEFT methods | 24 | ||
| ATP bioluminescence and impedance method (as enumeration method) |
Pharmaceuticals and cosmetic products (like dextrose, potassium dihydrogen phosphate, magnesium phosphate heptohydrate, ammonium sulphate) | Neither ATP bioluminescence nor DEFT methods gave a satisfactory dose/response curve while impedance method presented a satisfactory dose/response results. | |
| Biosensor techniques (used as enumeration method) |
Food Products | Good correlation between the methods and total colony counts | 47,48 |
| RT-PCR method (as diagnostic method) |
Pharmaceutical product |
Advantages: A precise and accurate method with ability to provide accurate detection nucleic acids with a low concentration of starting amount. Disadvantages: It is an indirect method for detection of amikacin. It is also a complex method that involves expensive equipments, as well as very labor-intensive procedures. |
28-31 |
| Multiplex PCR (as diagnostic method) |
Pharmaceutical products | Ability to determine low levels of microbial contamination less than 10 cfu per milliliter or gram of product. The microbial quality control of nonsterile pharmaceutical products can be performed in a cost-effective and timely manner in pharmaceutical industry. |
40 |
| Multiplex PCR (as diagnostic method) |
Clinical, dairy and poultry samples | Rapidly identify the entrococci in clinical, dairy and poultry samples | 41-44 |
Conclusion
The field of rapid microbial detection and automation has been converted to essential area in pharmaceutical, cosmetics, clinical and food industries. The use of miniaturized systems, bioluminescence processes, biosensors, etc. allows for extensive developments in the microorganism detection. The ease of use and the flexibility of these methods leads to significant savings in labor and time compared to the conventional methods. Rapid method in microbiology should continue to grow as a valuable tool for pharmaceutical, food and clinical applications. The type and number of microbiological rapid tests will rise in the future because of the ever-increasing concerns on food and pharmaceuticals safety as well as public health. The future of rapid methodologies and automation in microbiology will be so hopeful and optimistic due to the great budding in many exciting new advanced procedures. The field of biochips and microarrays for microbial detection in pharmaceutical is a potential area but more attention is needed to make this technology more applicable in microbiology. This field can be more important in the genomics area that involve large amount of data to produce valuable information. Definitely, developments in pharmaceutical microbiological areas will be clearer and more recognized in the near future.
Ethical Issues
Not applicable.
Conflict of Interest
The authors report no conflicts of interest.
References
- 1.Mugoyela V, Mwambete KD. Microbial contamination of nonsterile pharmaceuticals in public hospital settings. Ther Clin Risk Manag. 2010;6:443–8. doi: 10.2147/TCRM.S12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi H, Rittmann BE. Microbial removal of hazardous organic compounds. Environ Sci Technol. 1982;16(3):170A–83A. doi: 10.1021/es00097a002. [DOI] [Google Scholar]
- 3.Garrett TR, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18(9):1049–56. doi: 10.1016/j.pnsc.2008.04.001. [DOI] [Google Scholar]
- 4.Ratajczak M, Kubicka MM, Kaminska D, Sawicka P, Dlugaszewska J. Microbiological quality of non-sterile pharmaceutical products. Saudi Pharm J. 2015;23(3):303–7. doi: 10.1016/j.jsps.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sandle T. Approaching the selection of rapid microbiological methods. 2014 [12 Jun 2014]; Available from: http://www.ivtnetwork.com/article/approaching-selection-rapid-microbiological-methods.
- 6.Connolly P, Bloomfield SF, Denyer SP. A study of the use of rapid methods for preservative efficacy testing of pharmaceuticals and cosmetics. J Appl Bacteriol. 1993;75(5):456–62. doi: 10.1111/j.1365-2672.1993.tb02802.x. [DOI] [PubMed] [Google Scholar]
- 7.Fung DY. Rapid methods and automation in food microbiology: a review. Food Rev Int. 1994;10(3):357–5. doi: 10.1080/87559129409541006. [DOI] [Google Scholar]
- 8.Wawerla M, Stolle A, Schalch B, Eisgruber H. Impedance microbiology: applications in food hygiene. J food Prot. 1999;62(12):1488–96. doi: 10.4315/0362-028x-62.12.1488. [DOI] [PubMed] [Google Scholar]
- 9.Fung DY. Rapid methods and automation in microbiology. Berlin: Springer; 2002. [DOI] [PubMed] [Google Scholar]
- 10.Hodges NA, Baird R, Denyer S. Handbook of microbiological quality control. New York: Taylor & Francis; 2000. [Google Scholar]
- 11.Cady P, Dufour SW, Shaw J, Kraeger SJ. Electrical impedance measurements: rapid method for detecting and monitoring microorganisms. J Clin Microbiol. 1978;7(3):265–72. doi: 10.1128/jcm.7.3.265-272.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denyer SP, Hodges NA, Gorman SP, Gilmore BF. Hugo and Russell's Pharmaceutical Microbiology. 8th ed. New York: John Wiley & Sons; 2011. [Google Scholar]
- 13.Andrews W, Hammack T. Bacteriological Analytical Manual (BAM) New York: Silver Spring; 2003. [Google Scholar]
- 14.Cesarpastor GC, Mariajosefina NA, Omarelind AH. Fungal and bacterial contamination on indoor surfaces of a hospital in Mexico. Jundishapur J Microbiol. 2012;2012(3):460–4. doi: 10.5812/jjm.2625. [DOI] [Google Scholar]
- 15.Prezant B. Recognition, evaluation, and control of indoor mold. Fairfax: AIHA; 2008. [Google Scholar]
- 16.Buttner MP, Cruz P, Stetzenbach LD, Cronin T. Evaluation of two surface sampling methods for detection of Erwinia herbicola on a variety of materials by culture and quantitative PCR. Appl Environ Microbiol. 2007;73(11):3505–10. doi: 10.1128/AEM.01825-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fung DY, Thompson LK, Crozier-Dodson BA, Kastner CL. Hands-Free, “POP-UP,” Adhesive Tape Method For Microbial Sampling Of Meat Surfaces. J Rapid Method Automat Microbiol. 2000;8(3):209–17. doi: 10.1111/j.1745-4581.2000.tb00218.x. [DOI] [Google Scholar]
- 18.Napoli C, Marcotrigiano V, Montagna MT. Air sampling procedures to evaluate microbial contamination: a comparison between active and passive methods in operating theatres. BMC Public Health. 2012;12:594. doi: 10.1186/1471-2458-12-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andon BM. Active air vs. passive air (settle plate) monitoring in routine environmental monitoring programs. PDA J Pharm Sci Technol. 2006;60(6):350–5. [PubMed] [Google Scholar]
- 20.Sarker SD, Nahar L, Kumarasamy Y. Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods. 2007;42(4):321–4. doi: 10.1016/j.ymeth.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J Clin Microbiol. 2008;46(4):1281–4. doi: 10.1128/JCM.01687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama A, Sunamura M. Simultaneous direct counting of total and specific microbial cells in seawater, using a deep-sea microbe as target. Appl Environ Microbiol. 2000;66(5):2211–5. doi: 10.1128/aem.66.5.2211-2215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blodgett R. Most probable number from serial dilutions, bacteriological analytical manual. 2010 [October 2010]; Available from: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm109656.htm.
- 24.Irwin P, Tu S, Damert W, Phillips J. A Modified Gauss‐Newton Algorithm and Ninety‐Six Well Micro‐Technique For Calculating MPN Using Excel Spreadsheets. J Rapid Method Automat Microbiol. 2000;8(3):171–91. doi: 10.1111/j.1745-4581.2000.tb00216.x. [DOI] [Google Scholar]
- 25.Newby P. Analysis of high‐quality pharmaceutical grade water by a direct epifluorescent filter technique microcolony method. Lett Appl Microbiol. 1991;13(6):291–3. doi: 10.1111/j.1472-765X.1991.tb00631.x. [DOI] [Google Scholar]
- 26.Nakajima K, Nonaka K, Yamamoto K, Yamaguchi N, Tani K, Nasu M. Rapid monitoring of microbial contamination on herbal medicines by fluorescent staining method. Lett Appl Microbiol. 2005;40(2):128–32. doi: 10.1111/j.1472-765X.2004.01643.x. [DOI] [PubMed] [Google Scholar]
- 27.Turner DE, Daugherity EK, Altier C, Maurer KJ. Efficacy and limitations of an ATP-based monitoring system. J Am Assoc Lab Anim Sci. 2010;49(2):190–5. [PMC free article] [PubMed] [Google Scholar]
- 28.Squirrell DJ, Price RL, Murphy MJ. Rapid and specific detection of bacteria using bioluminescence. Anal Chim Acta. 2002;457(1):109–14. doi: 10.1016/S0003-2670(01)01495-7. [DOI] [Google Scholar]
- 29.Beckers B, Lang HR, Schimke D, Lammers A. Evaluation of a bioluminescence assay for rapid antimicrobial susceptibility testing of mycobacteria. Eur J Clin Microbiol. 1985;4(6):556–61. doi: 10.1007/BF02013394. [DOI] [PubMed] [Google Scholar]
- 30.Mafu AA, Roy D, Savoie L, Goulet J. Bioluminescence assay for estimating the hydrophobic properties of bacteria as revealed by hydrophobic interaction chromatography. Appl Environ Microbiol. 1991;57(6):1640–3. doi: 10.1128/aem.57.6.1640-1643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Girotti S, Ferri EN, Bolelli L, Sermasi G, Fini F. Applications of bioluminescence in analytical chemistry. In: Garcia-Campana AM, Baeyens WRG, editors. Chemiluminescence in analytical chemistry. Switzerland: Dekker; 2001. P. 247-84.
- 32.Fritzler MJ. Advances and applications of multiplexed diagnostic technologies in autoimmune diseases. Lupus. 2006;15(7):422–7. doi: 10.1191/0961203306lu2327oa. [DOI] [PubMed] [Google Scholar]
- 33.Yakub GP, Stadterman-Knauer KL. Immunomagnetic separation of pathogenic organisms from environmental matrices. Methods Mol Biol. 2004;268:189–97. doi: 10.1385/1-59259-766-1:189. [DOI] [PubMed] [Google Scholar]
- 34. Fung DY. Rapid methods and automation for seafood microbiology. In: Martin AM, editor. Fisheries Processing. New York: Springer; 1994. P. 18-50.
- 35.Nollet LM, Toldra F. Handbook of muscle foods analysis. New York: CRC Press; 2008. [Google Scholar]
- 36.Hui Y, Nollet LM, Toldra F. Advances in food diagnostics. New York: John Wiley & Sons; 2008. [Google Scholar]
- 37.Adzitey F, Huda N, Ali GRR. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech. 2013;3(2):97–107. doi: 10.1007/s13205-012-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amani J, Mirhosseini SA, Fooladi AA. A Review Approaches to Identify Enteric Bacterial Pathogens. Jundishapur J Microbiol. 2015;8(2):e17473. doi: 10.5812/jjm.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lotfipour F, Yeganeh F, Tamizi E, Zahedi A, Asefi M. Study of the Efficacy of Real Time-PCR Method for Amikacin Determination Using Microbial Assay. Adv Pharm Bull. 2015;5(2):181–8. doi: 10.15171/apb.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farajnia S, Hassan M, Hallaj Nezhadi S, Mohammadnejad L, Milani M, Lotfipour F. Determination of indicator bacteria in pharmaceutical samples by multiplex PCR. J Rapid Method Automat Microbiol. 2009;17(3):328–38. doi: 10.1111/j.1745-4581.2009.00154.x. [DOI] [Google Scholar]
- 41.Hassan M, Diep DB, Javadzadeh Y, Dastmalchi S, Nes IF, Sharifi Y. et al. Prevalence of bacteriocin activities and bacteriocin-encoding genes in enterococcal clinical isolates in Iran. Can J Microbiol. 2012;58(4):359–68. doi: 10.1139/w11-136. [DOI] [PubMed] [Google Scholar]
- 42.Javaherzadeh V, Milani M, Jamshidian M, Zahraei M, Lotfipour F. Evaluation of bacteriocin activities among enterococcal dairy isolates from North-West Iran. Int J Biol Pharm Appl Sci. 2014;3(10):2424–40. [Google Scholar]
- 43.Javaherzadeh V, Jamshidian M, Zahraei M, Youseftabar A, Milani M, Hassan M. et al. Evaluation of Bacteriocin Activities among Enterococcal Poultry Isolates from East Azarbaijan Iran. Pharm Sci. 2015;21(2):72–76. doi: 10.15171/PS.2015.20. [DOI] [Google Scholar]
- 44.Hassan M, Brede DA, Diep DB, Nes IF, Lotfipour F, Hojabri Z. Efficient Inactivation of Multi-Antibiotics Resistant Nosocomial Enterococci by Purified Hiracin Bacteriocin. Adv Pharm Bull. 2015;5(3):393–401. doi: 10.15171/apb.2015.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricke SC, Jones FT. Perspectives on food-safety issues of animal-derived foods. USA: University of Arkansas Press; 2010. [Google Scholar]
- 46.Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;14(5):1310–6. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 47.Luque de Castro MD, Herrera MC. Enzyme inhibition-based biosensors and biosensing systems: questionable analytical devices. Biosens Bioelectron. 2003;18(2-3):279–94. doi: 10.1016/s0956-5663(02)00175-6. [DOI] [PubMed] [Google Scholar]
- 48.Buhl A, Metzger JH, Heegaard NH, Von Landenberg P, Fleck M, Luppa PB. Novel Biosensor-Based Analytic Device for the Detection of Anti-Double-Stranded DNA Antibodies. Clin Chem. 2007;53(2):334–41. doi: 10.1373/clinchem.2006.077339. [DOI] [PubMed] [Google Scholar]
- 49.Zelles L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol Fertil Soils. 1999;29(2):111–29. doi: 10.1007/s003740050533. [DOI] [Google Scholar]
- 50.Frostegård Å, Bååth E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soil. 1996;22(1):59–65. doi: 10.1007/s003740050076. [DOI] [Google Scholar]


